Abstract
Germination of endospermic seeds is partly regulated by the micropylar endosperm, which acts as constraint to radicle protrusion. Gibberellin (GA) signalling pathways control coat-dormancy release, endosperm weakening, and organ expansion during seed germination. Three GIBBERELLIN INSENSITIVE DWARF1 (GID1) GA receptors are known in Arabidopsis thaliana: GID1a, GID1b, and GID1c. Molecular phylogenetic analysis of angiosperm GID1s reveals that they cluster into two eudicot (GID1ac, GID1b) groups and one monocot group. Eudicots have at least one gene from each of the two groups, indicating that the different GID1 receptors fulfil distinct roles during plant development. A comparative Brassicaceae approach was used, in which gid1 mutant and whole-seed transcript analyses in Arabidopsis were combined with seed-tissue-specific analyses of its close relative Lepidium sativum (garden cress), for which three GID1 orthologues were cloned. GA signalling via the GID1ac receptors is required for Arabidopsis seed germination, GID1b cannot compensate for the impaired germination of the gid1agid1c mutant. Transcript expression patterns differed temporarily, spatially, and hormonally, with GID1b being distinct from GID1ac in both species. Endosperm weakening is mediated, at least in part, through GA-induced genes encoding cell-wall-modifying proteins. A suppression subtraction hybridization (SSH) cDNA library enriched for sequences that are highly expressed during early germination in the micropylar endosperm contained expansins and xyloglucan endo-transglycosylases/hydrolases (XTHs). Their transcript expression patterns in both species strongly suggest that they are regulated by distinct GID1-mediated GA signalling pathways. The GID1ac and GID1b pathways seem to fulfil distinct regulatory roles during Brassicaceae seed germination and seem to control their downstream targets distinctly.
Keywords: Arabidopsis thaliana, endosperm weakening, expansin, GIBBERELLIN INSENSITIVE DWARF1, Lepidium sativum, seed germination, xyloglucan endo-transglycosylase/hydrolase
Introduction
Germination of endospermic seeds is a complex developmental process. It starts with water uptake by imbibition of the dry seed and ends when the radicle has protruded through all seed covering layers (Bewley, 1997a). In mature seeds of most angiosperm species, the embryo is encased by the endosperm and testa (seed coat) as covering layers (Linkies et al., 2010a). The endosperm is therefore localized between the embryo and the testa and, in most mature seeds, the endosperm is a living tissue. Traditionally the endosperm was recognized primarily as a nutrient source during seed development and seedling growth (Lopes and Larkins, 1993). However, evidence has accumulated over time (Linkies et al., 2010a) that shows that the endosperm in mature seeds, in particular the micropylar endosperm covering the radicle also plays a major role in regulating germination timing (Ni and Bradford, 1993; Bewley, 1997b; Leubner-Metzger, 2003; Linkies et al., 2009). For the completion of germination, the tissue resistance of the micropylar endosperm must be overcome by the growth potential of the radicle. For several species it has been shown that, during germination, weakening of the micropylar endosperm takes place, which decreases the threshold of the force needed for radicle protrusion through the endosperm layer to complete germination. This is the case in species with a thick endosperm, such as tomato (Lycopersicon esculentum), tobacco (Nicotiana tabacum), and coffee (Coffea arabica) and also for seeds with a thin endosperm, such as lettuce (Lactuca sativa) and garden cress (Lepidium sativum) (Ni and Bradford, 1993; Bewley, 1997b; Toorop et al., 2000; Leubner-Metzger, 2003; da Silva et al., 2004; Müller et al., 2006). This supports the view that the endosperm, in particular the micropylar endosperm, participates in regulating germination timing.
Seed germination is inhibited by abscisic acid (ABA) and promoted by gibberellins (GA) (Kucera et al., 2005). It has been shown that the induction of endosperm weakening requires an embryo signal which consists, at least in part, of bioactive GA or GA-biosynthesis precursors (Bewley, 1997b; Ogawa et al., 2003). Treatment of ‘isolated micropylar endosperm caps’ with GA can replace the embryo signal to induce the onset and accelerate L. sativum endosperm weakening (Müller et al., 2006). ABA-insensitive mutants like Arabidopsis thaliana (Arabidopsis) abi3 and ABA-deficient mutants like Arabidopsis aba1-1 and tomato sitiens show reduced seed dormancy (Groot and Karssen, 1992; Nambara et al., 2000; Bassel et al., 2006). Inhibition of endosperm weakening by ABA has been shown for L. sativum and other seeds (Bewley, 1997a; Müller et al., 2006; Linkies et al., 2009). Contrary to that, high GA contents and sensitivities lead to dormancy release and promotion of the germination process. GA-deficient mutants, such as ga1-3 (Arabidopsis) and gib1 (tomato), need an external supply of GA to induce and complete germination (Koornneef and Veen, 1980; Groot et al., 1987). Endogenous GA is perceived by the GA receptors of the GID1 (GIBBERELLIN INSENSITIVE DWARF1) family, first identified in rice (OsGID1, Oryza sativa) (Ueguchi-Tanaka et al., 2005). In Arabidopsis, three GID1-type GA receptors are known: AtGID1a, AtGID1b, and AtGID1c. After binding to its receptor, the GA–GID1 complex interacts with DELLA proteins which are negative regulators of the GA signalling pathway (Richards et al., 2001). The F-Box protein SLY1/GID2, part of the SCFSLY1/GID2 E3 ubiquitin ligase complex, recognizes the GA–GID1–DELLA complex and targets the DELLA repressors for degradation through the 26S proteasome. This leads to a de-repression and allows transcription of GA-responsive genes (Ueguchi-Tanaka et al., 2007; Yamaguchi, 2008). GA treatment of the GA-deficient Arabidopsis mutant ga1-3 results in the disappearance of the DELLA protein RGL2 (Tyler et al., 2004) for which SLY1 is required (Ariizumi et al., 2007). GA-insensitive Arabidopsis mutants, for example, gid1-receptor triple and sly1-10 knockouts, and gain-of-function DELLA-mutants such as gai, show impaired germination (Steber et al., 1998; McGinnis et al., 2003; Dill et al., 2004; Griffiths et al., 2006; Iuchi et al., 2007; Willige et al., 2007). By contrast, loss-of-function DELLA-mutants such as gai-t6 show enhanced germination (Kucera et al., 2005). L. sativum endosperm weakening and its promotion by GA as a replacement for the embryo signal (Müller et al., 2006) is thought to be achieved by cell-wall-modifications. The plant cell wall is a highly complex composite composed mainly of cellulose microfibrils embedded in a matrix of hemicellulosic and pectic polysaccharides (Cosgrove, 2005; Knox, 2008). Cell expansion growth is driven by water uptake and is restricted by the cell wall, whose structural properties and mechanical strength determine the shape, rate, and direction of growth of individual cells as well as of whole tissues (Cosgrove, 2005; Schopfer, 2006). Cell wall loosening is therefore an important process in all stages of plant development requiring elongation growth or tissue weakening, as it is the case during seed germination, where radicle growth and endosperm weakening take place. Some cell-wall modifying proteins are known to accumulate in a germination-specific and GA-induced manner. For endospermic Solanaceae seeds, these include β-1,3-glucanase (Leubner-Metzger et al., 1995; Petruzzelli et al., 2003) and β-1,4-mannanase (Bewley, 1997b; Nonogaki et al., 2000). For endospermic Brassicaceae seeds, differential regulation of β-1,4-mannanase in the L. sativum micropylar endosperm and radical tissue has been shown by Morris et al. (2011) and Arabidopsis β-1,4-mannanase knockout mutants have a germination phenotype (Iglesias-Fernandez et al., 2011). Isoforms of both enzymes accumulate specifically in the micropylar endosperm, covering the radicle tissue, but not in the non-micropylar endosperm. One of the most important stabilizing and tension-bearing interactions in the primary cell wall of eudicots is between cellulose microfibrils and the hemicellulose xyloglucan, on which xyloglucan endo-transglycosylases/hydrolases (XTHs) can act by transglycosylation (cleaving and reconnecting) or hydrolysis (Rose et al., 2002). Another group of cell-wall-modifying proteins are expansins, thought to act by loosening of non-covalent bonds (Sampedro and Cosgrove, 2005). Both XTHs and expansins are induced by GA in the micropylar endosperm during tomato seed germination and proposed to facilitate endosperm weakening (Chen and Bradford, 2000; Chen et al., 2002).
The bigger seeds of L. sativum, a close relative of Arabidopsis, were used to construct an endosperm-specific subtractive suppression hybridization (SSH) cDNA library to clone candidate genes involved in the early germination processes. Genes encoding cell-wall-modifying proteins including XTHs and expansins were over-represented in this library. Since GA is known to promote endosperm weakening, this prompted us to investigate the transcript co-expression pattern of the GA receptors GID1a, b, and c and putative cell-wall-modifying genes. A combined approach was carried out with Arabidopsis mutants and seed and seed-tissue-specific analysis of Arabidopsis and L. sativum transcript expression. Our work shows that the eudicot GID1 receptors group into a GID1ac and a GID1b group, which, during seed germination, are regulated differentially and suggest two separate, but partially redundant pathways for GA responsiveness that have distinct downstream cell-wall-loosening genes as targets.
Materials and methods
Plant materials and germination assays
Mature seeds of Arabidopsis thaliana (L.) Heyhn. were harvested and stored for after-ripening at 25 °C, 51–54% relative humidity. The seeds were incubated on 1/10 Murashige-Skoog salts (medium) pH 7.0, solidified with 1% (w/v) agar-agar in continuous white light (c. 100 μmol s−1 m−2) at 24 °C. Homozygous gid1 mutant seeds were kindly provided by Professor Dr Claus Schwechheimer, ZMBP, Tübingen, Germany (Willige et al., 2007) and by Shiori Ota, Bio Resource Centre RIKEN, Iberaki, Japan (Iuchi et al., 2007). Testa rupture and endosperm rupture were scored using a binocular microscope as described by (Müller et al. (2006). After-ripened seeds of Lepidium sativum FR14 (Juliwa, Germany) were incubated in Petri dishes on two layers of filter paper with 6 ml 1/10 Murashige-Skoog salts medium in the light. Where indicated, gibberellin A4+7 (GA4+7; Duchefa, The Netherlands) or cis-S(+)-abscisic acid (ABA; Duchefa) were added in the concentrations indicated.
RNA extraction and creation of the subtractive cDNA library
Total RNA from endosperm caps dissected from seeds after 8 h and 18 h imbibition was prepared as described by Cadman et al. (2006). A Suppression Subtraction Hybridization (SSH) library was constructed using the PCR Select™ cDNA Subtraction kit (Clontech, USA) as described in Linkies et al. (2010b). cDNA from micropylar endosperm dissected after 8 h imbibition of the whole seed was used as tester, subtracted with cDNA from endosperm caps dissected after 18 h imbibition as a driver. The resulting cDNA fragments were cloned in pUC19 and pCR®4-TOPO, multiplied in E. coli Top10 and sequenced (GATC, Germany). 184 cDNAs were sequenced and have been deposited as ESTs to GenBank, their accession numbers are listed in Supplementary Table S2 at JXB online.
Cloning of cDNA sequences from L. sativum
First strand cDNA was synthesized in 50 μl reactions from 5 μg total RNA from L. sativum FR14 seedlings, 2.5 μM oligo(dT)16, 2.5 μM random hexamers, using the Superscript III reverse transcriptase kit (Invitrogen, Germany) according to its instructions. PCR was performed using primers designed based on known A. thaliana sequences. At least three independent cDNA clones were sequenced to verify L. sativum sequences.
Analyses of transcript levels by quantitative real-time RT-PCR (qRT-PCR)
Total RNA was extracted from 200 L. sativum radicles and 1000 micropylar endosperm caps which were dissected from seeds at the times indicated, frozen in liquid nitrogen, and stored at –20 °C. RNA extraction of Arabidopsis was done using 200 whole seeds of WT or the gid1 mutants. RNA extraction was carried out as described by Chang et al. (1993). Four biological replicate RNA samples of each time point and treatment were used for downstream applications. qRT-PCR was performed with first-strand cDNAs as templates that were obtained using the Superscript III reverse transcriptase kit (Invitrogen, Germany) with 0.3 nmol random 15-mers for reverse transcription of 5 μg RNA. Aliquots of 1 μl were then used for each quantitative RT-PCR reaction. Absolute QPCR SYBR Green ROX Mix (ABgene; UK) was used according to its instructions for quantification with the ABI PRISM 7300 Sequence Detection System (Applied Biosystems, UK). A melting curve confirmed single product amplification. Analysis of the raw data and calculation of the efficiency (E) for every single well was done using the software PCR miner (Zhao and Fernald, 2005). Relative expression for each well was calculated as (1+E)–CT. Expression data for Arabidopsis and L. sativum was normalized by using the geometric mean (geomean) of validated housekeeping genes. Mean values ±SE shown were calculated from four biological replicates. Primers were designed using the bioinformatics software Geneious 4.7.5 (Biomatters, New Zealand) and are listed in Supplementary Table S3 at JXB online.
Histochemical GUS-staining and visualization
pAtGID1a::AtGID1a-GUS, pAtGID1b::AtGID1b-GUS, and pAtGID1c::AtGID1c-GUS reporter lines were kindly provided by Masatoshi Nakajima, University of Tokyo, Japan (Suzuki et al., 2009). Seeds were placed in Petri dishes on two layers of filter paper with 1/10 Murashige-Skoog (MS) salts and imbibed for 20 h at 24 °C in continuous white light (c. 100 μmol s−1 m−2). Where indicated, 5 μM ABA or 10 μM GA4+7 were added to the medium. 10 seeds with ruptured testa but intact endosperm were dissected into the embryo and the seed ‘coats’ (testa plus endosperm). Tissue fixation was done with 90% acetone for 20 min at room temperature. Afterwards, the seed tissues were incubated in 100 μl staining solution (50 mM phosphate-buffer (pH 7.2), 0.2% (v:v) Triton X-100, 10 mM EDTA (pH 8.0), and 2 mM X-gluc). Images were taken after 20 h of staining with the software IM1000 (Leica Microsystems, Wetzlar, Germany) and a Leica DCF480 digital camera attached to a Leica 12.5 binocular microscope.
Tetrazolium assay testing embryo viability
Testa and endosperm of imbibed seeds were removed and embryos were stained according to the procedure described by Graeber et al. (2010) in 1% (w/v) 2,3,5-triphenyltetrazolium chloride (Sigma, Germany) in phosphate buffer (pH 7) at room temperature for the time indicated. As negative controls, heat-killed seeds (dry seeds were incubated at 100 °C for 1 h) were used; wild-type seed served as positive controls. Two biological replicates of several seeds each were analysed per line.
Sequence alignments and molecular phylogenetic analysis
For sequence analysis, the bioinformatics software Geneious 4.7.5 (Biomatters, New Zealand) was used. Geneious Align was used for sequence alignments, with the BLOSUM62 matrix for alignments of protein sequences. Geneious Tree Builder was used for construction of the phylogenetic tree using the sequences of Supplemenatry Table S1 at JXB online.
Results
Three putative orthologues of the Arabidopsis GID1a, GID1b, and GID1c genes are expressed in L. sativum
Three L. sativum GID1 cDNAs were cloned and analysed, including the complete coding sequences (cds) and the 5′ and 3′ untranslated regions (UTRs). Based on detailed analysis (Fig. 1; see Supplementary Figs S1 and S2 at JXB online), these sequences of L. sativum were named LesaGID1a, LesaGID1b, and LesaGID1c according to the putative orthologous AtGID1a, AtGID1b, and AtGID1c genes of Arabidopsis, respectively, and have been submitted to GenBank (accession numbers HQ003455, HQ003456, HQ003457). Comparative sequence analysis between the cds of GID1a, GID1b, and GID1c of Arabidopsis and L. sativum showed 91.4, 89.4, and 91% pairwise identity (see Supplementary Fig. S1 at JXB online), which are values similar to known orthologues of L. sativum and Arabidopsis (Linkies et al., 2009, 2010b; Graeber et al., 2010). Pairwise comparative analysis on amino acid level showed that the putative GID1a, GID1b, and GID1c orthologous proteins of L. sativum and Arabidopsis displayed 96.2, 92.8, and 94.8% pairwise identity, respectively. In most cases, the observed amino acid changes did not affect protein hydrophobicity or polarity (see Supplementary Fig. S2 at JXB online). GID1 belongs to the family of hormone-sensitive lipases which are characterized by two conserved amino acid motifs: HGG and GXSXG (Østerlund, 2001; Hirano et al., 2008). It was demonstrated that all three Arabidopsis GID1s bind GA and interact with DELLA proteins (Nakajima et al., 2006; Suzuki et al., 2009). From the crystal structures of the GA–GID1 and GA–GID1–DELLA complexes (Murase et al., 2008; Shimada et al., 2008) conserved amino acids for GA binding and DELLA repressor binding are known (for details see Supplementary Fig. S2 at JXB online). These conserved amino acids, as well as the HGG and GXSXG motifs are all present at the expected positions in the predicted GID1 proteins of L. sativum (see Supplementary Fig. S2 at JXB online). It is therefore proposed that LesaGID1a, LesaGID1b, and LesaGID1c are fully functional GA receptors.
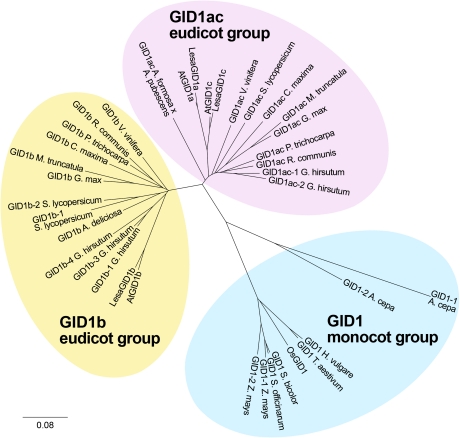
Fig. 1.
Molecular phylogenetic analysis of the angiosperm GID1 receptor family reveals individual clustering into three groups: eudicot GID1ac, eudicot GID1b, monocot GID1. Translated protein sequences of 37 available GID1 full-length or near full-length cDNA sequences from different species were aligned into a phylogenetic unrooted tree as described in the methods. A. cepa=Allium cepa; A. deliciosa=Actinidia deliciosa; A. formosa=Aquilegia formosa; A. pubescens=Aquilegia pubescens; At=Arabidopsis thaliana; Lesa=Lepidium sativum; C. maxima=Cucurbita maxima; G. hirsutum=Gossypium hirsutum; G. max=Glycine max; H. vulgare=Hordeum vulgare; M. truncatula=Medicago truncatula; Os=Oryza sativa; P. trichocarpa=Populus trichocarpa; R. communis=Ricinus communis; S. bicolor=Sorghum bicolor; S. lycopersicum=Solanum lycopersicum; S. officinarum=Saccharum officinarum; T. aestivum=Triticum aestivum; V. vinifera=Vitis vinifera; Z. mays=Zea mays. Sequence accession numbers are listed in Supplementary Table S1 at JXB online. Note that, in addition to tomato, various other asterids (Lactuca and Helianthus species) have at least one GID1ac and one GID1b gene. They are listed in Supplementary Table S1 at JXB online, but were not included in the phylogeny as no full-length sequences are available. (This figure is available in colour at JXB online.)
Angiosperm GID1 gene family members fall into three distinct phylogenetic groups: eudicot GID1ac, eudicot GID1b, and monocot GID1
As Arabidopsis and L. sativum have several GID1 genes, it was investigated whether this is a general feature of the eudicots and compared it with the monocots, in which, for example, the rice genome contains only one GID1 gene (OsGID1). Thirty-seven angiosperm GID1 cds were used to carry out a molecular phylogenetic analysis (Fig. 1). Alignment of all predicted proteins showed that the eudicot GID1 genes cluster into two distinct groups: the ‘GID1ac’ and the ‘GID1b’ group, which were named according to the corresponding Arabidopsis GID1 genes that they contain. Almost all of the included core eudicot species (rosids and asterids) have at least one GID1 member of each group. An exception is Actinidia deliciosa for which only one GID1 was found, which may be due to insufficient sequence availability. In addition to the asterid species included in the phylogenetic analysis (Fig. 1), EST sequences were also found that demonstrate that several Lactuca and Helianthus species have at least one GID1 from each group (see Supplementary Table S1 at JXB online). It is an interesting finding that the monocot GID1 proteins (in contrast to the core eudicots) cluster into one separate group with no further split, even when they contain more than one GID1 gene (Fig. 1).
Comparison of GID1 transcript secondary structures and stability motifs suggests high mRNA turnover, and GID1 transcript expression during Arabidopsis seed germination is differentially regulated by GA and ABA
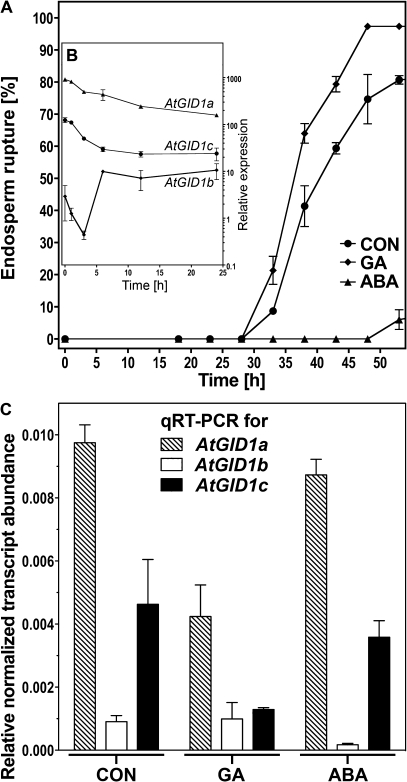
The predicted secondary structures differed between the six GID1 mRNA molecules of L. sativum and Arabidopsis, but in all cases one intron splice site just downstream the start of the coding sequence and relatively long 5' and 3' UTRs are evident (see Supplementary Figs S1 and S3 at JXB online). The average 5' and 3' UTR lengths from all annotated genes of the Arabidopsis genome are 0.13 kb and 0.24 kb, respectively (Haas et al., 2003). With 0.35–0.49 kb, all the GID1 3' UTRs except LesaGID1c are longer than average (see Supplementary Fig. S1 at JXB online) and this is often associated with high mRNA turnover (Garneau et al., 2007). mRNA half-lives of AtGID1a and AtGID1b determined by Narsai et al. (2007) in Arabidopsis cell cultures are short with t1/2 <1 h, whereas AtGID1c transcripts are of moderate stability. Together with mRNA secondary structure (see Supplementary Fig. S3 at JXB online; Hofacker and Stadler 2006), various global motifs associated with mRNA destabilization, identified by Narsai et al. (2007) and Jiao et al. (2008) are prominent in the GID1 3' and 5’ UTRs (see Supplementary Figs S3 and S4 at JXB online). Therefore, GID1 transcripts of both species may not only be highly regulated, but also possess conserved regulatory patterns as well as differences between the two species. As considerable amounts of bioactive GA4 are already present in dry Arabidopsis seeds (Ogawa et al., 2003), it is expected that GA responsiveness of the seed tissues is induced immediately upon imbibition and that its regulation during germination depends, at least in part, on the expression of the three AtGID1 genes. The AtGID1a, AtGID1b, and AtGID1c transcript expression patterns were analysed during the germination of unstratified Arabidopsis seeds by quantitative real-time RT-PCR (qRT-PCR, Fig. 2): The transcript abundance at 30 h imbibition of AtGID1a and AtGID1c was high compared with AtGID1b, with AtGID1a transcripts being twice as abundant as AtGID1c transcripts (CON, Fig. 2C). This difference in transcript abundance, AtGID1a>AtGID1c>>AtGID1b, is already evident in dry seeds and during the early germination phase, as shown in Fig. 2B, based on in silico analysis of microarray expression data (Preston et al., 2009) with the Arabidopsis eFP browser (Winter et al., 2007). Furthermore, the hormonal regulation of the AtGID1 transcript expression was analysed by qRT-PCR (Fig. 2C): GA exerts a negative feedback mechanism causing about a 2-fold down-regulation of the transcript levels of AtGID1a and probably also AtGID1c, but did not affect AtGID1b expression. In contrast to GA, ABA did not exert any significant effect on the transcript levels of AtGID1a, AtGID1b or AtGID1c (Fig. 2C). The differential transcript regulation in seeds, especially the higher abundance and the GA-triggered negative feedback loop associated only with the two GID1ac group members, but not with GID1b, suggests that the two groups of GID1 receptors may play distinct roles during the germination process.
Fig. 2.
Transcript abundances in whole seeds of the three Arabidopsis thaliana GID1 genes during germination. (A) The time-course of endosperm rupture of A. thaliana (Col) seeds on medium without (CON) or with 10 μM GA4+7 (GA) or 1 μM ABA added. Afterripened seeds were incubated at 24 °C in continuous light without preceding stratification. Mean values ±SE of 3×50 seeds. (B) In silico analysis on GID1 transcript abundance during early germination of unstratified Arabidopsis (Col) seeds based on eFP-Browser microarray data; Relative expression values ±SD for AtGID1a (At3g05120), AtGID1b (At3g63010), and AtGID1c (At5g27320). (C) GID1 transcript abundance pattern determined by qRT-PCR in whole seeds of Arabidopsis (Col) at 30 h, i.e. at 0% endosperm rupture for CON (see A). ddCt expression values relative to validated constitutive transcripts are presented. Medium additions as in (A). Mean values ±SE of 4× >1000 seeds.
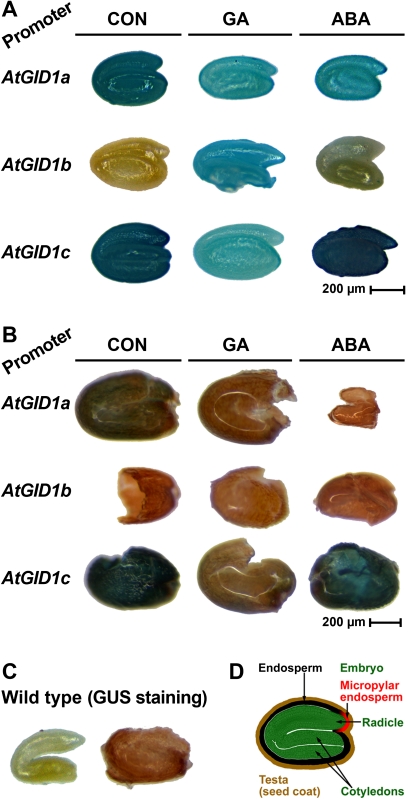
The pAtGID1::AtGID1-GUS reporter gene lines from Suzuki et al. (2009) were used to investigate the spatially and hormonally regulated expression patterns of the three individual GID1 genes in different Arabidopsis seed tissues, i.e. the embryo and the endosperm, during germination. Transcription of the three pAtGID1::AtGID1-GUS reporter constructs is controlled by c. 3 kb of AtGID1a, AtGID1b, and AtGID1c 5' regulatory region containing the corresponding GID1 gene promoters, and in the following text the three transgenic lines are referred to with the abbreviations ‘PGID1a’, ‘PGID1b’, and ‘PGID1c’, respectively. In agreement with the qRT-PCR results on whole Arabidopsis seeds imbibed in medium without the addition of hormones (control, CON), the histological GUS staining of PGID1a and PGID1c embryos was much stronger compared with PGID1b (CON in Fig. 3A). As the GID1 gene promoter activity is an approximate estimate for GID1 gene transcription and, based on the roughly equal distribution of blue staining over the entire embryos, it is concluded that GID1 gene transcription is not confined to a highly localized zone. There are, however, clear differences in the strength of the three GID1 promoter activities that suggest that the high GID1ac and low GID1b transcript abundances in CON-seeds (Fig. 2) are the result of higher-level GID1ac and low-level GID1b gene transcription in CON-embryos (Fig. 3A). To investigate the effects of ABA and GA, seeds of the different transgenic lines were also imbibed with the hormones added. In agreement with a GA-mediated down-regulation of the GID1ac transcript abundances in the qRT-PCR analysis (Fig. 2), GA addition decreased the staining intensity of the PGID1a and PGID1c embryos (Fig. 3A). In contrast to the roughly equal transcript abundances for GID1b in CON and GA (Fig. 2), the blue staining is stronger in embryos from GA-treated seeds compared with CON (Fig. 3A). This suggests as a possible explanation that, for GID1b, the transcript degradation is of utmost importance for the GA-mediated down-regulation of the transcript abundances. Neither GA nor ABA changed the spatial expression patterns in embryos for the three GID1 genes. While GA affected the staining intensities, ABA did not appreciably change the staining intensities of the embryos compared with CON (Fig. 3A). The staining was slightly stronger for PGID1c- compared to PGID1a-embryos for all three treatments (Fig. 3A), whereas, for the transcript abundances, it was the other way around. This implies that transcript degradation may be more important for regulating the GID1c transcript abundances. As for the embryo, the staining intensities in the endosperm were PGID1c>>PGID1a> PGID1b for the CON-series (Fig. 3B). Interestingly, the effects of the hormones on the staining intensities differed between embryo and endosperm: In contrast to the embryo, GA-treatment did not cause increased staining in the PGID1b endosperm. But, as in the embryo, GA-treatment caused decreased staining for PGID1a and PGID1c. For the ABA-treatment the decreasing effect on the PGID1a staining in the embryo was even more pronounced in the endosperm, while for PGID1b and PGID1c the ABA effects did not differ between embryo and endosperm (Fig. 3). Taken together, the embryo and the endosperm of Arabidopsis seeds (Fig. 3D) exhibited similar and distinct aspects regarding the spatial and hormonal regulation of the three GID1 gene promoter activities. This regulation was more similar for GID1a and GID1c, but considerably different between GID1b and GID1ac., and further support the view that the two groups of GID1 receptors may play distinct roles during the germination process.
Fig. 3.
Histochemical analysis of gene promoter activities in of the three Arabidopsis thaliana GID1 genes during seed germination. Seeds of GUS reporter lines in which the three GID1 gene promoters, AtGID1a, AtGID1b, and AtGID1c control the transgene transcription were imbibed in medium without (CON) or with 10 μM GA4+7 (GA) or 5 μM ABA added. After seed dissection into embryo and ‘coat’ (testa plus endosperm) at 20 h, histochemical GUS analysis was performed. (A) Histochemical GUS analysis of isolated embryos from GID1 reporter gene seeds. (B) Histochemical GUS analysis of isolated ‘coats’ from GID1 reporter gene seeds. (C) Negative control for the histochemical GUS staining with isolated wild-type embryo and ‘coat’. (D) Cross-section of A. thaliana seed indicating the specific seed tissues analysed used in the experiment.
Germination kinetics of Arabidopsis gid1 mutants reveal different relevance of the three GID1 receptors during seed germination
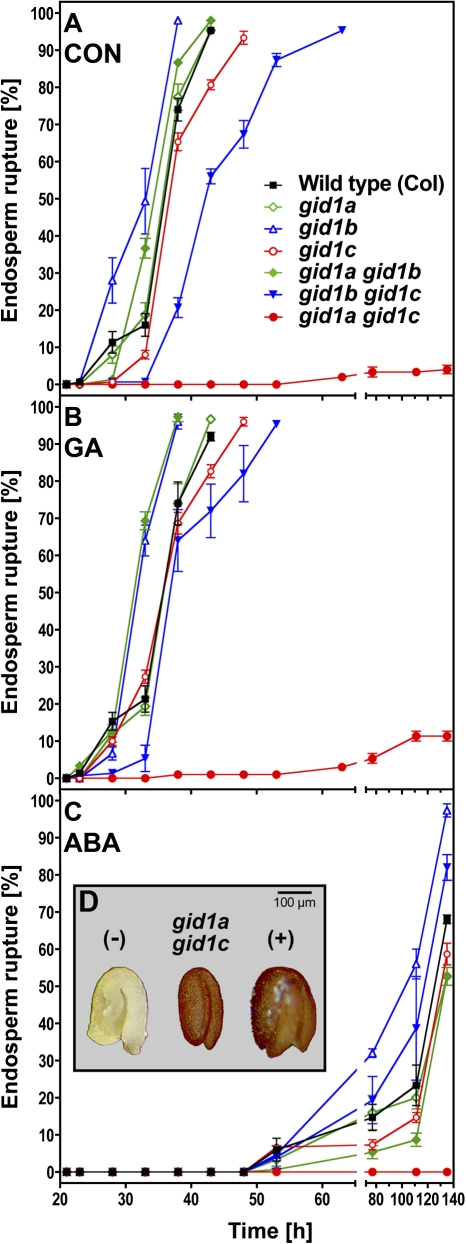
To investigate the individual roles and importance of the three GID1 receptors in the germination process, phenotypic analyses of Arabidopsis gid1a, gid1b, and gid1c single- and double-knockout mutants were carried out. Endosperm rupture of the seed populations was scored over time on medium without (CON) and with GA or ABA added (Fig. 4). Without hormonal addition none of the single-knockout mutants exhibited an appreciable phenotype, although gid1c completed germination slightly later compared with the wild type (WT) and the other single mutants (Fig. 4A). A very striking phenotype was observed for gid1agid1c: only 4% of the seeds completed germination, and this happened only very late (Fig. 4A). The viability of the gid1agid1c seeds was confirmed using the tetrazolium assay (Fig. 4D). The tetrazolium assay is a standard test for seed viability and is widely used for this purpose (Graeber et al., 2010). Also gid1bgid1c completed germination later than the WT and the single mutants.
Fig. 4.
The effect of the three GID1 receptors on the seed germination of Arabidopsis thaliana studied with mutants for AtGID1a, AtGID1b, and AtGID1c. Time-course analysis of endosperm rupture of after-ripened seeds of Arabidopsis single (gid1a, gid1b, gid1c) and double (gid1agid1b, gid1bgid1c, gid1agid1c) mutants imbibed at 24 °C in continuous light without preceding stratification. The results presented were obtained with the mutant set by Iuchi et al. (2007), the same results were obtained with an independent mutant set by Willige et al. (2007). (A) Medium without hormones added (control, CON). (B) 10 μM GA4+7 (GA). (C) 1 μM ABA. (D) Tetrazolium-assay to confirm viability of gid1agid1c; (–) and (+): negative (seed killed by preceding heat treatment, 1 h at 100 °C) and positive wild-type seed controls. Mean values ±SE of 3×50 seeds are presented.
Treatment with GA did not appreciably alter the germination kinetics of the knockout mutants (Fig. 4B). Only 11% of the gid1agid1c seeds completed germination, demonstrating that most seeds are GA-insensitive and that AtGID1b cannot replace the AtGID1ac group receptors that appear to be required for Arabidopsis seed germination. Treatment with ABA strongly delayed endosperm rupture and completely inhibited gid1agid1c seed germination (Fig. 4C). The strong inhibition of gid1agid1c germination and delayed germination of seeds containing the gid1c mutation was also evident with an independent second set of T-DNA insertion single and double knockout mutants (data not shown). Taken together, the three GID1 genes of Arabidopsis seem to have distinct importance during seed germination: At least one GID1ac group member is required with AtGID1c being more important than AtGID1a, while AtGID1b seems to play a minor role.
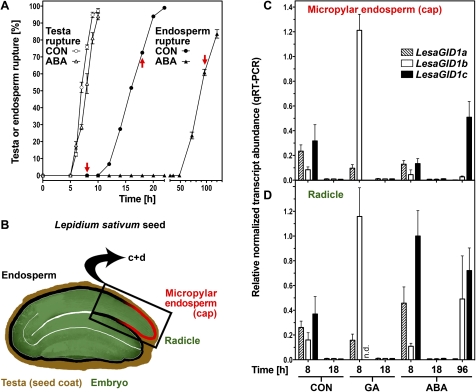
Tissue-specific analysis of L. sativum seeds reveals spatial, temporal, and hormonal differences of GID1 transcript expression
The different GID1 GA receptors seem to play distinct roles during plant development. For germination their function during the process of endosperm weakening is of special interest and was examined in individual tissues for this purpose. The bigger seed size and the temporal and ABA-related separation of testa and endosperm rupture of L. sativum facilitate analysis of defined seed tissues (Fig. 5), such as the micropylar endosperm and the radicle. Preparation of samples was done by removing the testa covering the hypocotyl- and radicle part of the seed, cutting off the radicle (lower 1/3 of the hypocotyl-radicle axis) and detaching the micropylar endosperm and the radicle tissue. Transcript expression patterns of LesaGID1a, LesaGID1b, and LesaGID1c in the micropylar endosperm and in the radicle after 8 h and 18 h of imbibition of L. sativum were analysed by qRT-PCR (Fig. 5C, D). Those times were selected in order to capture the early germination phase of Lepidium sativum, just after testa rupture at the onset of endosperm weakening, and the late germination phase, just prior to endosperm rupture. Without hormone treatment (CON), the transcript expression levels at 8 h of LesaGID1a and LesaGID1c were similar and about twice that compared with LesaGID1b. The levels of all three transcripts decline to very low and roughly equal levels from the early to the late germination phase in both tissues (Fig. 5C, D).
Fig. 5.
Transcript abundance patterns of the LesaGID1a, LesaGID1b, and LesaGID1c receptor genes in specific seed tissues of Lepidium sativum during germination. A, Time course analysis of testa and endosperm rupture of L. sativum FR14 seeds in medium without (CON) and with ABA added. Continuous light, 18 °C. Mean values ±SE of 3×50 seeds are presented. Red arrows indicate sampling time points for the qPCR. (B) Cross-section of L. sativum seed indicating the specific seed tissues used in the experiment: micropylar endosperm and radicle (lower 1/3 of the hypocotyl-radicle axis). (C, D) qRT-PCR analysis of LesaGID1a, LesaGID1b, and LesaGID1c transcript abundances for micropylar endosperm (C) and radicles (D) at 8, 18, and 96 h imbibition. ddCt expression values relative to validated constitutive transcripts are presented. Medium additions, as indicated: 10 μM GA4+7, 10 μM ABA. Mean values ±SE of 4×1000 micropylar endosperms and 4×200 radicles are presented; n.d.=not detectable. (This figure is available in colour at JXB online.)
GA treatment slightly promoted L. sativum seed germination and triggered a negative feedback loop that resulted in reduced transcript levels of LesaGID1a and LesaGID1c at 8 h compared with CON (Fig. 5C, D). While transcript abundance of LesaGID1a was reduced to about 30% upon GA treatment, LesaGID1c mRNA became undetectable in both tissues at 8 h. In contrast to the transcripts of the L. sativum GID1ac group members, GA treatment caused a 14.3-fold (micropylar endosperm) and a 7.2-fold (radicle) increase in LesaGID1b transcript abundance at 8 h compared with CON. LesaGID1b transcript levels declined from the early (8 h) to the late (18 h) germination phase to low levels upon GA treatment as in CON.
ABA treatment delays endosperm rupture (Fig. 5). Comparison of CON and ABA at 8 h shows in the radicle about a 2-fold increase of the LesaGID1a and LesaGID1c transcript levels, but no difference for LesaGID1b. As in CON and GA, upon ABA treatment the expression of all three transcripts was also down-regulated to very low levels at 18 h. However, at 96 h ABA, where the percentage of germinated seeds is comparable with 18 h CON (Fig. 5A), in the micropylar endosperm the LesaGID1c transcript levels were up-regulated roughly 3-fold compared with 8 h ABA (Fig. 5C). In the radicle, LesaGID1b and LesaGID1c transcripts levels (96 h versus 8 h ABA) were up-regulated 5-fold to roughly equal values (Fig. 5D). By contrast, the LesaGID1a transcript levels remained low at 96 h ABA in both tissues (Fig. 5C, D). Taken together, a GA-triggered negative feedback loop on transcript expression of the GID1ac group members is evident in the L. sativum micropylar endosperm and the radicle. In addition, a GA-triggered positive feedback loop on LesaGID1b was found during L. sativum seed germination, and the ABA regulation of the GID1 transcripts showed complex temporal and tissue-specific patterns.
A subtractive suppression hybridization (SSH) cDNA library from micropylar endosperm tissue of germinating seeds of L. sativum contains candidate genes for endosperm weakening
An SSH library from micropylar endosperm tissue was constructed enriched for transcripts at 8 h (early germination) compared with 18 h (late germination). This library contains transcripts that are more highly abundant or unique during the early germination phase and, therefore, are likely to be important for the germination process. 184 clones were sequenced and the partial sequences were given a putative function based on their highest BLAST hit (see Supplementary Table S2 at JXB online). About 10% of the transcripts were coding for ribosomal proteins, indicating the importance of protein biosynthesis for germination. This is further supported by the presence of the universal translation initiation and elongation factors. The subtractive cDNA library also contained transcripts of proteins involved in mRNA splicing, binding, and turnover. Furthermore, a clone was found for an RCAR/PYR/PYL-type ABA receptor and many of the library clones are GA regulated (see Supplementary Table S2 at JXB online). An analysis of Gene Ontology (GO) classes in our subtractive cDNA library compared with the whole set of GOs annotated on TAIR by the software GOstat (Beißbarth and Speed, 2004) showed that cDNAs annotated as ‘cell wall’ (GO:0005681) or ‘external encapsulating structure’ (GO:0030312) were significantly over-represented (P <0.001). They contained genes such as the putative orthologues of EXLA1 (an expansin-like A, At3g45970) and EXPA9 (an α-expansin, At5g02260) from the expansin superfamily, and XTH18 (At4g30280) and XTH19 (At4g30290) from the xyloglucan endo-transglycosylases/hydrolases (XTHs) family. They are potential candidates to be involved in early GA-induced weakening of the endosperm tissue and their transcript expression was therefore analysed further.
Analysis of expansin and XTH transcript expression patterns in individual seed tissues of L. sativum and in the gid1 single and double mutants of Arabidopsis during germination assigns them to distinct regulatory GID1 pathways
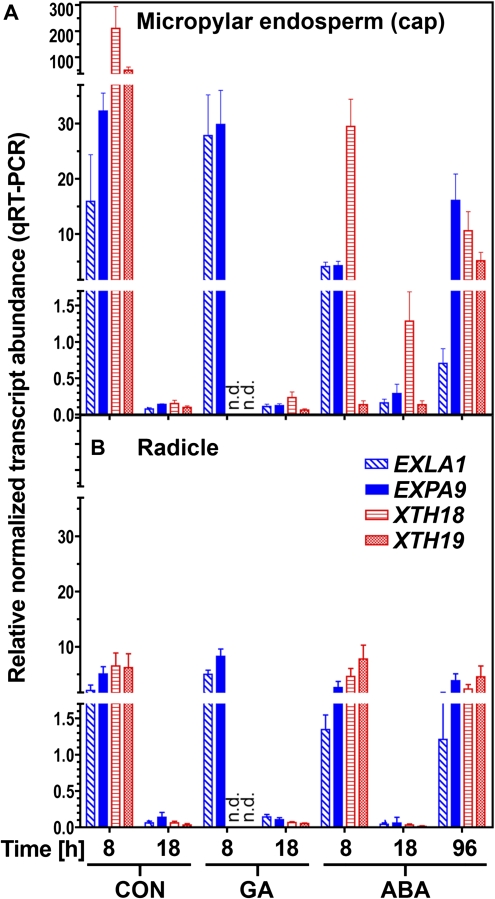
The transcript expression of EXLA1, EXPA9, XTH18, and XTH19 in the micropylar endosperm and the radicle of L. sativum during germination was determined by qRT-PCR (Fig. 6). All genes showed higher expression at 8 h compared with 18 h (the two time points used for library construction) in the micropylar endosperm (Fig. 6A), which confirms the reliability of the data obtained from the SSH library. At 8 h, the transcript abundances of all four genes was higher in the micropylar endosperm compared witho the radicle (Fig. 6). The expression levels of all four transcripts declined from the early (8 h) to the late germination (18 h) phase to very low and roughly equal levels in both tissues. Interestingly, upon GA treatment EXLA1 and EXPA9 displayed a distinct pattern compared to both XTH genes during early germination. The transcript levels of XTH18 and XTH19 declined below the detection limit in 8 h GA-imbibed seed tissues, while transcript levels of EXLA1 and EXPA9 remained high upon GA treatment during early (8 h) germination. At 18 h with GA treatment all four transcripts showed similar low transcript abundance. This distinct effect of GA on the 8 h transcript expression levels, with a GA-triggered down regulation affecting the XTHs, but not the expansins, shows that, during early germination, they are regulated by distinct pathways regarding GA, but during late germination equally regarding the down-regulation. Since ABA strongly delays endosperm rupture, an additional time point very late during germination was investigated (96 h) which, in terms of endosperm rupture, corresponds to 18 h CON and for which transcripts of LesaGID1c and LesaGID1b accumulated (Fig. 6). Upon ABA treatment there is an up-regulation of all four candidate cell-wall-modifying protein transcripts in the micropylar endosperm and the radicle very late during germination (at 96 h; Fig. 6). Taken together, this shows that the XTHs and expansins in L. sativum are regulated differentially: Both XTHs appear to be in a GID1ac pathway, since LesaGID1a, LesaGID1c, as well as XTH18 and XTH19 are down-regulated by GA during early germination (Figs 5C, 6). Contrary to that, both expansins are not down-regulated by GA during early Lepidium germination, which assigns them to a different pathway.
Fig. 6.
Transcript abundance patterns of putative cell-wall-loosening genes (expansins and XTHs) in specific seed tissues of Lepidium sativum during germination. qRT-PCR analysis of EXLA1, EXPA9, XTH18, and XTH19 transcript abundances for (A) micropylar endosperm and (B) radicles at 8, 18, and 96 h imbibition. ddCt expression values relative to validated constitutive transcripts are presented. For time-courses of testa and endosperm rupture in continuous light (18 °C) see Fig. 5A. Medium additions, as indicated: 10 μM GA4+7, 1 μM ABA. Mean values ±SE of 4×1000 micropylar endosperms and 4×200 radicles are presented; n.d.=not detectable. (This figure is available in colour at JXB online.)
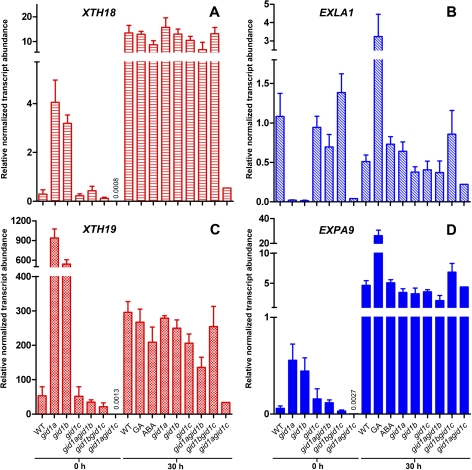
The XTH18 and XTH19 transcript levels are up-regulated upon imbibition of Arabidopsis wild-type (WT) seeds, based on comparing dry seeds with 30 h-imbibed seeds (Fig. 7), i.e. a timepoint during germination that corresponds physiologically to the 8 h CON timepoint in L. sativum. Treatment of Arabidopsis WT with GA did not affect the relative transcript abundances of both XTHs in whole seeds as it was evident in specific seed tissues in L. sativum. If, in Arabidopsis, the XTH18 and XTH19 transcripts are regulated by the GID1ac pathway, their regulation should be impaired in the gid1agid1c mutant which also exhibits a germination phenotype (Fig. 4). To test if, in Arabidopsis, the XTH18 and XTH19 transcripts are also regulated by the GID1ac pathway, their expression was compared in seeds of the gid1 single and double mutants (Fig. 7). In contrast to the WT and most gid1 mutants, the XTH18 and XTH19 transcript levels remained low in imbibed gid1agid1c mutant seeds (Fig. 7). As the relative transcript abundances of both XTHs were up-regulated during germination in WT and in the gid1 single and double knockout mutants, except for the gid1agid1c mutant, XTH18 and XTH19 regulation in Arabidopsis can be assigned to the GID1ac pathway.
Fig. 7.
Transcript abundances of XTH18, XTH19, EXLA1, and EXPA9 in Arabidopsis gid1 mutant seeds. qRT-PCR analysis of (A) XTH18 (At4g30280), (B) EXLA1 (At3g45970), (C) XTH19 (At4g30290), and (D) EXPA9 (At5g02260) in wild-type (WT) and gid1 single and double mutants in dry seeds and after 30 h imbibition. ddCt expression values relative to validated constitutive transcripts are presented. Medium additions, as indicated: 10 μM GA4+7, 1 μM ABA. Mean values ±SE of 4×200 seeds are presented. (This figure is available in colour at JXB online.)
In Arabidopsis it was also found that, as in L. sativum, the expansins exhibited a distinct regulation of their transcript expression during seed germination for WT and the individual gid1 single- and double-mutants (Fig. 7). It is known that transcripts of α-expansins like EXPA9 accumulate strongly during the early phase of Arabidopsis seed germination and that this induction is promoted by GA, is not inhibited by ABA, and is mainly localized in the endosperm (Holdsworth et al., 2008a; Preston et al., 2009). In agreement with this, it was found that EXPA9 transcripts accumulate in whole seeds of Arabidopsis during early germination, and that this induction is promoted by GA, but not inhibited by ABA (Fig. 7). GA treatment also promoted the transcript expression of the expansin-like A gene EXLA1. GA addition therefore leads to higher transcript levels of both expansins but does not affect the XTH transcript levels. Based on the decrease in AtGID1A and AtGID1c, but not AtGID1b, transcript abundance through the addition of GA (Fig. 2) this suggests that both expansins in Arabidopsis are regulated in a pathway involving GID1b. In contrast to the two XTHs, relative transcript abundances of EXPA9 were not affected in the gid1agid1c mutant and was roughly constant in imbibed seeds of WT and all of the gid1 mutants (Fig. 7). In summary, evidence is provided from two Brassicaceae species that, during seed germination, the GID1ac pathway and the GID1b pathway have different roles and regulate different downstream genes.
Discussion
Eudicots contain at least one GID1 gibberellin receptor from each of the two distinct groups, GID1ac and GID1b, which are associated with two distinct regulatory pathways
Our molecular phylogenetic analysis showed that the known angiosperm GID1 receptors fall into three distinct groups: eudicot GID1ac, eudicot GID1b, and monocot GID1 (Fig. 1). The finding that the core eudicot species always have at least one GID1 gene from each of the two eudicot groups supports the view that the two distinct groups fulfil some distinct developmental functions required for all angiosperms and suggests distinct regulatory pathways. That the individual GID1 receptors of Arabidopsis (AtGID1a, AtGID1b, AtGID1c) display partial redundancy and have functional specificities for regulating the GA-responsiveness of different developmental processes has been postulated before (Griffiths et al., 2006; Iuchi et al., 2007). Hirano et al. (2007) investigated GA signalling components in a broad evolutionary context by phylogenetic comparison of the GID1 proteins of the two angiosperms, Arabidopsis and rice, to pine as a gymnosperm, Selaginella moellendorffii as a lycophyte and Physcomitrella patens as a moss representative. They showed that the GID1 proteins of pine, the lycophyte, and the moss, cluster further away from the GID1 group with the two angiosperms. In agreement with this, when putative GID1 coding sequences of Selaginella moellendorffii, Physcomitrella patens, and the three gymnosperms Picea glauca, Picea sitchensis, and Pinus taeda are included in our phylogenetic analysis, they also clustered as a distinct group (data not shown) apart from our comprehensive eudicot GID1ac, eudicot GID1b, and monocot-GID1 clusters that are based on comparing the sequences of 36 angiosperm species (Fig. 1; see Supplementary Table S1 at JXB online). The distinction between two eudicot groups (GID1ac and GID1b) and the monocot group is not only supported by the molecular phylogeny, but also by biochemical evidence in studies with the three Arabidopsis GID1s: Nakajima et al. (2006) showed that AtGID1a and AtGID1c bind GA4 and GA3 with a c. 10-fold lower affinity compared with AtGID1b. All three Arabidopsis GID1 receptors can interact with all five Arabidopsis DELLA proteins (GAI, RGA, RGL1, RGL2, RGL3) targets (Nakajima et al., 2006; Willige et al., 2007; Suzuki et al., 2009). However, Suzuki et al. (2009) showed that differential in vivo interaction affinities exist: the major preference of AtGID1b is GAI and RGA, while the major preferences of AtGID1a and AtGID1c are RGL1 and RGL3; AtGID1a has, in addition, RGL2 as a second major preference. These biochemical features, combined with distinct tissue-specific and hormonal expression patterns of the different GID1 receptors and DELLA repressors, can therefore define distinct GID1ac and GID1b pathways in eudicots. In contrast, in monocot species, only one group of GID1 receptors seems to exist, even when more than one GID1 homologue is present within a species (Fig. 1). This might be linked to the fact that, in monocot species, fewer DELLA proteins exist: one DELLA protein in rice (Oryza sativa; SLR1) and barley (Hordeum vulgare; SLN1) and a putative second homologue in maize (Zea mays; D8 and D9), and, therefore, less specialization of the GID1s is needed (Peng et al., 1999; Ikeda et al., 2001; Gubler et al., 2002; Weston et al., 2008). Differential expression and distinct patterns of degradation of different DELLA repressors has been shown in seeds (Bassel et al., 2004; Ariizumi et al., 2008; Piskurewicz et al., 2008, 2009; Piskurewicz and Lopez-Molina, 2009). In an evolutionary context a separation between an GID1ac-type (interacting preferentially with the RGL1/RGL2/RGL3 group of DELLA repressors) and a GID1b-type (interacting preferentially with the GAI/RGA group of DELLA repressors) pathway hints at a greater specialization of eudicot GID1-mediated GA signalling, while such a partial functional separation has not occurred within the monocots. It has to be noted, though, that less work has been carried out on monocot species seed germination so far, and that the degree of partial redundancy between the two eudicot pathways may differ depending on the developmental process.
The regulation of the GID1 pathways and its roles during seed germination differ from other developmental processes and may be conserved among the Brassicaceae
Our analysis of the Arabidopsis knockout mutants for the three GID1 receptors clearly showed that the AtGID1b receptor is not able to compensate for the phenotype of the gid1agid1c double mutant, i.e. GA signalling via the GID1ac receptors is required for seed germination (Fig.4). By contrast, the AtGID1c and AtGID1a receptors are partially redundant and can compensate for a lack of AtGID1b. Based on the seed germination phenotypes of the Arabidopsis knockout mutants and the ABA-related transcript expression pattern in the micropylar endosperm and the radicle of L. sativum, the GID1c receptors may be more important for Brassicaceae seed germination compared with the GID1a receptors. While transcript abundances in the order AtGID1a>AtGID1c>AtGID1b were evident in dry and imbibed seeds (Cadman et al., 2006; Preston et al., 2009), the order AtGID1a>AtGID1b>AtGID1c was evident throughout plant development in many other tissues and in seedlings (Griffiths et al., 2006). This further indicates that there is a different importance of the individual GID1 receptors during seed germination compared with other developmental processes. The hypothesis of the different importance of the individual GID1 genes during seed germination, with GID1b being distinct from GID1ac and having less importance, is also supported by the stronger GUS staining of the PGID1ac reporter gene lines compared with PGID1b. This finding is further supported by the fact that Griffiths et al. (2006) found negative feedback-regulation by GA for all three GID1 transcripts in seedlings, while in our transcript analyses and GUS reporter line staining results, combined with in silico analysis with the eFP browser, down-regulation by GA during the germination of unstratified Arabidopsis seeds was evident only for the AtGID1a and AtGID1c transcripts, but not for the AtGID1b transcripts. It is therefore proposed that, in germinating seeds, AtGID1b transcript abundance is not negatively regulated by GA. An alternative interpretation would be a higher GA sensitivity of the negative feedback mechanism on AtGID1b transcript abundance compared with AtGID1a and AtGID1c as it was proposed by Iuchi et al. (2007). The endogenous GA content would then already be sufficient to decrease AtGID1b transcript abundance. However, in support of our proposal, the GA-triggered negative feedback loop on AtGID1a and AtGID1c, but not on AtGID1b, was also evident in 9 h-imbibed ga1-3 Ler seeds (Ogawa et al., 2003). In agreement with this, during L. sativum seed germination, a GA-triggered negative feedback loop in the micropylar endosperm and the radicle was only evident for the LesaGID1a and LesaGID1c transcripts. This strongly suggests that a GA-triggered negative feedback loop during seed germination exists for GID1a and GID1c in Brassicaceae seeds, while GID1b-type transcripts are not down-regulation targets. Besides that, in both species expression patterns are similar regarding transcript abundance during early germination (8 h in L. sativum versus 30 h Arabidopsis), in which the GID1ac transcript levels are usually higher compared to GID1b, and no significant regulation by ABA during early germination takes place.
A subtractive cDNA library identified candidate genes for transcript splicing and turnover, translation and proteolysis, hormone signalling, and weakening in the micropylar endosperm
Weakening of the micropylar endosperm of L. sativum progresses between the early (8 h) and late (18 h) time points used in our germination experiments (CON), and is promoted by GA and inhibited by ABA (Müller et al., 2006). Using a subtractive cDNA library of the micropylar endosperm, transcripts that are more highly expressed or unique during early germination were identified. The overrepresentation of the GO categories ‘ribosomal protein’ (GO:0003735) and ‘ribosome’ (GO:0005840) (GOstat; P-value <0.001; Beißbarth and Speed, 2004) indicates intense protein biosynthesis during early germination. The importance of de novo protein biosynthesis for the germination process of Arabidopsis has been shown by Rajjou et al. (2004). It seems that de novo synthesis of ribosomal components is needed for germination and subsequent seedling growth (Holdsworth et al., 2008b). The importance of early translation is further indicated by the occurrence of a translation initiation factor (EIF3G) and a translation elongation factor (EF1-α) in our library.
In addition to protein synthesis, UBQ11 and three protease clones (see Supplementary Table S2 at JXB online), support our earlier finding that proteolysis in the micropylar endosperm is important (Müller et al., 2010). The micropylar endosperm library also contains transcription and splicing factors indicating de novo mRNA biosynthesis and processing, as well as RNA binding proteins and putative ribonucleases that may be involved in diverse pathways of mRNA decay, in particular, the polyA-binding protein RBP31 (Ni et al., 2010) and a core protein of the evolutionarily conserved CCR4-NOT complex (At2g32070), both involved in the deadenylation pathway of mRNA decay (Denis et al., 2003; Garneau et al., 2007).
The SSH library also contains a clone encoding PYL6, an RCAR/PYR/PYL-type ABA receptor (Nambara et al., 2010; Raghavendra et al., 2010). In germinating Arabidopsis seeds (eFP browser; Toufighi et al., 2005); PYL6 transcripts accumulate during imbibition, are down-regulated by ABA, and up-regulated by GA. In our tissue-specific transcriptome analysis of L. sativum seed germination (Linkies et al., 2009) transcripts of the putative PYL6 orthologue are far more abundant in the micropylar endosperm compared to the non-micropylar endosperm and the radicle. The RCAR/PYR/PYL-type and the GID1-type receptors may, therefore, be crucial components in regulating ABA and GA responsiveness important for endosperm weakening.
Two XTHs and two expansins are regulated differentially with expression patterns that assign them to distinct hormonal regulation
The two XTHs that were found in our subtractive library are putative orthologues of AtXTH18 and AtXTH19, two phylogenetically closely related group-2-type XTHs (Rose et al., 2002; Osato et al., 2006). Several members of the XTH-family are known to be positively regulated on both the transcriptional and activity levels by growth-promoting plant hormones such as auxin and GA. For example, GA stimulates leaf elongation and XTH expression and activity in barley, while dwarf mutants with dysfunctional GA signal transduction show reduced XTH activities and shorter leaves (Smith et al., 1996). XTHs are implicated in cell-wall hemicellulose remodelling leading to loosening (Van Sandt et al., 2007) or stiffening (Maris et al., 2009). The two expansins in our subtractive library, putative orthologues of AtEXLA1 (an expansin-like A) and AtEXPA9 (an α-expansin), may be involved in cell expansion and/or weakening of the micropylar endosperm during germination. Expansins loosen cell walls at acidic pH of around 4.5–6, which corresponds to the pH in the apoplast (Cosgrove, 2005; Sampedro and Cosgrove, 2005). No enzymatic activity has been found for expansins, and it has been hypothesized that they act on non-covalent bonds such as hydrogen bridges between cellulose microfibrils and hemicelluloses. Cell-wall loosening activity of α-expansins, but not of expansin-like proteins, has been demonstrated. In the seed, tissue-specific transcriptome analysis of L. sativum transcripts of the putative EXPA9 orthologue are far more abundant in the micropylar endosperm and the radicle compared with the non-micropylar endosperm (Linkies et al. 2009; Morris et al. 2011). This suggests that the EXPA9 proteins are involved in micropylar endosperm weakening and in radicle growth.
Expansins and XTHs have both been described in the context of GA and ABA regulation during seed germination (Finkelstein et al., 2008; Holdsworth et al., 2008a). In our tissue-specific analysis of the XTH and expansin transcript expression in L. sativum, their higher expression during early (8 h) germination indicates the necessity of early transcription of cell-wall-modifying genes before the onset of endosperm weakening. The temporal, spatial, and hormonal (GA and ABA) co-expression patterns (Figs 5, 6) of the XTH18, XTH19, and LesaGID1 transcripts suggest that these XTHs are regulated in a LesaGID1ac involving pathway. Both during seed germination of Arabidopsis (whole seeds; Fig. 2) and L. sativum (specific seed tissues; Fig. 5) the GA-triggered negative feedback loop affected only the GID1ac orthologues, while transcripts of the GID1b orthologues are not down-regulated by GA in Arabidopsis and are strongly up-regulated in L. sativum. By contrast, the expression patterns of the EXPA9 and EXLA1 transcripts assign them to a different pathway, not involving LesaGID1ac. This is supported by the fact that, in 30 h-imbibed Arabidopsis seeds, AtGID1b expression is not affected by GA, while AtGID1a and AtGID1c transcripts are down-regulated by GA (Fig. 2). At the same time, GA induces expression of EXLA1 and EXPA9, but not of the two XTH genes (Fig. 7), which indicates that the main regulation of both expansins during Arabidopsis germination is in a GID1b-involving pathway, while the two XTHs are in another pathway. In support of the association of the two XTHs with a GID1ac signalling pathway, the Arabidopsis gid1agid1c double mutant is impaired in the accumulation of transcripts upon imbibition for the two XTHs, but EXPA9 transcripts accumulate as in WT. The gid1agid1c double mutant also has a seed germination phenotype (Fig. 4). Thus, although for Arabidopsis whole seeds and for L. sativum specific seed tissues were analysed and not every aspect of the regulation is equal in both species, as a general conclusion the cross-species approach provided support for the hypothesis that, during germination of both Brassicaceae species, the two XTHs appear to be regulated by the GID1ac pathway, while the expansins seem to be associated with the GID1b pathway.
In conclusion, eudicot species contain at least one GID1ac and one GID1b group member. GA responsiveness is similarly important and regulated during seed germination of different Brassicaceae species [e.g. Arabidopsis thaliana: Ogawa et al. (2003), Lepidium sativum: Müller et al. (2006), Sisymbrium officinale: Hilhorst et al. (1986); Iglesias-Fernández and Matilla (2010)] and in seeds from other eudicot families [e.g. Solanaceae: Ni and Bradford (1993); Toorop et al. (2000); Leubner-Metzger (2002, 2003); Petruzzelli et al. (2003); Asteraceae: Argyris et al. (2008); Rubiaceae: da Silva et al. (2004)]. Distinct, but partially overlapping roles are proposed for the GID1ac and GID1b pathways and their downstream target genes during seed germination that may be conserved among Brassicaceae (this work) or even among eudicots.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. GID1 transcript sequence alignments of L. sativum and Arabidopsis.
Supplementary Fig. S2. GID1 protein sequence alignments of L. sativum and Arabidopsis.
Supplementary Fig. S3. Comparison of L. sativum and Arabidopsis GID1 transcript secondary structures and stability motifs in 5' and 3' UTRs.
Supplementary Fig. S4. L. sativum and Arabidopsis GID1 5' and 3' UTRs motifs associated with mRNA stability.
Supplementary Table S1. Angiosperm phylogeny and GID1 receptor sequence accession.
Supplementary Table S2. cDNA clones obtained of the early micropylar endosperm SSH library.
Supplementary Table S3. Primer sequences used for quantitative PCR analyses.
Acknowledgments
Our work is funded by grants from the Deutsche Forschungsgemeinschaft (Grant DFG LE720/6 and MU3114/1-1) and the Deutscher Akademischer Austauschdienst (Grant DAAD D/0628197) to GL-M and KM, and the Wissenschaftliche Gesellschaft Freiburg to AL and AV. Seeds of the gid1 mutants in Arabidopsis were provided by Claus Schwechheimer, ZMBP, Tübingen, Germany and by Shiori Ota, Bio Resource Centre RIKEN, Iberaki, Japan, seeds of the pAtGID1::AtGID1-GUS reporter lines in Arabidopsis were provided by Masatoshi Nakajima, University of Tokyo, Japan, which is gratefully acknowledged.
References
- Argyris J, Dahal P, Hayashi E, Still DW, Bradford KJ. Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiology. 2008;148:926–947. doi: 10.1104/pp.108.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Murase K, Sun T-P, Steber CM. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. The Plant Cell. 2008;20:2447–2459. doi: 10.1105/tpc.108.058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Steber CM. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. The Plant Cell. 2007;19:791–804. doi: 10.1105/tpc.106.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Mullen RT, Bewley JD. ABI3 expression ceases following, but not during, germination of tomato and Arabidopsis seeds. Journal of Experimental Botany. 2006;57:1291–1297. doi: 10.1093/jxb/erj101. [DOI] [PubMed] [Google Scholar]
- Bassel GW, Zielinska E, Mullen RT, Bewley JD. Down-regulation of DELLA genes is not essential for germination of tomato, soybean, and Arabidopsis seeds. Plant Physiology. 2004;136:2782–2789. doi: 10.1104/pp.103.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beißbarth T, Speed TP. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. The Plant Cell. 1997a;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends in Plant Science. 1997b;2:464–469. [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seed during cycling through dormant and non-dormant states indicate a common underlying dormancy control mechanism. The Plant Journal. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Chen F, Bradford KJ. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiology. 2000;124:1265–1274. doi: 10.1104/pp.124.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Nonogaki H, Bradford KJ. A gibberellin-regulated xyloglucan endotransglycosylase gene is expressed in the endosperm cap during tomato seed germination. Journal of Experimental Botany. 2002;53:215–223. doi: 10.1093/jexbot/53.367.215. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- da Silva EAA, Toorop PE, van Aelst AC, Hilhorst HWM. Abscisic acid controls embryo growth potential and endosperm cap weakening during coffee (Coffea arabica cv. Rubi) seed germination. Planta. 2004;220:251–261. doi: 10.1007/s00425-004-1344-0. [DOI] [PubMed] [Google Scholar]
- Denis CL, Chen J, Kivie M. Progress in Nucleic Acid Research and Molecular Biology. Academic Press; 2003. The CCR4–NOT complex plays diverse roles in mRNA metabolism; pp. 221–250. [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling receptors for gibberellin-induced degradation. The Plant Cell. 2004;16:1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annual Reviews of Plant Biology. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nature Reviews Molecular Cell Biology. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Müller K, Wunchova A, Rott A, Leubner-Metzger G. Cross-species approaches to seed dormancy and germination: conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Molecular Biology. 2010;73:67–87. doi: 10.1007/s11103-009-9583-x. [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Bruinsma J, Karssen CM. The role of endogenous gibberellin in seed and fruit development of tomato: studies with a gibberellin-deficient mutant. Physiologia Plantarum. 1987;71:184–190. [Google Scholar]
- Groot SPC, Karssen CM. Dormancy and germination of abscisic acid-deficient tomato seeds. Plant Physiology. 1992;99:952–958. doi: 10.1104/pp.99.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV. Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiology. 2002;129:191–200. doi: 10.1104/pp.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Research. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HWM, Smitt AI, Karssen CM. Gibberellin-biosynthesis and -sensitivity mediated stimulation of seed germination of Sisymbrium officinale by red light and nitrate. Plant Physiology. 1986;67:285–290. [Google Scholar]
- Hirano K, Nakajima M, Asano K, et al. The GID1-mediated gibberellin perception mechanism is conserved in the lycophyte Selaginella moellendorffii but not in the bryophyte Physcomitrella patens. The Plant Cell. 2007;19:3058–3079. doi: 10.1105/tpc.107.051524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Ueguchi-Tanaka M, Matsuoka M. GID1-mediated gibberellin signaling in plants. Trends in Plant Science. 2008;13:192–199. doi: 10.1016/j.tplants.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytologist. 2008a;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends in Plant Science. 2008b;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Matilla A. Genes involved in ethylene and gibberellins metabolism are required for endosperm-limited germination of Sisymbrium officinale L. seeds. Planta. 2010;231:653–664. doi: 10.1007/s00425-009-1073-5. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernandez R, Rodriguez-Gacio MC, Barrero-Sicilia C, Carbonero P, Matilla A. Three endo-β-mananase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta. 2011;233:25–36. doi: 10.1007/s00425-010-1257-z. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. The Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim Y-C, et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. The Plant Journal. 2007;50:958–966. doi: 10.1111/j.1365-313X.2007.03098.x. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Riechmann JL, Meyerowitz EM. Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. The Plant Cell. 2008;20:2571–2585. doi: 10.1105/tpc.108.062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP. Revealing the structural and functional diversity of plant cell walls. Current Opinion in Plant Biology. 2008;11:308–313. doi: 10.1016/j.pbi.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theoretical and Applied Genetics. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G. Plant hormone interactions during seed dormancy release and germination. Seed Science Research. 2005;15:281–307. [Google Scholar]
- Leubner-Metzger G. Seed after-ripening and over-expression of class I β-1,3-glucanase confer maternal effects on tobacco testa rupture and dormancy release. Planta. 2002;215:959–968. doi: 10.1007/s00425-002-0837-y. [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G. Functions and regulation of β-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Science Research. 2003;13:17–34. [Google Scholar]
- Leubner-Metzger G, Fründt C, Vögeli-Lange R, Meins F. Class I β-1,3-glucanases in the endosperm of tobacco during germination. Plant Physiology. 1995;109:751–759. doi: 10.1104/pp.109.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Gräber K, Knight C, Leubner-Metzger G. The evolution of seeds. New Phytologist. 2010a;186:817–831. doi: 10.1111/j.1469-8137.2010.03249.x. [DOI] [PubMed] [Google Scholar]
- Linkies A, Müller K, Morris K, et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. The Plant Cell. 2009;21:3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Schuster-Sherpa U, Tintelnot S, Leubner-Metzger G, Müller K. Peroxidases identified in a subtractive cDNA library approach show tissue-specific transcript abundance and enzyme activity during seed germination of Lepidium sativum. Journal of Experimental Botany. 2010b;61:491–502. doi: 10.1093/jxb/erp318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA. Endosperm origin, development, and function. The Plant Cell. 1993;5:1383–1399. doi: 10.1105/tpc.5.10.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A, Suslov D, Fry SC, Verbelen J-P, Vissenberg K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. Journal of Experimental Botany. 2009;60:3959–3972. doi: 10.1093/jxb/erp229. [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T, Steber CM. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. The Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, Linkies A, Müller K, Oracz K, Wang X, Lynn JR, Leubner-Metzger G, Finch-Savage WE. Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: a global tissue-specific transcript analysis. Plant Physiology. 2011 doi: 10.1104/pp.110.169706. doi: 10.1104/pp.110.169706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Job C, Belghazi M, Job D, Leubner-Metzger G. Proteomics reveal tissue-specific features of the cress (Lepidium sativum L.) endosperm cap proteome and its hormone-induced changes during seed germination. Proteomics. 2010;10:1–11. doi: 10.1002/pmic.200900548. [DOI] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:864–877. doi: 10.1093/pcp/pcj059. [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun T-P, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, et al. Identification and characterization of Arabidopsis gibberellin receptors. The Plant Journal. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Developmental Biology. 2000;220:412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y. Abscisic acid and the control of seed dormancy and germination. Seed Science Research. 2010;20:55–67. [Google Scholar]
- Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. The Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni BR, Bradford KJ. Germination and dormancy of abscisic acid- and gibberellin-deficient mutant tomato (Lycopersicon esculentum) seeds (sensitivity of germination to abscisic acid, gibberellin, and water potential) Plant Physiology. 1993;101:607–617. doi: 10.1104/pp.101.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni R-J, Shen Z, Yang C-P, Wu Y-D, Bi Y-D, Wang B- C. Identification of low abundance polyA-binding proteins in Arabidopsis chloroplast using polyA-affinity column. Molecular Biology Reports. 2010;37:637–641. doi: 10.1007/s11033-009-9478-6. [DOI] [PubMed] [Google Scholar]
- Nonogaki H, Gee OH, Bradford KJ. A germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiology. 2000;123:1235–1246. doi: 10.1104/pp.123.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. The Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. Journal of Plant Research. 2006;119:153–162. doi: 10.1007/s10265-006-0262-6. [DOI] [PubMed] [Google Scholar]
- Østerlund T. Structure–function relationships of hormone-sensitive lipase. European Journal of Biochemistry. 2001;268:1899–1907. doi: 10.1046/j.1432-1327.2001.02097.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. Green revolution genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Petruzzelli L, Müller K, Hermann K, Leubner-Metzger G. Distinct expression patterns of β-1,3-glucanases and chitinases during the germination of Solanaceous seeds. Seed Science Research. 2003;13:139–153. [Google Scholar]
- Piskurewicz U, Lopez-Molina L. The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signaling and Behaviour. 2009;4:63–65. doi: 10.4161/psb.4.1.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Tureckova V, Lacombe E, Lopez-Molina L. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO Journal. 2009;28:2259–2271. doi: 10.1038/emboj.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. The Plant Cell. 2008;20:2729–2745. doi: 10.1105/tpc.108.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J, Tatematsu K, Kanno Y, Hobo T, Kimura M, Jikumaru Y, Yano R, Kamiya Y, Nambara E. Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: a comparative study on dormant and non-dormant accessions. Plant and Cell Physiology. 2009;50:1786–1800. doi: 10.1093/pcp/pcp121. [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends in Plant Science. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D. The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiology. 2004;134:1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd NP. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annual Reviews of Plant Physiology. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove D. The expansin superfamily. Genome Biology. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P. Biomechanics of plant growth. American Journal of Botany. 2006;93:1415–1425. doi: 10.3732/ajb.93.10.1415. [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- Smith RC, Matthews PR, Schünmnn PHD, Chandler PM. The regulation of leaf elongation and xyloglucan endotransglycosylase by gibberellin in Himalaya™ barley (Hordeum vulgare L.) Journal of Experimental Botany. 1996;47:1395–1404. [Google Scholar]
- Steber CM, Cooney SE, McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149:509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Park SH, Okubo K, et al. Differential expression and affinities of Arabidopsis gibberellin receptors can explain variation in phenotypes of multiple knock-out mutants. The Plant Journal. 2009;60:48–55. doi: 10.1111/j.1365-313X.2009.03936.x. [DOI] [PubMed] [Google Scholar]
- Toorop PE, van Aelst AC, Hilhorst HWM. The second step of the biphasic endosperm cap weakening that mediates tomato (Lycopersicon esculentum) seed germination is under control of ABA. Journal of Experimental Botany. 2000;51:1371–1379. [PubMed] [Google Scholar]
- Toufight K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: eNorthern, expression angling, and promoter analyses. The Plant Journal. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun T. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiology. 2004;135:1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signalling in plants. Annual Reviews of Plant Biology. 2007;58:183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- Van Sandt VST, Stieperaere H, Guisez Y, Verbelen J-P, Vissenberg K. XET activity is found near sites of growth and cell elongation in bryophytes and some green algae: new insights into the evolution of primary cell wall elongation. Annals of Botany. 2007;99:39–51. doi: 10.1093/aob/mcl232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston DE, Elliott RC, Lester DR, Rameau C, Reid JB, Murfet IC, Ross JJ. The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiology. 2008;147:199–205. doi: 10.1104/pp.108.115808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. The Plant Cell. 2007;19:1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An electronic fluorescent pictograph browser for exploring and analyzing large-scale biological data sets. PloS one. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annual Reviews of Plant Biology. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. Journal of Computational Biology. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.