Abstract
A total of 355 simple sequence repeat (SSR) markers were developed, based on expressed sequence tag (EST) and bacterial artificial chromosome (BAC)-end sequence databases, and successfully used to construct an SSR-based genetic linkage map of the apple. The consensus linkage map spanned 1143 cM, with an average density of 2.5 cM per marker. Newly developed SSR markers along with 279 SSR markers previously published by the HiDRAS project were further used to integrate physical and genetic maps of the apple using a PCR-based BAC library screening approach. A total of 470 contigs were unambiguously anchored onto all 17 linkage groups of the apple genome, and 158 contigs contained two or more molecular markers. The genetically mapped contigs spanned ∼421 Mb in cumulative physical length, representing 60.0% of the genome. The sizes of anchored contigs ranged from 97 kb to 4.0 Mb, with an average of 995 kb. The average physical length of anchored contigs on each linkage group was ∼24.8 Mb, ranging from 17.0 Mb to 37.73 Mb. Using BAC DNA as templates, PCR screening of the BAC library amplified fragments of highly homologous sequences from homoeologous chromosomes. Upon integrating physical and genetic maps of the apple, the presence of not only homoeologous chromosome pairs, but also of multiple locus markers mapped to adjacent sites on the same chromosome was detected. These findings demonstrated the presence of both genome-wide and segmental duplications in the apple genome and provided further insights into the complex polyploid ancestral origin of the apple.
Keywords: Genetic map, genome duplication, Malus×domestica, physical map, segmental duplication, simple sequence repeat
Introduction
Apple is one of the most important fruit crops in the world. The apple belongs to the Rosaceae family. This family is composed of >100 genera and 3000 species, and has been traditionally divided into four subfamilies: Prunoideae (x=8), Spiraeoideae (x=9), Rosoideae (x=7, or 8, or 9), and Maloideae (x=17) (Potter et al., 2002). The taxonomic diversification within the Rosaceae family has led the Rosaceae community to select three species including apple, peach (Prunus persica), and diploid strawberry (Fragaria vesca) as model systems for genomics studies (Shulaev et al., 2008, 2011). To date, genomic studies have been extensively conducted for these three key Rosaceae fruit species. For example, genetic resources such as genetic linkage maps, bacterial artificial chromosome (BAC) libraries, expressed sequence tags (ESTs), microarrays, and genome-wide physical maps have been developed for apple (Xu et al., 2001; Newcomb et al. 2006; Silfverberg-Dilworth et al., 2006; Han et al., 2007; N'Diaye et al., 2008; Schaffer et al., 2008; Gasic et al., 2009; Soria-Guerra et al., 2011), peach (Georgi et al., 2002; Dirlewanger et al., 2006; Zhebentyayeva et al., 2008), and strawberry (Davis et al., 2010).
The domesticated apple, Malus×domestica Borkh., is self-incompatible, highly heterozygous, and displays a juvenile period of 6–10 years or more (Korban and Tartarini, 2009). The apple is a diploid (2n=34) with a relatively small genome size of ∼750 Mb per haploid. More recently, a physical map of the apple genome has been constructed based on fingerprinting analysis of a total of 74 281 BAC clones representing ∼10.5× haploid genome equivalents (Han et al., 2007). This physical map consists of 2702 contigs, and it is estimated to span ∼927 Mb in physical length. However, anchoring these contigs onto chromosomes requires a combination of genetic and physical maps. A high-density genetic map is essential for generating such an integrated physical and genetic map. Moreover, such a genetic linkage map is not only useful for assigning loci to physical positions on chromosomes, but it can also reveal those duplicated areas of a genome. If markers can detect two different loci within a given genome, this may be used to investigate further whether duplicated sequences are randomly arranged in a genome or whether pairs of loci are found in a collinear pattern. An ordered arrangement of duplicated sequences along pairs of chromosomes points to the common origin of these chromosomal segments. Such patterns could be attributed to duplication of a segment of a chromosome or could be due to the polyploid origin of a genome (Schmidt, 2002). In apple, duplication of the apple genome has been previously reported by Xu and Korban (2004), Han and Korban (2008), and, more recently, by Velasco et al. (2010) following the release of the first draft of the apple genome sequence. It is likely that there are several mechanisms responsible for these segmental duplications, but they are usually derived from unequal crossing-over or retroposition. Segmental duplications are important in the structural evolution and functional diversity of plant and animal genomes. However, little is known as to how these duplications are generated or their impact on gene expression and recombination. Due to peculiarities and frequency of occurrence, unequal crossing-over produces tandemly arrayed sets of segmental duplications (Sanzol, 2010). Segmental duplications can involve multiple small-scale local duplications or large duplications of several genes in a single event by affecting larger chromosomal regions or a less frequent event of whole genome duplication, namely polyploidization (Sanzol, 2010).
In this study, a high-density genetic map was constructed for the apple genome using 449 apple simple sequence repeat (SSR)-based markers, including EST-SSRs, BAC-end sequence (BES)-SSRs, and sequence tagged sites (STS). All these markers and their primers are freely available online (http://titan.biotec.uiuc.edu/apple/resources.shtml). This collection of SSR markers was also used to screen apple BAC libraries to anchor the physical map onto the genetic map of the apple genome, yielding an integrated map spanning ∼421 Mb in cumulative physical length for the draft of the apple genome. This integrated physical and genetic map revealed duplication of various homoeologous linkage group pairs, thus confirming genome-wide duplication in the apple genome. Moreover, multilocus markers were mapped to adjacent sites on the same chromosomes, thus identifying segmental duplications in the apple genome.
Materials and methods
Plant material
A segregating F1 population derived from a cross between ‘Co-op 17’ and ‘Co-op 16’ was used for linkage map construction. ‘Co-op 17’ and ‘Co-op 16’ are advanced selections from the Purdue–Rutgers–Illinois (PRI) scab (Venturia inaequalis) resistance cooperative apple breeding programme initiated back in 1945 (Williams et al., 1975). The segregating population consisted of 142 individual seedlings. Young leaves of these apple seedlings and their parents were collected for DNA isolation.
Fluorescent microsatellite genotyping
Genotyping was carried out using a three-primer strategy, including a forward primer with an M13 tail (5′-GTAAAACGACGGCCAGT-3′) at the 5′ end, a regular reverse primer, and a universal M13 primer labelled with one of the following dyes: 6FAM, VIC, NED, or PET (Applied Biosystems, Foster City, CA, USA). The tailed primer provided a complementary sequence to the fluorescent labelled M13 primer, leading to the amplification of fluorescent PCR products.
The PCR was prepared in 10 μl containing 1× Colorless GoTaq® Flexi PCR buffer (Promega, Madison, WI, USA), 1.5 mM MgCl2, 0.2 μM dNTP, 0.0125 μM tailed forward primer, 0.25 μM reverse primer, 0.15 μM labelled M13 primer, 0.5 U of GoTaq® Flexi DNA Polymerase (Promega), and 50 ng of genomic DNA. PCR amplification was conducted as follows: 5 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 56 °C, and 1 min at 72 °C, and a final extension at 72 °C for 30 min. A total of 12 PCR products of different sizes and/or labelled with different fluorescent dyes were mixed, and the mixture was further diluted 1:10. A 2.0 μl aliquot of the PCR product mixture was precipitated using ethanol and resuspended in 10 μl of deionized-distilled water. A 3 μl aliquot of desalted PCR products was added to 8 μl of formamide containing 0.8 μl of GeneScan 500 LIZ standard (Applied Biosystems), and the mixture was run on an ABI3730 DNA analyzer (Applied Biosystems). Data were collected and analysed using GeneMapper v3.7 software (Applied Biosystems).
Construction of a genetic linkage map
A total of 142 F1 seedlings of the cross Co-op 17×Co-op 16 were used for the construction of a genetic linkage map for the apple. The linkage analysis was performed using JoinMap version 4.0 (Van Ooijen, 2006). Two heterozygous SSR alleles from either parent were expected to segregate at a 1:1 ratio, and all SSR loci were tested by χ2 test. Markers were excluded from the linkage analysis when their distorted segregation conflicted with the segregation pattern of neighbouring markers. A LOD score of 8.0 was initially set as the linkage threshold to determine markers belonging to the same linkage groups. Once 17 linkage groups corresponding to the known haploid chromosome number of the apple genome were determined, the rest of the markers were added to their corresponding groups using a less stringent criterion of LOD score of 4.0. Maternal and paternal data sets were created using the function of ‘Create Maternal and Paternal Node’ in the JoinMap program. The regression mapping algorithm was used for map construction. Once the female and male maps were established, a consensus map was built using the CP population model. Marker orders in the female and male maps were used as preferred orders (the ‘fixed order’ function) for the construction of a consensus map. Map distances were calculated using Kosambi's mapping function, and denoted in centiMorgans (cM).
BAC library screening
A total of 46 080 BAC clones, representing 6× haploid genome equivalents, were subjected to screening. Of these BAC clones, 23 808 and 22 272 clones were derived from BamHI- and HindIII-restricted BAC libraries of apple cv. GoldRush, respectively. To facilitate screening of the BAC library using the PCR-based protocol, BAC clones were mixed to generate plate, row, and column pools. Each plate pool consisted of a preparation of all BAC clones from a single 384-well plate. For row and column pools, 384-well plates were divided into units of 10 plates. Each unit of 10 plates was then stacked. Each row or column of stacked plates was combined, yielding row and column pools, respectively. As a result, a total of 600 pools, comprising 120 plate pools, 192 row pools, and 288 column pools, were constructed. Positive BAC clones were identified using a two-step PCR-based screening procedure. The first step involved screening plate pools. A total of 40 pools, 24 column pools and 16 row pools, corresponding to each positive plate pool, were then selected and subjected to screening. Positive clones were individually screened if determined to be derived from the same 384-well plate. The PCR program consisted of 3 min at 94 °C, followed by 34 cycles of 30 s at 94 °C, 30 s at 58 °C, 60 s at 72 °C, and a final extension for 5 min at 72 °C.
Results
Development of candidate EST-SSRs and BAC-end sequence-derived SSRs
The apple EST database was mined for the presence of SSRs, and the results, such as SSR repeat type and length, and suggested primers were deposited in a public database at the Genomic Facility of the University of California-Davis web site (http://cgf.ucdavis.edu/home/). Based on these results, a total of 1160 non-redundant EST sequences containing SSR motifs were identified, and their designed primers were synthesized to amplify genomic DNA of two parents of the mapping population Co-op 17×Co-op 16. PCR products were separated on a 2% MetePhor® agarose gel (Cambrex Bio Science Rockland Inc., Rockland, ME, USA). Of 1160 pairs of primers, 323 (27.8%) were identified to be polymorphic between ‘Co-op 17’ and ‘Co-op 16’, and their names had prefixes ‘CTG’ or ‘CN’.
Similarly, a total of 187 pairs of primers were designed for candidate BES-SSRs based on previous analysis of the BES database (Han and Korban, 2008). Of 187 pairs of primers, 32 (17.1%) were determined to be polymorphic between ‘Co-op 17’ and ‘Co-op 16’, and their names had a prefix ‘BACSSR’. Primer sequences of all candidate EST-SSRS and BES-SSRs were deposited in a University of Illinois online database (http://titan.biotec.uiuc.edu/apple).
Construction of an SSR-based genetic linkage map
To map the newly developed EST- and BES-SSRs described above genetically, a total of 279 previously published SSR markers (258 single locus and 21 multilocus) were further selected from a public domain of apple molecular markers (http://www.hidras.unimi.it/), and used to screen the parents ‘Co-op 17’ and ‘Co-op 16’. The results indicated that 87 out of 279 published SSR markers were polymorphic between the two parents. Of these 87 SSRs, six belonged to EST-SSRs and 81 were developed from genomic DNA sequences, referred to as G-SSRs. Moreover, a total of 12 STS markers derived from BAC-end sequences (BES-STSs, unpublished) and three gene-linked G-SSRs or STSs, namely F3H-1-SSR, F3H-2-SSR, and F3H-2-indel (Han et al., 2010), were also identified to be polymorphic between ‘Co-op 17’ and ‘Co-op 16’. Those BES-STSs markers are identified with a ‘KB’ prefix.
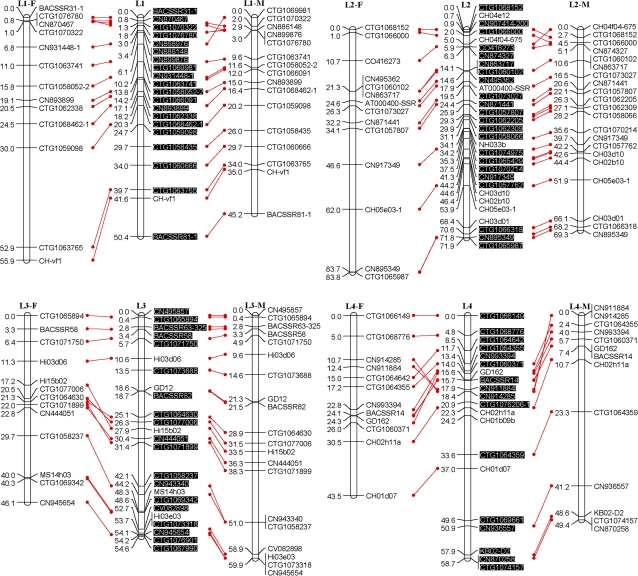
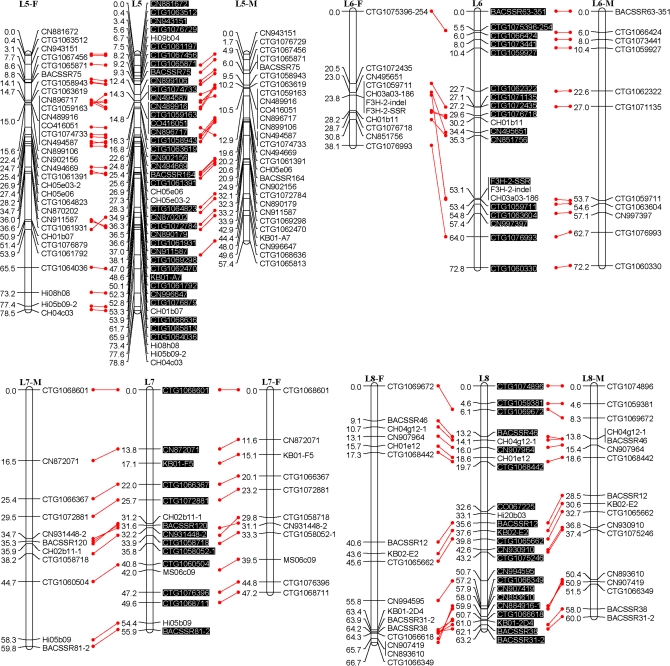
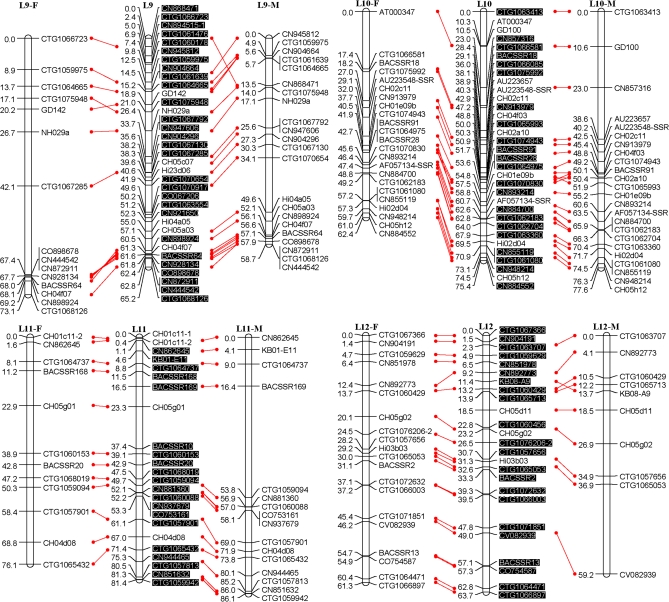
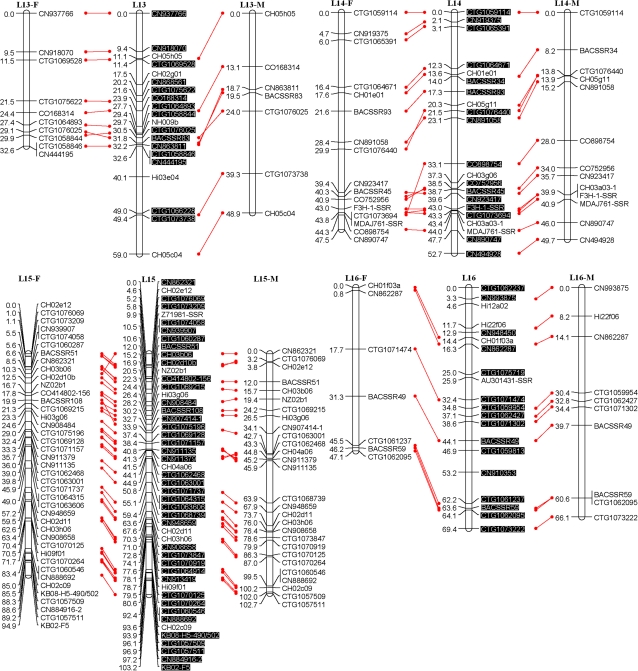
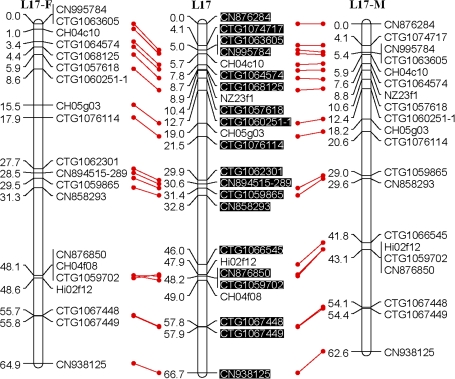
All the above polymorphic markers, namely 329 EST-SSRs, 115 G-SSRs/BES-SSRs, and 13 STSs, were then used to screen all seedlings of the mapping population, Co-op 17×Co-op 16. This screening revealed the presence of a total of 472 loci; however, 23 loci, CTG1072937, CTG1072509, CTG1071564, CTG1068197, CTG1066180, CN870190, CN848860, CN914459, CO723439, Z71980-SSR, MS01a03, CH01c08, CH01c09, CH04d11, BACSSR104, BACSSR153, CN889061, CN896269, CN948094, CN947990, KB01-2D6L, KB01-A9, and KB09-E2, were later excluded from linkage analysis as they either failed to link with any of the linkage groups or their distorted segregation conflicted with the segregation patterns of neighbouring markers. Moreover, 15 seedlings were found to carry several double recombination events, and were thus excluded from further linkage analysis. Finally, a consensus linkage map consisting of 449 loci or markers on 17 linkage groups was successfully generated (Fig. 1). All markers mapped onto linkage groups have been summarized in Table 1. Each linkage group had 16–49 markers with an average of 26.4 (Table 2). The consensus linkage map spanned 1143 cM with an average density of 2.5 cM per marker.
Fig. 1.
An SSR-marker based genetic linkage map of the apple genome. Female (F) and male (M) maps are shown on the left and right, respectively, and the consensus map is shown in the centre. Homologous loci are connected with solid lines. New SSR markers developed in this study are shaded in black and are placed only on the consensus map. (This figure is available in colour at JXB online.)
Table 1.
SSR markers used to construct the genetic linkage map of the apple genome
| Marker type | EST-SSR |
G-SSR |
STS |
|||
| Single-locus | Multilocus | Single-locus | Multilocus | Single-locus | Multilocus | |
| Newly developed | 303 | 9 | 26 | 5 | 9 | 2 |
| Previously published | 6 | – | 79 | 5 | – | – |
| Total | 309 | 8 | 105 | 10 | 9 | 2 |
Table 2.
Numbers of SSR markers, indels, and cumulative physical lengths of BAC contigs genetically mapped onto different linkage groups in apple
| Chromosomea | Linkage group |
Physical map |
|||
| No. of markers | Length (cM) | No. of contigs | No. of markers | Length (Mb) | |
| 1 | 21 | 50.4 | 24 | 32 | 26.96 |
| 2 | 30 | 71.9 | 39 | 50 | 30.01 |
| 3 | 24 | 54.6 | 23 | 38 | 22.21 |
| 4 | 20 | 58.7 | 21 | 37 | 22.59 |
| 5 | 43 | 78.8 | 37 | 64 | 30.36 |
| 6 | 20 | 72.8 | 17 | 29 | 15.51 |
| 7 | 16 | 55.9 | 19 | 20 | 17.00 |
| 8 | 24 | 63.2 | 23 | 34 | 18.52 |
| 9 | 35 | 65.2 | 24 | 43 | 20.35 |
| 10 | 33 | 75.4 | 42 | 55 | 33.78 |
| 11 | 24 | 81.4 | 42 | 46 | 34.23 |
| 12 | 25 | 63.7 | 31 | 40 | 23.64 |
| 13 | 20 | 59.0 | 28 | 35 | 23.82 |
| 14 | 21 | 52.7 | 19 | 39 | 18.17 |
| 15 | 49 | 103.2 | 40 | 62 | 37.73 |
| 16 | 20 | 69.4 | 24 | 38 | 22.26 |
| 17 | 24 | 66.7 | 20 | 37 | 23.62 |
| Total | 449 | 1143.0 | 473 | 699 | 420.76 |
The order of chromosome numbers is consistent with that of the genetic linkage groups.
Integration of physical and genetic maps
Previously, a BAC-based physical map of the apple genome was constructed (Han et al., 2007). To anchor the physical map onto the genetic map, two approaches were attempted. One approach involved developing BAC-end sequence-based single nucleotide polymorphisms (SNPs). Briefly, BAC clones along ends of contigs were selected, and sequenced at both ends. The BAC-end sequences were then used to develop SNPs using a bin mapping population (Han et al., 2009). As a result, a total of 52 SNPs were successfully developed, and 51 contigs were subsequently anchored onto the genetic map (Supplementary Table S1 available at JXB online). Another PCR-based approach involved screening of the BAC library using 649 genetic markers. Of these 649 markers, 279 were previously published SSRs and 370 were newly developed markers, including 323 EST-SSRs, 32 BES-SSRs, 12 BES-STSs, and three gene tags. Results of this screening indicated that 20 SSRs (six published and 14 newly developed) failed to detect any positive BAC clones as all BAC DNA pools were either negative or positive, whereas each of the remaining 599 single-locus markers identified ∼7.0 positive BAC clones, ranging from one to 16. As regards 30 multilocus markers, each detected 5–25 positive BAC clones with an average of ∼9.5.
The physical location of positive BAC clones was determined based on the BAC-based physical map. As a result, 308 single-locus markers were identified in single contigs, and 291 single-locus markers were identified in two or three contigs. Contigs were deemed overlapping if their ends consisted of positive clone(s) from the same makers, and a total of 231 contigs were later merged. A total of 539 single-locus markers were then anchored onto single contigs. Out of the remaining 60 single-locus markers, 32 were located in two contigs on the same chromosome and probably represent different haplotypes (Table 3). A total of 24 single-locus markers were identified in two contigs on different chromosomes, representing homologous regions that will be described in further detail (Table 4). Among 30 multilocus markers, seven markers, CH04f04, CH01c11, CH05e03, CN884916, CN894515, CTG1065432, and KB01-A9, were identified in single contigs. Twenty-three multilocus markers were identified in either two or three contigs that were subsequently distinguished and anchored onto linkage groups according to allele sizes of these multilocus markers and/or their neighbouring molecular markers located on the same contigs. In addition, 33 sequence-tagged connectors (STCs) used to screen the apple BAC library were anchored onto different contigs (STCs are highlighted with a yellow background in Supplementary Table S1 at JXB online).
Table 3.
Markers anchored onto different contigs using PCR-based BAC library screening
| Marker | Linkage group | Contiga |
Contiga |
||
| Name | Length (kb) | Name | Length (kb) | ||
| CN899876 | 1 | 364 | 2577 | 397 | 823 |
| CH05g08 | 1 | 42 | 1259 | 1172 | 121 |
| CH03b01 | 2 | 400 | 487 | 4151 | 363 |
| AJ251116-SSR | 2 | 1326 | 542 | 1550 | 156 |
| CH03d10 | 2 | 84 | 1950 | 1663 | 160 |
| CTG1066149 | 4 | 1733 | 1010 | 1344 | 125 |
| CH02h11a | 4 | 1100 | 1938 | 1773 | 211 |
| AU223670-SSR | 5 | 22 | 2125 | 3650 | 265 |
| CTG1063512 | 5 | 88 | 1938 | 2571 | 168 |
| CTG1076729 | 5 | 172 | 1064 | 1886 | 199 |
| MS06c09 | 7 | 1950 | 1396 | 3125 | 152 |
| CH04e05 | 7 | 449 | 3552 | 2135 | 1489 |
| CH01f12 | 10 | 3280 | 1232 | 3573 | 764 |
| CH02c11 | 10 | 3993 | 889 | 733 | 148 |
| CTG1075992 | 10 | 395 | 1392 | 2545 | 97 |
| AF057134-SSR | 10 | 3796 | 975 | 3437 | 316 |
| Hi02d04 | 10 | 1296 | 1918 | 4197 | 226 |
| CN851632 | 11 | 2202 | 682 | 3202 | 144 |
| CH03d02 | 11 | 2174 | 780 | 4168 | 620 |
| CO753161 | 11 | 4188 | 975 | 1763 | 230 |
| CO754587 | 12 | 660 | 476 | 3909 | 464 |
| Hi02b07 | 12 | 207 | 862 | 1515 | 136 |
| AU223486-SSR | 13 | 1647 | 351 | 2313 | 129 |
| CH05g11 | 14 | 73 | 986 | 649 | 312 |
| Hi02d11 | 14 | 73 | 986 | 732 | 554 |
| NZ22c6 | 14 | 892 | 296 | 1163 | 436.8 |
| MDAJ761-SSR | 14 | 490 | 1220 | 414 | 877 |
| Hi11a01 | 15 | 2890 | 1661 | 737 | 636 |
| Hi09f01 | 15 | 2890 | 1661 | 737 | 636 |
| CH02a03 | 16 | 505 | 1041 | 3377 | 682 |
| AT000174-SSR | 17 | 85 | 3724 | 3719 | 444 |
| CH04b10 | 17 | 3977 | 990 | 2201 | 335 |
The two contigs derived from the same marker represent different haplotypes or homologous regions within the apple genome.
Table 4.
Markers anchored onto contigs from different chromosomes using PCR-based BAC library screening
| Marker | Contig 1 |
Contig 2 |
|||
| Name | LG | Name | Markera | LG | |
| CO416273 | 284 | 2 | 435 | CTG1062468 | 15 |
| CH02c02a | 335 | 2 | 2369 | CTG1071737 | 15 |
| CTG1067990 | 1581 | 3 | 2202 | CN851632 | 11 |
| CN945654 | 1581 | 3 | 2155 | CN944465 | 11 |
| CH01c08 | 516 | 3 | 112 | CN491050-SSR | 11 |
| CTG1065813 | 1159 | 5 | 3796 | AF057134-SSR | 10 |
| CTG1065871 | 876 | 5 | 495 | CH02b03b | 10 |
| CH04g09 | 2425 | 5 | 3911 | BACSSR18 | 10 |
| CH05a02 | 3408 | 8 | 329 | NZ02b1 | 15 |
| BACSSR103 | 352 | 10 | 92 | CTG1072784 | 5 |
| CN944465 | 2155 | 11 | 1581 | CN945654 | 3 |
| CTG1059094 | 870 | 11 | 64 | CTG1069342 | 3 |
| CH03d02 | 2174 | 11 | 91 | Hi15h12 | 3 |
| CH01b12 | 532 | 12 | 763 | CTG1074157 | 4 |
| AU1223486-SSR | 1647 | 13 | 3355 | CH02d10a | 16 |
| CH05h05 | 749 | 13 | 505 | CH02a03 | 16 |
| CN494928 | 246 | 14 | 712 | CH03c01 | 6 |
| MDAJ761-SSR | 490 | 14 | 2917 | CTG1059711 | 6 |
| CTG1074058 | 1332 | 15 | 637 | CTG1068442 | 8 |
| Z71981-SSR | 1332 | 15 | 637 | CH01e12 | 8 |
| CTG1073222 | 3182 | 16 | 1876 | Hi08f06 | 13 |
| CTG1067448 | 3395 | 17 | 1625 | CH01h02 | 9 |
| CTG1067449 | 3395 | 17 | 1625 | CH01h02 | 9 |
| CTG1057618 | 85 | 17 | 1334 | CN868471 | 9 |
The nearest marker to positive BAC clone(s). LG corresponds to the linkage group.
To merge potentially overlapping contigs, the Ctg→Ends function at a relaxed threshold of the Sulston score (3e-6) was used. BAC clones comprised of terminal ends of contigs were allowed to join other contigs following manual review. Contig pairs were merged if their terminal clones shared >10 bands and their overall fingerprint patterns supported the junction. As a result, a total of 664 contigs were merged to generate a genome-wide physical map for the apple genome. The physical map consisted of 1806 contigs, and it was estimated to span ∼828.03 Mb in physical length. Of 1806 contigs, 484 were anchored by one or more molecular marker(s) with contig 85 on LG17 covering a maximum of 11 molecular markers (Supplementary Table S1 at JXB online).
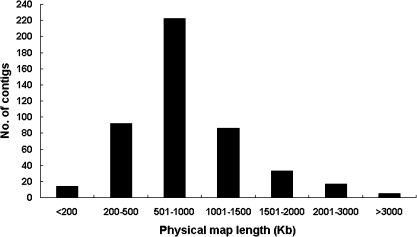
The physical length distribution of these anchored contigs is shown in Fig. 2. Most of the contigs (47%) are 500–1000 kb in length, and 141 contigs (30%) are longer than 1 Mb. The average size of these anchored contigs is 897.4 kb, ranging from 97 kb to 4.0 Mb. Among 484 contigs, 470 contigs spanning ∼420.76 Mb in cumulative physical length have been unambiguously anchored onto 17 linkage groups (Table 2). Given the estimated 700 Mb size of the apple genome (Velasco et al., 2010), the physical length of anchored contigs covers 60% of the genome. The average physical length of the integrated map on each linkage group is ∼24.8 Mb, ranging from 17.0 Mb to 37.73 Mb. Fourteen contigs, spanning 6.4 Mb in cumulative physical length, are tagged with molecular markers or STCs. However, they could not be definitively assigned to any of the apple linkage groups as their corresponding markers or STCs have not yet been genetically mapped (Supplementary Table S1 at JXB online).
Fig. 2.
Distribution of physical lengths of BAC contigs anchored onto the apple genetic map.
Integration of physical and genetic maps confirms duplication signatures within the apple genome
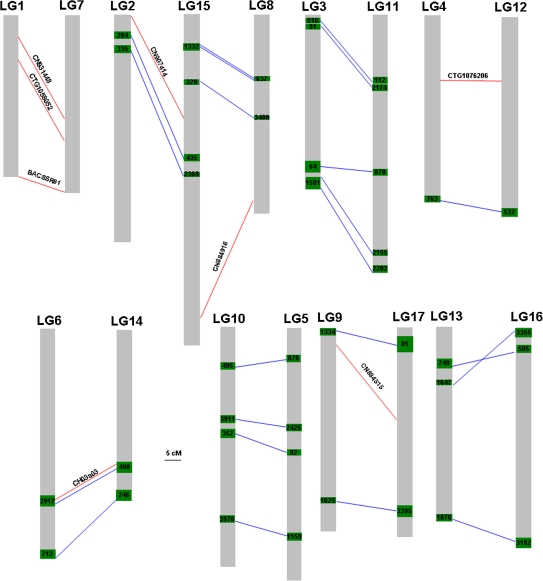
It is well known that the apple genome was derived from a polyploid origin (Phipps et al., 1991; Evans and Campbell, 2002). In this study, integrating physical and genetic maps revealed the presence of homoeologous pairs of chromosomes in apple. Initially, it was found that positive clones screened by PCR using the same primers were identified in two contigs that were unambiguously anchored onto different linkage groups. For example, the EST-SSR marker CTG1065813 was mapped onto LG5, and its corresponding positive clones were located in the following two contigs, 1159 and 3796. Contig 1159 consisted of two markers, CTG1064036 and BSNP74, both of which were mapped onto LG5, whereas contig 3796 contained the marker AF057134-SSR that anchored onto LG10. This finding suggested that PCR probably amplified fragments of highly homologous sequences from homoeologous chromosomes when using BAC DNA as templates. Thus, a total of 24 SSR markers were later found to identify positive clones in two contigs from different linkage groups (Table 4). The two linkage groups were deemed to be homoeologous if they shared two or more homologous sequences that were detected by PCR screening of the BAC library as mentioned above. These homoeologous linkage group pairs are presented in Fig. 3.
Fig. 3.
Multilocus SSR markers and genetically anchored BAC contigs reveal duplication within the apple genome. The red lines indicate homologous sites detected by multilocus SSR markers with their names above the line. Contigs from different chromosomes consisting of positive BAC clones identified with the same SSR markers are indicated by blue lines. The numbers in green boxes represent genetically anchored BAC contigs.
Those multilocus SSR markers that have been genetically mapped also provided insights into the homoeologous pairs of chromosomes. A total of eight multilocus SSRs were added to the list of markers used for screening the BAC library, and these revealed additional homoeologous pairs of linkage groups (Fig. 3). Thus, integrating physical and genetic maps revealed duplication of linkage group pairs, including the following: 1–7, 2–15, 3–11, 4–12, 5–10, 6–14, 8–15, 9–17, and 13–16 (Fig. 3). These homoeologous linkage groups clearly demonstrated genome-wide duplication in the apple genome.
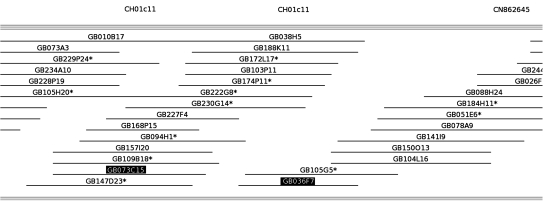
In addition to the observed genome-wide duplication, segmental duplications at adjacent sites on the same chromosomes were also identified. For example, the SSR marker CH01c11 genetically mapped to two sites that were in close proximity to each other on LG11 (Fig. 1). PCR screening of the apple BAC library revealed that CH01c11 hit the two BAC clones GB073C15 and GB036F7. These two clones were assembled into contig 2, and they did not physically overlap with each other (Fig. 4). This demonstrated that the physical positions of the multilocus marker CH01c11 were consistent with its genetic position. Further, Fernández-Fernández et al. (2008) also mapped a multilocus SSR marker CH04h02 in the same region of LG11 at a similar position to CH01c11. These findings strongly suggested that the homologous regions surrounding CH01c11-1 and CH01c11-2 were derived from a tandem segmental duplication.
Fig. 4.
An example of a BAC contig genetically anchored using two SSR markers, CH01c11 and CN862645. GB073C15 and GB036F7 in black boxes are positive BAC clones of the same SSR marker CH01c11.
Discussion
Integration of genome-wide physical and genetic maps has been reported in a wide variety of model plant species such as Arabidopsis and rice. However, for fruit trees, integrated physical and genetic maps have only been reported in peach (Zhebentyayeva et al., 2008) and papaya (Yu et al., 2009). In this study, an integrated physical and genetic map spanning ∼421 Mb in cumulative physical length and representing 60% of the apple genome has been developed. The framework physical map of the apple will serve as a valuable resource for various other studies such as effective positional cloning of genes and quantitative trait locus (QTL) fine-mapping.
Developing apple SSR markers and construction of an apple linkage map
SSR markers can be divided into two groups, G-SSRs and EST-SSRs, based on their origin. G-SSRs are developed from genomic DNA, whereas EST-SSRs are derived from expressed sequences of the genome. G-SSRs are usually predicted to have a higher level of polymorphism than EST-SSRs as DNA sequences are conserved in expressed regions (Cho et al., 2000). However, EST-SSRs have been reported to have the same level of polymorphism as G-SSRs in loblolly pine (Liewlaksaneeyanawin et al., 2004). This phenomenon has also been observed in this study. For example, apple EST-SSRs have revealed higher levels of polymorphism between the two parents ‘Co-op 17’ and ‘Co-op 16’ than BES-SSRs (27.8% versus 17.1%) (Han et al., 2009). Moreover, out of 254 published G-SSRs (Liebhard et al., 2003; Silfverberg-Dilworth et al., 2006; N'Diaye et al., 2008), only 81 (31.9%) have displayed polymorphism between ‘Co-op 17’ and ‘Co-op 16’. It is worth noting that the low frequency of detecting G-SSRs may be partially attributed to primer site variation as 99% of EST-SSR primers generated PCR products. However, ∼10% of primers from published G-SSRs and BBES-SSRs have either failed to amplify DNA or their PCR products are faint on agarose gels. The observed failure of PCR amplification may be primarily due to primer site variation as the apple genome is highly heterozygous. Therefore, primers from genomic sequences have a higher tendency to uncover allele(s) than those from expressed sequences, thus leading to a biased estimation of the level of polymorphism detected by G-SSRs.
Of 355 EST-SSRs and BES-SSRs developed in this study, 337 have been used to construct an SSR-based genetic linkage map of the apple. In general, SSR markers are not evenly distributed across all apple chromosomes (Fig. 1). To check for reliability of this linkage group map, genetic map positions of published SSR markers (mostly prefixed with ‘CH’ or ‘Hi’) have been compared with those of previously published linkage maps (http://users.unimi.it/hidras/). Out of 87 published SSRs, 83 have been mapped to positions similar to those reported previously. A single SSR, CH04f04, has been mapped to LG2, whereas it has been previously reported to be located on LG5 (Liebhard et al. 2002). This inconsistency suggests that CH04f04 is probably a multilocus marker as its PCR products obtained in this study are 675 bp in size, whereas its alleles reported previously ranged from 146 bp to 166 bp in size. In addition, two SSRs, CH03a03 and CH05e03, previously presumed to be single-locus markers, are found to be multilocus markers in this study (Fig. 1). Altogether, these results suggest that the linkage map in this study is reliable.
PCR screening of the BAC library and homoeologous chromosome pairs in apple
BAC library screening is a critical and substantial component of any effort to develop a genome-wide integrated physical and genetic map, and it is commonly undertaken using a hybridization strategy (Chen et al., 2002; Han and Korban, 2010). In this study, a total of 629 SSR markers have been used to screen the apple BAC library using a PCR-based protocol, and these markers have been anchored onto 470 BAC contigs. Among those genetically anchored contigs, 158 (33.7%) consist of two or more molecular markers, suggesting that the integrated physical and genetic map of the apple is reliable. Interestingly, this integration has also revealed that single-locus markers are identified in two contigs that are anchored onto different linkage groups. For example, SSR marker CH03d02 has identified 11 positive BAC clones and, of these, four are located on contig 91 that have also been identified by SSR marker Hi15h12. The remaining seven clones are assembled within contig 2174. SSR markers CH03d02 and Hi15h12 are genetically mapped onto LG11 and LG3, respectively (N'Diaye et al., 2008). LG3 and LG11 have been reported to be a homoeologous pair (Dirlewanger et al., 2004; Velasco et al., 2010). It is thus speculated that PCR screening of the apple BAC library is likely to amplify homologous sequences from homoeologous chromosomes, and these findings of PCR-based BAC library screening have been subsequently verified. Thus, a total of 24 single-locus markers have been identified and subsequently used to establish homoeologous pairs of linkage groups (Table 4, Fig. 3).
The apple genome has been sequenced using a whole-genome shotgun sequence strategy, and the sequence comparison reveals homoeologous chromosome pairs, namely 1–7, 2–15, 3–11, 4–12, 5–10, 6–14, 8–15, 9–17, and 13–16 (Velasco et al., 2010). These homoeologous chromosome pairs have also been detected in this present study. It is worth pointing out the homoeologous relationships among LG2, LG8, and LG15. In this study, LG8 is homologous to both the upper and lower regions of LG15; moreover, the upper region of LG2 is homologous to the middle region of LG15. This finding is quite consistent with the observation noted from sequence comparisons of apple chromosomes 2, 8, and 15 (Velasco et al., 2010). Therefore, the results presented here indicate that PCR screening of the apple BAC library can be used not only for anchoring physical and genetic maps, but also for identifying homoeologous pairs of linkage groups in apple. However, it is important to ponder the question of why single-locus markers can amplify multiple targets when they are used in screening BAC libraries. PCR screening of a BAC library is performed using pooled BAC DNA as a template. The pooled BAC clones represent only a small portion of the apple genome, thus increasing the likelihood that primers bind to alternative site(s) and amplify homologous sequences. However, the possibility that some of the presumed single-locus markers may actually be multilocus markers cannot be excluded. In addition, it has been reported that BAC library screening can generate false positives (Mahairas et al., 1999). The recovery of false positives must be attributed to the presence of homoeologous chromosomes and/or regions.
Polyploid origin of the apple genome
Allopolyploidization and autopolyploidization are two main hypotheses proposed for the origin of the apple genome. The allopolyploidization hypothesis claims that the apple genome is derived from a wide hybridization between spireoid (n=9) and amygdaloid (n=8) progenitors (Chevreau et al., 1985; Phipps et al., 1991; Shulaev et al., 2008). The autopolyploidization hypothesis has been recently proposed and states that the apple genome has probably evolved from a cross between two ancestors of Gillenia (n=9), followed by aneuploidization and loss of a single chromosome (Evans and Campbell, 2002). In this study, 17 linkage groups have been assembled into nine homoeologous pairs, namely 1–7, 2–15, 3–11, 4–12, 5–10, 6–14, 8–15, 9–17, and 13–16. This finding clearly demonstrates that whole-genome duplication must be involved in the origin of the apple genome.
Out of 17 linkage groups in apple, LG15 represents the largest chromosome, and it is very interesting. Its upper and lower regions are homoeologous to LG8, whereas its middle region is homologous to the upper region of LG2 (Fig. 3). Therefore, LG8 and LG15 are more likely to be a homoeologous pair. Homology between LG2 and LG15 suggests that LG2 probably has a homoeologous counterpart, LG18, during the early evolutionary stages of apple. LG18, or at least a part of it, has been inserted into LG15, leading to the observed aneuploidization, n=17, in the apple genome. Thus, the origin of the apple has probably evolved from autopolyploidization and subsequent aneuploidization events.
In this study, duplications across a whole chromosome have also been identified between homoeologous linkage group pairs such as 3–11, 5–10, 9–17, and 13–16, whereas duplications between homoeologous linkage group pairs 1–7, 2–15, 4–12, 6–14, and 13–16 are restricted to specific chromosome regions. These findings are consistent with the reported presence of large-scale collinearity between chromosomes 3 and 11, 5 and 10, 9 and 17, and 13 and 16, and the small-scale collinearity found between chromosomes 1 and 7, 2 and 15, 4 and 12, 12 and 14, 6 and 14, and 8 and 15 (Velasco et al., 2010). It is worth pointing out that small-scale homology between chromosome pairs 2–7 and 12–14 has not been identified in this study. This may be attributed to either the limited number of SSR markers used for BAC library screening or the relatively low sequence identity between paired homologous segments in chromosomes 2 and 7 or 12 and 14.
In addition to whole-genome duplication, segmental duplication has also been detected on chromosome 11 in this study. Duplication events within the same chromosome have also been previously reported. For example, the two SSR markers CH05c07 and CH02h02 have been mapped to two loci on LG9 (Gao et al., 2005). Transposable elements comprise 42.4% of the total apple genome (Velasco et al., 2010). Transposable elements involved in segmental duplications have been proposed in several studies (Fiston-Lavier et al., 2007; Lin et al., 2010). Therefore, segmental duplications in the apple genome are probably related to the presence of transposable elements, and it is likely that more segmental duplications are present in the apple genome.
Haplotype assembly and integrating physical and genetic maps in apple
It has been reported that a high level of heterozygosity could lead to independent assembly of haplotypes in hypervariable genomic regions of Populus (Kelleher et al., 2007). The domesticated apple is self-incompatible and it is highly heterozygous. Independent assembly of haplotypes has also been observed during construction of the apple physical map. For example, the SSR marker CO754587 has two alleles; one allele has been identified in contig 660, while another allele has been identified on contig 3909. In this study, a total of 32 markers have been anchored onto two contigs (Table 3). These contigs have been identified by a single marker. As mentioned above, PCR screening of the apple BAC library is likely to amplify homologous sequences from homoeologous chromosomes. Therefore, two paired contigs identified by the same marker may represent different haplotypes or homologous regions within the apple genome. Moreover, a total of 599 single-locus markers have been used to screen the apple BAC library using a PCR-based protocol. Of the 599 markers, 539 (89.9%) have been anchored onto single contigs, and 24 have been successfully used to establish homoeologous chromosome pairs. Taken together, the results suggest that independent assembly of haplotypes is not an obstacle for the construction of a physical map in apple.
PCR screening of a BAC library is a time-consuming effort and, as mentioned, it can amplify highly homologous sequences from homoeologous chromosomes. In contrast, genetic markers derived from BESs can anchor either corresponding BAC clones or contigs onto a genetic map without any need for BAC library screening. Therefore, development of BES-based markers is a promising tool for generating integrated physical and genetic maps. In a previous study, 52 BES-SNPs have been developed and used to anchor 51 contigs onto the apple genetic map using a bin mapping strategy (Han et al., 2009). Of these anchored contigs, 21 have been further anchored with SSR markers in the present study. Analysis of the genetic mapping approach used has indicated that SSR and SNP markers within single contigs are mapped onto the same genetic region. This finding strongly demonstrates that the previous integration of the apple physical and genetic maps using BES-SNPs in conjunction with a bin mapping strategy is a reliable strategy. In addition, BAC-end sequence-derived microsatellite markers have been proven to be useful for integrating physical and genetic maps in soybean (Shultz et al., 2007). In this study, BES-SSR markers have also been evaluated, and 24 of these have been successfully used to integrate physical and genetic maps of the apple (Fig. 1). However, the level of polymorphism of BES-SSRs is low (17.1%). About 16.2% of BES-SSRs are multiple locus markers, and thus their corresponding contigs could not be directly anchored onto the genetic map without BAC library screening. Therefore, development of BES-SNPs seems to be more suitable for integrating physical and genetic maps of the apple.
The draft sequence of the apple genome has been recently reported (Velasco et al., 2010). However, it covers ∼70% of the apple genome. The remaining 30% of the apple genome probably contains repetitive sequence. Therefore, the integrated physical and genetic map will be very helpful for the development of a high-quality draft sequence of the apple genome in the future. Moreover, the integrated map with anchored SSR markers is an invaluable resource for the Rosaceae research community.
Supplementary Material
Acknowledgments
This project was supported by the USDA Cooperative State Research, Education and Extension Service-National Research Initiative-Plant Genome Program grant no. 2005-35300-15538. Also, support was received from the Illinois C-FAR grant 06I-003-3-SEN and the National Natural Science Foundation of China under grant no. 30971987.
References
- Chen M, Presting G, Barbazuk W, et al. An integrated physical and genetic map of the rice genome. The Plant Cell. 2002;14:537–545. doi: 10.1105/tpc.010485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreau E, Lespinasse Y, Gallet M. Inheritance of pollen enzymes and polyploid origin of apple. (Malus×domestica Borkh. Theoretical and Applied Genetics. 1985;71:268–277. doi: 10.1007/BF00252066. [DOI] [PubMed] [Google Scholar]
- Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) Theoretical and Applied Genetics. 2000;100:713–722. [Google Scholar]
- Davis TM, Shields ME, Zhang Q, Tombolato-Terzic, Bennetzen JL, Pantaroli AC, Wang H, Yao Q, San Miguel P, Folta KM. An examination of targeted gene neighborhoods in strawberry. BMC Plant Biology. 2010;10:81e. doi: 10.1186/1471-2229-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirlewanger E, Cosson P, Boudehri K, Renaud C, Capdeville G, Tauzin Y, Laigret F, Moing A. Development of a second generation genetic linkage map for peach [ Prunus persica (L.) Batsch] and characterization of morphological traits affecting flower and fruit. Tree Genetics and Genomes. 2006;3:1–13. [Google Scholar]
- Dirlewanger E, Graziano E, Joobeur T, Garriga-Calderé F, Cosson P, Howad W, Arús P. Comparative mapping and marker assisted selection in Rosaceae fruit crops. Proceedings of the National Academy of Sciences, USA. 2004;101:9891–9896. doi: 10.1073/pnas.0307937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RC, Campbell CS. The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. American Journal of Botany. 2002;89:1478–1484. doi: 10.3732/ajb.89.9.1478. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández F, Evans K, Clarke J, Govan C, James C, Marič S, Tobutt K. Development of an STS map of an interspecific progeny of Malus. Tree Genetics and Genomes. 2008;4:469–479. [Google Scholar]
- Fiston-Lavier AS, Anxolabehere D, Quesneville H. A model of segmental duplication formation in Drosophila melanogaster. Genome Research. 2007;17:1458–1470. doi: 10.1101/gr.6208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, van de Weg WE, Schaart JG, van Arkel G, Breiteneder H, Hoffmann-Sommergruber K, Gilissen LJWJ. Genomic characterization and linkage mapping of the apple allergen genes Mal d 2 (thaumatin-like protein) and Mal d 4 (profilin) Theoretical and Applied Genetics. 2005;111:1087–1097. doi: 10.1007/s00122-005-0034-z. [DOI] [PubMed] [Google Scholar]
- Gasic K, Gonzalez DO, Thimmapuram J, et al. Comparative analysis and functional annotation of a large expressed sequence tag collection of apple. Plant Genome. 2009;2:23–38. [Google Scholar]
- Georgi LL, Wang Y, Yverggniaux D, Ormsbee T, Inigo M, Reighard GL, Abbott AG. Construction of a BAC library and its application to the identification of simple sequence repeats in peach [ Prunus persica (L.) Batsch] Theoretical and Applied Genetics. 2002;105:1151–1158. doi: 10.1007/s00122-002-0967-4. [DOI] [PubMed] [Google Scholar]
- Han Y, Chagné D, Gasic K, Rikkerink EHA, Beever JE, Gardiner SE, Korban SS. BAC-end sequence-based SNPs and Bin mapping for rapid integration of physical and genetic maps in apple. Genomics. 2009;93:282–288. doi: 10.1016/j.ygeno.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Han Y, Gasic K, Marron B, Beever JE, Korban SS. A BAC-based physical map of the apple genome. Genomics. 2007;89:630–637. doi: 10.1016/j.ygeno.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Han Y, Korban SS. An overview of the apple genome through BAC end sequence analysis. Plant Molecular Biology. 2008;67:581–588. doi: 10.1007/s11103-008-9321-9. [DOI] [PubMed] [Google Scholar]
- Han Y, Korban SS. Map-based cloning strategies in apple. Critical Revies in Plant Science. 2010;29:165–284. [Google Scholar]
- Han Y, Vimolmangkang S, Soria-Guerra RE, Rosales-Mendoza S, Zheng D, Lygin AV, Korban SS. Ectopic expression of apple F3'H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiology. 2010;153:806–820. doi: 10.1104/pp.109.152801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher CT, Chiu R, Shin H, et al. A physical map of the highly heterozygous Populus genome: integration with the genome sequence and genetic map and analysis of haplotype variation. The Plant Journal. 2007;50:1063–1078. doi: 10.1111/j.1365-313X.2007.03112.x. [DOI] [PubMed] [Google Scholar]
- Korban SS, Tartarini S. Apple structural genomics. In: Folta KM, Gardiner SE, editors. Genetics and genomics of rosaceae, plant genomics and genomics: crops and models. Vol. 6. New York: Springer-Science; 2009. pp. 85–119. [Google Scholar]
- Liebhard R, Gianfranceschi L, Koller B, Ryder CD, Tarchini R, Weg E, van de Gessler C. Development and characterization of 140 new microsatellites in apple (Malus×domestica Borkh.) Molecular Breeding. 2002;10:217–241. [Google Scholar]
- Liebhard R, Koller B, Gianfranceschi L, Gessler C. Creating a saturated reference map for the apple (Malus× domestica Borkh.) genome. Theoretical and Applied Genetics. 2003;106:1497–1508. doi: 10.1007/s00122-003-1209-0. [DOI] [PubMed] [Google Scholar]
- Liewlaksaneeyanawin C, Ritland CE, El-Kassaby YA, Ritland K. Single-copy, species-transferable microsatellite markers developed from loblolly pine ESTs. Theoretical and Applied Genetics. 2004;109:361–369. doi: 10.1007/s00122-004-1635-7. [DOI] [PubMed] [Google Scholar]
- Lin JY, Stupar RM, Hans C, Hyten DL, Jackson SA. Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris. The Plant Cell. 2010;22:2545–2561. doi: 10.1105/tpc.110.074229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahairas GG, Wallace JC, Smith K, et al. Sequence-tagged connectors: a sequence approach to mapping and scanning the human genome. Proceedings of the National Academy of Sciences, USA. 1999;96:9739–9744. doi: 10.1073/pnas.96.17.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N'Diaye A, Van de Weg WE, Kodde LP, Koller B, Dunemann F, Thiermann M, Tartarini S, Gennari F, Durel CE. Construction of an integrated consensus map of the apple genome based on four mapping populations. Tree Genetics and Genomes. 2008;4:727–743. [Google Scholar]
- Newcomb RE, Crowhurst RN, Gleave AP, et al. Analysis of expressed sequence tags from apple. Plant Physiology. 2006;141:147–166. doi: 10.1104/pp.105.076208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps JB, Robertson KR, Rohrer JR, Smith PG. Origins and evolution of subfam. Maloideae (Rosaceae) Systematic Botany. 1991;16:303–332. [Google Scholar]
- Potter D, Gao F, Bortiri PE, Oh SH, Baggett S. Phylogenetic relationships in Rosaceae inferred from chloroplast matK and trnLtrnF nucleotide sequence data. Plant Systematics and Evolution. 2002;231:77–89. [Google Scholar]
- Sanzol J. Dating and functional characterization of duplicated genes in the apple (Malus domestica Borkh.) by analyzing EST data. BMC Plant Biology. 2010;10:87. doi: 10.1186/1471-2229-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer RJ, Friel EN, Souleyre EJ, et al. A genomics approach reveals that aroma production in apple is controlled by ethylene predominantly at the final step in each biosynthetic pathway. Plant Physiology. 2007;144:1899–1912. doi: 10.1104/pp.106.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. Plant genome evolution: lessons from comparative genomics at the DNA level. Plant Molecular Biology. 2002;48:21–37. [PubMed] [Google Scholar]
- Shulaev V, Korban SS, Sosinski B, et al. Multiple models for Rosaceae genomics. Plant Physiology. 2008;147:985–1003. doi: 10.1104/pp.107.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, et al. The genome of woodland strawberry (Fragaria vesca) Nature Genetics. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz JL, Kazi S, Bashir R, Afzal JA, Lightfoot DA. The development of BAC-end sequence-based microsatellite markers and placement in the physical and genetic maps of soybean. Theoretical and Applied Genetics. 2007;114:1081–1090. doi: 10.1007/s00122-007-0501-9. [DOI] [PubMed] [Google Scholar]
- Silfverberg-Dilworth E, Matasci CL, Van de Weg WE, et al. Microsatellite markers spanning the apple (Malus× domestica Borkh.) genome. Tree Genetics and Genomes. 2006;2:202–224. [Google Scholar]
- Soria-Guerra RE, Rosales-Mendoza S, Gasic K, Wisniewski ME, Band M, Korban SS. Gene expression is highly regulated in early developing fruit of apple. Plant Molecular Biology Reports. 2011 DOI: 10.1007/s11105-011-0300-y. [Google Scholar]
- Van Ooijen JW. JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations. Wageningen, The Netherlands: Kyazma BV; 2006. [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus× domestica Borkh.) Nature Genetics. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- Williams EB, Janick J, Emerson FH, Dayton DF, Mowry JB, Hough LF, Bailey C. Co-op 12–18: seven scab-resistant apple selections released for advanced testing. 1975 Purdue AES Station Bulletin No. 69. [Google Scholar]
- Xu M, Korban SS. Somatic variation plays a key role in the evolution of the Vf gene family residing in the Vf locus that confers resistance to apple scab disease. Molecular Phylogenetics and Evolution. 2004;32:57–65. doi: 10.1016/j.ympev.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Xu M, Song J, Cheng Z, Jiang J, Korban SS. A bacterial artificial chromosome (BAC) library of Malus floribunda 821 and contig construction for positional cloning of the apple scab resistance gene Vf. Genome. 2001;44:1104–1113. [PubMed] [Google Scholar]
- Yu Q, Tong E, Skelton RL, et al. A physical map of the papaya genome with integrated genetic map and genome sequence. BMC Genomics. 2009;10:371. doi: 10.1186/1471-2164-10-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhebentyayeva TN, Swire-Clark G, Georgi LL, et al. A framework physical map for peach, a model Rosaceae species. Tree Genetics and Genomes. 2008;4:745–756. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.