Abstract
Dahlias (Dahlia variabilis) exhibit a wide range of flower colours because of accumulation of anthocyanin and other flavonoids in their ray florets. Two lateral mutants were used that spontaneously occurred in ‘Michael J’ (MJW) which has yellow ray florets with orange variegation. MJOr, a bud mutant producing completely orange ray florets, accumulates anthocyanins, flavones, and butein, and MJY, another mutant producing completely yellow ray florets, accumulates flavones and butein. Reverse transcription–PCR analysis showed that expression of chalcone synthase 1 (DvCHS1), flavanone 3-hydroxylase (DvF3H), dihydroflavonol 4-reductase (DvDFR), anthocyanidin synthase (DvANS), and DvIVS encoding a basic helix–loop–helix transcription factor were suppressed, whereas that of chalcone isomerase (DvCHI) and DvCHS2, another CHS with 69% nucleotide identity with DvCHS1, was not suppressed in the yellow ray florets of MJY. A 5.4 kb CACTA superfamily transposable element, transposable element of Dahlia variabilis 1 (Tdv1), was found in the fourth intron of the DvIVS gene of MJW and MJY, and footprints of Tdv1 were detected in the variegated flowers of MJW. It is shown that only one type of DvIVS gene was expressed in MJOr, whereas these plants are likely to have three types of the DvIVS gene. On the basis of these results, the mechanism regulating the formation of orange and yellow ray florets in dahlia is discussed.
Keywords: Anthocyanin, bHLH, CACTA superfamily, dahlia, transcription factor
Introduction
Dahlias (Dahlia variabilis) are popular ornamental plants cultivated in many countries. They belong to the Asteraceae and are autoallooctoploids with chromosome number 2n=64 (Lawrence, 1929, 1931b; Lawrence and Scott-Honcrieff, 1935; Gatt et al., 1998). Because of high polyploidy, dahlias exhibit various inflorescence shapes, colours, and sizes, and ∼50 000 cultivars have been identified and named in the past century (McClaren, 2009). In particular, dahlias exhibit a wide range of ray floret colours, such as ivory, yellow, pink, red, purple, and black. The pigments accumulated in ray florets are flavonoids, mainly anthocyanins, butein, and flavones and their derivatives that produce red, yellow, and ivory colours, respectively (Bate-Smith and Swain, 1953; Nordström and Swain, 1953, 1956, 1958; Bate-Smith et al., 1955; Harborne et al., 1990; Yamaguchi et al., 1999). In previous studies, four elements were believed to explain the inheritance of ray floret colours in dahlia: A (pale anthocyanin), B (deep anthocyanin), I (flavone), and Y (yellow) (Lawrence, 1931a; Lawrence and Scott-Honcrieff, 1935; Bate-Smith et al., 1955; Broertjes and Ballego, 1967; Singh et al., 1970). Despite its importance, few studies use molecular analysis techniques such as gene isolation and enzyme analysis (Fischer et al., 1988; Wimmer et al., 1998; Yamaguchi et al., 1999; Ogata et al., 2001; Suzuki et al., 2002). Flavonoid synthesis pathway genes have not yet been isolated, except for flavonoid 3′-hydroxylase (Schlangen et al., 2009, 2010a).
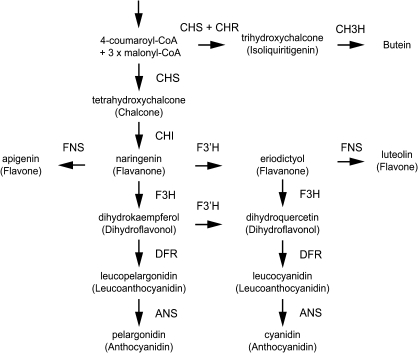
The flavonoid synthesis pathway is the most well-studied secondary metabolite synthesis pathway in plants. Since flavonoids determine flower colours, regulation of the flavonoid synthesis pathway is very important in ornamental plants. In particular, anthocyanins are found in the flowers of many species and have many functions, such as recruiting pollinators and seed dispersers and scavenging active oxygen species (Yamasaki et al., 1996; Winkel-Shirley, 2001). Anthocyanidin, the aglycone of anthocyanin, is formed in the anthocyanin synthesis pathway by the condensation of three molecules of malonyl-CoA with one molecule of 4-coumaroyl-CoA (Fig. 1); the enzymes involved in this pathway are as follows: chalcone synthase (CHS), chalcone isomerase (CHI), flavonoid 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), and anthocyanidin synthase (ANS). The synthesized anthocyanidins are glycosylated by glucosyltransferase and modified through processes such as acylation and malonylation (Tanaka et al., 2008). The genes encoding the above-mentioned enzymes involved in the anthocyanin synthesis pathway are regulated by transcription factors such as basic helix–loop–helix (bHLH), R2R3-MYB, and WD40 repeats (WDRs); these three proteins function as transcription factors by forming complexes or acting alone (Koes et al., 2005). In D. variabilis, first, three molecules of malonyl-CoA and one molecule of 4-coumaroyl-CoA, the same substrate as that in chalcone synthesis, are converted to trihydroxychalcone by CHS and chalcone reductase (Bomati et al., 2005) and then hydroxylated by chalcone 3-hydroxylase or a particular copy of flavonoid 3′-hydroxylase to form butein. Recently, the gene for chalcone 3-hydroxylase, which converts trihydroxychalcone to butein, was isolated from Cosmos sulphureus (Schlangen et al., 2010b), but whether the anthocyanin and butein synthesis pathways are regulated by the same regulator gene is unknown.
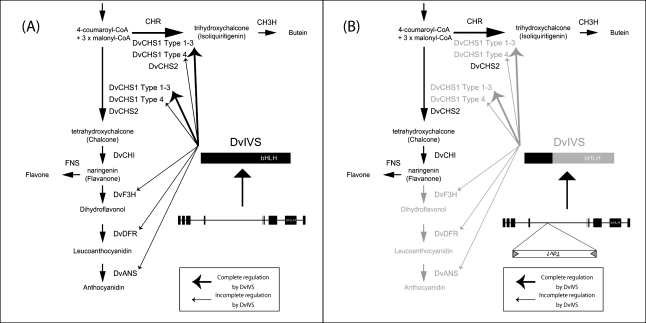
Fig. 1.
Simplified flavonoid synthesis pathways. Only compounds detected in ‘Michael J’ are shown. ANS, anthocyanidin synthase; CH3H, chalcone 3-hydroxylase; CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; DFR, dihydroflavonol 4-reductase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; FNS, flavone synthase.
To isolate the genes that impart colour to flowers, mutants formed by transposable elements have been widely used. This method of gene isolation, called transposon tagging, was used to isolate Antirrhinum majus pallida (DFR) (Martin et al., 1985), Zea mays bronze (UDP-glucose flavonoid 3-O-glucosyltransferase) (Fedoroff et al., 1984), Petunia hybrida ph6 (An1) (Chuck et al., 1993), and many more. Therefore, to clarify whether anthocyanin and butein synthesis in dahlia are regulated by the same or different mechanisms, two lateral mutants MJOr and MJY that spontaneously occurred in ‘Michael J’ (MJW), which has yellow ray florets with orange variegation, were used. MJOr, a bud mutant producing orange ray florets, accumulates anthocyanins, flavones, and butein, and MJY, another mutant producing yellow ray florets, accumulates flavones and butein. In this study, anthocyanin synthesis pathway genes and transcription factors were isolated and it was shown that DvIVS, belonging to the An1 subgroup bHLH transcription factor family, regulates anthocyanin synthesis in ray florets of dahlia. Finally, a putative model to explain the mechanism regulating expression of orange and yellow flower colours in dahlia is proposed.
Materials and methods
Plant material and developmental stages
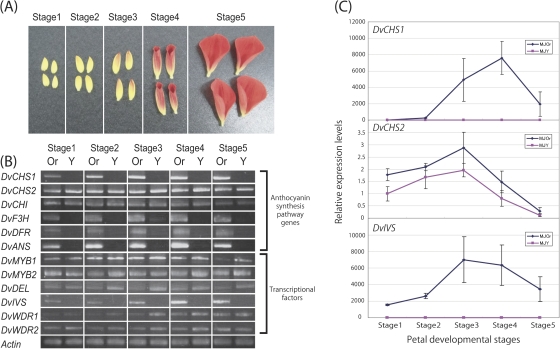
Dahlia variabilis cv. ‘Michael J’ (MJW) was obtained from Yukihiro Fukuda (Haibara, Nara, Japan). MJW has bright yellow ray florets with a small number of brilliant orange markings (Fig. 2A). It rarely produces completely orange (Fig. 2B) or completely yellow (Fig. 2C) ray florets as a bud mutation. The completely orange mutant line (MJOr) and the completely yellow ray floret mutant line (MJY) were isolated and obtained from Yukihiro Fukuda. The two lines and wild-type plants were grown under standard greenhouse conditions or in the experimental field of Kyoto University (Kyoto, Japan). To analyse temporal gene expression, the MJOr ray florets were classified into five stages on the basis of the degree of colouration (Fig. 3A): stage 1, an uncoloured ray floret; stage 2, the tip of a ray floret is coloured; stage 3, a ray floret coloured in the centre; stage 4, colouring is completed and the ray floret starts to unfold; and stage 5, a completely unfolded ray floret. The size and stage of MJY ray florets used were almost same.
Fig. 2.
Inflorescence phenotypes of ‘Michael J’ (MJW) and its bud mutants: (A) MJW, (B) MJOr, and (C) MJY. (A) MJW ray florets are bright yellow with a small number of brilliant orange markings. (B) MJOr ray florets are brilliant orange; this colour is mainly derived from anthocyanins and butein. (C) MJY ray florets are bright yellow; this colour is mainly derived from butein.
Fig. 3.
Temporal expression of anthocyanin synthesis pathway genes and transcription factors. (A) Developmental stages of ray florets. Developmental stages of MJOr ray florets were defined on the basis of the degree of colouration. (B) Semi-quantitative RT-PCR analysis of temporal expression of anthocyanin synthesis genes and transcription factors in MJOr and MJY. The constitutively expressed gene for actin in Dahlia variabilis was used as an internal control. (C) Relative expression levels of DvCHS1, DvCHS2, and DvIVS in MJOr compared with those in MJY at each ray floret developmental stage. The vertical bars are the standard error (±SE) of the means of three biological replicates. The constitutively expressed gene for actin in D. variabilis was used as an internal control.
HPLC analysis
Ray florets were soaked overnight in 5 ml of MAW solution (methanol:acetic acid:water, 4:1:5 v/v/v) to extract the pigments. For pigment hydrolysis, extracted solutions were evaporated, redissolved in 2 ml of 20% hydrochloric acid, boiled, and used as crude aglycones. HPLC analysis was performed using an LC10A system (Shimadzu, Kyoto, Japan) with a C18 column (Nihon Waters K.K., Tokyo, Japan) maintained at 40 °C and a photodiode array detector. The detection wavelength was 350 nm for flavones, 380 nm for chalcones, and 530 nm for anthocyanins. Elutant A was 1.5% phosphate dissolved in water and elutant B was 1.5% phosphate, 20% acetic acid, and 25% MeCN dissolved in water. Analysis was performed at a flow rate of 1 ml min−1 and column temperature of 40 °C, using a mobile phase gradient starting at 20% B to 85% B over 40 min with 5 min re-equilibration at 20% B. As standards for the determination of flavonoids, commercially available naringenin, apigenin, and luteolin (Wako Pure Chemical Industries Ltd, Osaka, Japan) as well as HPLC-separated and HPLC-purified hydrolysed cyanidin and pelargonidin from rose petals were used. To obtain hydrolysed standards of butein and isoliquiritigenin, extracts from orange ray florets of a seedling line, HywR7R, were separated by paper chromatography, and each band was eluted with methanol. Each eluate was dried and redissolved in a small amount of methanol. The colour, Rf value, and maximum wavelength of eluted compounds were measured, and the compounds were determined by comparing the data with those of authentic butein (kindly supplied by Dr Norio Saito) and previously reported data (Nordström and Swain, 1956; Saito et al., 1970).
Isolation of nucleic acids and sequencing
To clone the DvIVS genomic region and Tdv1 region, genomic DNA of ray florets was isolated using a modified cetyltrimethylammonium bromide method (Murray and Thompson, 1980) and purified with MagExtractor™-Plant Genome (Toyobo Co. Ltd, Osaka, Japan). Genomic DNA was also isolated using Qiagen Genomic-tip 100/G (Qiagen, Valencia, CA, USA) for genomic PCR analyses. Total RNA was isolated using the isothiocyanate/caesium chloride centrifugation method and an RNeasy Plant Mini Kit (Qiagen), a modified QuickGene RNA Cultured Cell Kit S (Fujifilm, Tokyo, Japan), or a Get pure RNA Kit (Dojindo, Kumamoto, Japan). All sequence analyses were performed using a BigDye® Terminator v 3.1 Cycle Sequencing Kit and a 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Gene isolation
DvCHS1, DvCHI, DvF3H, DvDFR, DvANS, DvIVS, DvDEL, and DvActin cDNAs were isolated using degenerate primers designed from conserved regions of each gene shown in Supplementary Table S1 available at JXB online. To isolate DvMYB1, DvMYB2, DvWDR1, and DvWDR2 cDNAs, dahlia cDNA libraries provided by Dr Yoshikazu Tanaka (Suntory, Shimamoto, Mishima, Osaka, Japan) were screened (Suzuki et al., 2002). Ipomoea nil MYB (AB232770) and WDR (AB232779) cDNAs were used as probes, and AlkPhos Direct (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA) was used for probe hybridization. To determine the complete sequence of mRNA of all genes, 3' rapid amplification of cDNA ends (RACE) and 5' RACE was performed using a GeneRacer™ Kit (Invitrogen, Carlsbad, CA, USA). Primers used are shown in Supplementary Table S2. Each end of the genes was sequenced for at least 16 clones for each gene to avoid single nucleotide polymorphisms (SNPs) because of dahlia’s high polyploidy. While performing 5' RACE of DvMYB1, a novel CHS-like fragment was obtained that differed from DvCHS1. This new CHS was named DvCHS2 and each cDNA end was determined by 5′ and 3′ RACE. For 5′ and 3′ RACE of the transcription factor genes, total RNA extracted from D. variabilis ‘Matsuribayashi’ was used.
RT-PCR and real-time RT-PCR
The total RNA of MJOr and MJY was subjected for reverse transcription using an oligo(dT)20 primer and ReverTra Ace (Toyobo). The obtained cDNA products served as templates for PCR performed using Blend Taq polymerase (Toyobo). DvActin was used as an internal standard. The primers used are shown in Supplementary Table S3 at JXB online. The PCR program was set at 94 °C for 2 min, followed by 30–35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2–3 min.
Total RNA extracted from stage 1–5 ray florets was used. Real-time RT-PCR was performed using SYBR® Premix Ex Taq™ II (Takara Bio Inc., Ohtsu, Japan) according to the manufacturer’s instructions. Real-time PCR was performed using a 7900HT Fast Real-Time PCR System (Applied Biosystems). The primers used are shown in Supplementary Table S4 at JXB online. The PCR program was set at 50 °C for 1 s, 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 15 s, 72 °C for 20 s, and the subsequent dissociation steps. Three different flowers were used as replicates for each stage, and DvActin was used as an internal standard. Because of the low transcription level, samples that did not reach the threshold were calculated as 40 cycles.
To test partial expression of DvIVS, three primer sets were used for RT-PCR: IVS-Full-F and IVS-Full-R, 35S-IVS-F and IVS-611R, and IVS-725F and IVS-Full-R (Supplementary Table S5 at JXB online). The first primer set amplifies from the start to the stop codon. The second primer set amplifies from 30 bp after the start codon to the third exon, and the third primer set amplifies from the fifth exon to the stop codon.
Cloning of the genomic DvIVS gene and the transposable element
Genomic PCR was performed with LA Taq (Takara) using 100 ng of genomic DNA as a template in a 10 μl volume. The PCR program was set at 94 °C for 1 min, followed by 35 cycles of 98 °C for 10 s, 55 °C for 10 s, and 68 °C for 15 min. Amplified PCR products were electrophoresed with 1× TAE and a 0.8% agarose gel, and cloned into pCR®-XL-TOPO using a TOPO® XL PCR Cloning Kit (Invitrogen) according to the manufacturer’s instructions. The DvIVS gene of MJOr was amplified using the primer set IVS-Full-F and IVS-Full-R (Supplementary Table S5 at JXB online). The insertion sequence of MJY was amplified with the primers IVS-G2241F and IVS-G2869R (Supplementary Table S5), which were designed on the basis of the upstream and downstream regions of the insertion sequence. Four plasmids with the DvIVS gene and six plasmids with inserted fragments were sequenced by primer walking using the primers shown in Supplementary Table S5.
Sequence analysis of cDNAs and the genomic DvCHS1 gene
To examine regulation of redundant copies of each anthocyanin synthesis gene, sequence analyses for multiple copies of these genes were performed. Extracted total RNAs of MJOr and MJY were subjected to reverse transcription with an oligo(dT)20 primer using ReverTra Ace (Toyobo). RT-PCRs of DvCHS1, DvCHI, DvF3H, and DvDFR were performed using KOD Plus polymerase (Toyobo), and PCR products were cloned into pCR®-Blunt II-TOPO® using a Zero Blunt® TOPO® PCR Cloning Kit (Invitrogen). The PCR program was set at 94 °C for 2 min, followed by 30 cycles of 94 °C for 50 s, 55 °C for 30 s, and 68 °C for 2 min. A total of 40 cloned plasmids of both lines with DvCHS1, DvCHI, DvF3H, and DvDFR were extracted, except for MJY DvCHS1 (only 20 plasmids were extracted). To determine the accurate sequence, plasmids with DvCHS1, DvF3H, and DvDFR of both lines were sequenced twice from both the 3′ and 5′ ends. The primers used are shown in Supplementary Table S6 at JXB online. To exclude misreading of sequences or misamplification in PCR, SNPs that were only observed in one clone were omitted.
To analyse the sequence of the genomic region of DvCHS1 in MJOr, MJY, and MJW, genomic PCR was performed using the primer set shown in Supplementary Table S3 at JXB online. A total of 30 clones of the DvCHS1 gene were sequenced for each line.
Denaturing gradient gel electrophoresis (DGGE) analysis
RT-PCR was performed with KOD Dash polymerase (Toyobo), using the cDNA of MJOr and MJY. The PCR program was set at 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 10 s, and 74 °C for 30 s. The primers used were DGGE-F (5′-CCCGGGCGGGCCCGCGATCTTGGA-3′) and DGGE-R (5′-GAATGTTTCAAGCACACATTGCTT-3′). After confirming a single band by agarose gel electrophoresis, perpendicular DGGE was performed using a DGGE Mini Electrophoresis System (NIHON EIDO Co. Ltd, Tokyo, Japan). To make the gradient gel, 35 ml of 0% denatured 6.5% acrylamide solution with 190 μl of 10% APS and 16 μl of TEMED was added to 35 ml of 80% denatured 6.5% acrylamide solution with 190 μl of 10% APS and 16 μl of TEMED in the gradient chamber. The mixed gel solution was gradually poured into the gradient gel electrophoresis tank (1.8–2 ml min−1) using a solution dispensing pump. An aliquot of 50 μl of the PCR product was applied to the gel and electrophoresed with 1× TBE buffer for 4 h at 80 V and 60 °C. The gel was stained with SYBR Gold (Invitrogen), and the band was confirmed by UV exposure.
GenBank accession numbers
Sequence data of these genes have been deposited in the DDBJ/GenBank data libraries under accession numbers AB576129, AB576130, AB576131, AB576132 (DvCHS1), AB576133 (DvCHS2), AB591827 (DvCHI), AB591828 (DvF3H), AB591829 (DvDFR), AB591830 (DvANS), AB601003 (DvMYB1), AB601004 (DvMYB2), AB601005 (DvIVS), AB601006 (DvDEL), AB601007 (DvWDR1), AB601008 (DvWDR2), AB601010 (DvIVS genomic region), and AB601009 (Tdv1).
Results
Pigment analysis
Wild-type MJW produces yellow ray florets with orange variegation, while MJOr and MJY produce completely orange and completely yellow ray florets, respectively, without any variegation. To clarify the difference in pigments between MJOr and MJY, aglycones were extracted from ray florets and analysed by HPLC. In both lines, peaks of chalcones (butein and isoliquiritigenin) and flavones (apigenin and luteolin) were detected. In addition to these, peaks of anthocyanidins (pelargonidin and cyanidin) were detected in MJOr but not in MJY (Table 1). These data showed that chalcones and flavones are synthesized in both lines, while anthocyanidins are synthesized only in MJOr.
Table 1.
Aglycones in the petals of MJOr and MJY
| Line | Anthocyanidin |
Flavone |
Chalcone |
|||
| Pelargonidin | Cyanidin | Apigenin | Luteolin | Butein | Isoliquiritigenin | |
| Orange | + | + | + | + | + | + |
| Yellow | – | – | + | + | + | + |
+, Abundant; –, not detected.
Temporal expression of anthocyanin synthesis pathway genes
To analyse genes associated with anthocyanin synthesis, partial sequences of DvCHS1, DvCHI, DvF3H, DvDFR, DvANS, DvIVS, and DvDEL were isolated using degenerate primers. DvMYB1, DvMYB2, DvWDR1, and DvWDR2 were isolated by cDNA library screening using cDNA fragments of InMYB1 and InWDR1 as probes (Morita et al., 2006). Full cDNA sequences of all genes including DvCHS2, another CHS gene, were isolated by 5′ and 3′ RACE. Partial sequences of DvActin were also isolated with degenerate primers by 3′ RACE.
To examine the temporal expression pattern of the anthocyanin synthesis pathway genes during ray floret development, the MJOr ray florets were divided into five developmental stages based on the degree of colouration and RT-PCR was performed. Although expression levels of DvCHS2 and DvCHI showed no difference, expression levels of DvCHS1, DvF3H, DvDFR, and DvANS were lower in MJY than in MJOr at all developmental stages (Fig. 3B). Down-regulation of multiple genes encoding the anthocyanin synthesis pathway enzymes suggested that transcription factor(s) do not function in MJY.
Temporal expression analysis by RT-PCR was also performed for transcription factors. Although expression levels of DvMYB1, DvMYB2, DvDEL, DvWDR1, and DvWDR2 did not differ significantly, the expression level of DvIVS was lower in MJY than in MJOr, which is similar to that of DvCHS1, DvF3H, DvDFR, and DvANS (Fig. 3B). This result suggested that DvIVS is a transcription factor that activates anthocyanin biosynthesis genes including DvCHS1, DvF3H, DvDFR, and DvANS.
To confirm these results, real-time RT-PCR of DvCHS1 and DvCHS2, which seem to be regulated and not regulated by DvIVS, respectively, was performed using total RNA extracted from stage 1–5 ray florets. In MJOr, the relative expression level of DvIVS and DvCHS1 compared with that of DvActin increased until stage 3 or 4 and then decreased, but consistently very low expression levels were maintained in MJY (Fig. 3C). In contrast, expression levels of DvCHS2 were slightly lower in MJY, but the expression pattern of DvCHS2 in MJOr and MJY was the same (Fig. 3C). This result supported the results of RT-PCR analyses and suggested that DvCHS1, but not DvCHS2, is regulated by DvIVS.
DvCHS1 (Supplementary Table S7 at JXB online), DvCHI (Supplemenary Table S8), DvF3H (Supplementary Table S9), and DvDFR (Supplementary Table S10) cDNA sequences isolated from MJOr and MJY ray florets were determined and the number of different sequences expressed in both lines was determined on the basis of SNPs. Based on sequence analysis of transcripts, 17 and 15 sequences of DvCHI (Supplementary Table S8), nine and 12 sequences of DvF3H (Supplementary Table S9), and nine and eight sequences of DvDFR (Supplementary Table S10) were observed in MJOr and MJY, respectively. Because drastic differences were not observed in the number of sequences expressed in both lines, whole copies were assumed to be suppressed in MJY. However, 12 different sequences of DvCHS1 were detected in MJOr, while only one sequence was detected in MJY (Supplementary Table S7). At least three major transcripts (DvCHS1 type 1, 2, and 4) were detected in MJOr, while only one transcript (DvCHS1 type 4) was detected in MJY (Supplementary Table S7). The genomic fragments of DvCHS1 were isolated and it was observed that not only MJOr and MJW, but also MJY has DvCHS1 genes that have coding sequences identical to these three major transcripts (Supplementary Tables S11, S12).
Characterization of DvIVS
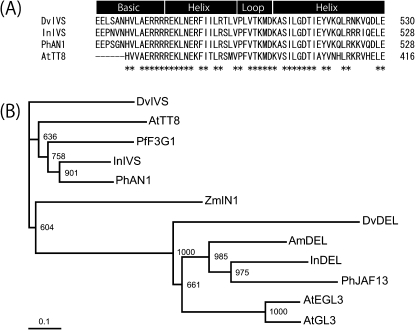
The full-length DvIVS cDNA was 2483 bp containing a 649 amino acid open reading frame (ORF). The DvIVS ORF retained the bHLH domain at 472–530 amino acids (Fig. 4A). BLASTP search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed that the deduced amino acid sequence was 48% identical to InIVS (I. nil), 50% identical to An1 (Petunia hybrida), and 48% identical to AtTT8 (Arabidopsis thaliana). To compare genetic distance among the bHLH proteins associated with anthocyanin synthesis in higher plants, a phylogenetic tree was constructed. The phylogenetic tree showed that DvIVS belonged to the An1 subgroup (Fig. 4B).
Fig. 4.
Phylogenetic analysis of DvIVS. (A) Amino acid comparison of the bHLH domain of DvIVS, InIVS, PhAN1, and AtTT8. Numbers (*) indicate amino acids that are fully conserved in each of the proteins. (B) Phylogenetic tree for bHLH transcription factors associated with anthocyanin synthesis pathways. The entire amino acid sequences were aligned using ClustalW, and the tree was constructed by the Neighbor–Joining method. Bootstrap values of 1000 retrials are indicated on each branch, and the scale shows 0.1 amino acid substitutions per site. The abbreviations shown in front of each protein indicate the plant species: Dv, Dahlia variabilis; Am, Antirrhinum majus; At, Arabidopsis thaliana; In, Ipomoea nil; Pf, Perilla frutescens; Ph, Petunia hybrida; Zm, Zea mays. The accession number of each protein is: DvIVS (BAJ33515), DvDEL (BAJ33516), AmDEL (AAA32663), AtEGL3 (NP_176552), AtGL3 (NP_680372), AtTT8 (CAC14865), InDEL (BAE94393), InIVS (BAE94394), PfF3G1 (BAC56998), PhAN1 (AAG25927), PhJAF13 (AAC39455), and ZmIN1 (AAB03841).
Analysis of the DvIVS genomic region
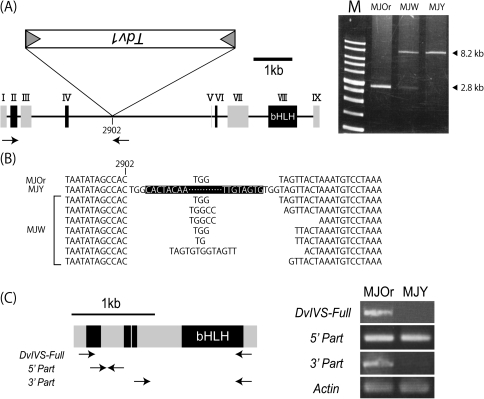
The DvIVS genomic region coding for full-length mRNA (AB601010) of MJOr was 8477 bp and contained nine exons and eight introns (Fig. 5A). Genomic PCR was performed to examine whether a transposable element was inserted in the DvIVS gene of MJY. An ∼8.2 kb amplified product was obtained from MJOr, and an ∼13.6 kb amplified product was obtained from MJY (data not shown). The 5385 bp insertion sequence (AB601009) was found in the fourth intron, 903 bp downstream of the fourth exon of the DvIVS gene of MJY. To characterize the structure of the DvIVS gene of MJW, genomic PCR was performed using the primer set IVS Full-F and IVS-G2869R. Not only the genomic fragment (8.2 kb) that should include the insertion sequence, but also a shorter fragment (2.8 kb) without the insertion sequence were detected in MJW (Fig. 5A).
Fig. 5.
Structure of the DvIVS gene. (A) Structure of the DvIVS gene and genomic PCR. Rectangles with roman figures indicate exons. The bHLH domain is located in the eighth exon. Tdv1 is inserted in the fourth intron in the antisense direction of the MJY genome. Arrows indicate the primer set used for genomic PCR. M indicates a 1 kb marker. (B) Footprints of Tdv1. The uppermost sequence is assumed to be the standard sequence. Sequences in the black box indicate the Tdv1 sequence. With respect to MJW, Tdv1-inserted sequences were not analysed. (C) Truncated transcripts of DvIVS in MJOr and MJY. Grey rectangles indicate odd numbered exons and black rectangles indicate even numbered exons. Arrows indicate primer sets used for RT-PCR.
The inserted element of MJY carried a terminal inverted repeat (TIR) starting with 5′-CACTA-3′ (Table 2), which is characteristic of a CACTA superfamily transposable element. The insertion generated a 3 bp (TGG) target site duplication (TSD), similar to the TSD generated by other CACTA-type transposable elements (Inagaki et al., 1994; Zabala and Vodkin, 2008; Xu et al., 2010). It also contained an ORF for antisense orientation (Fig. 5A) (nucleotides 1735–3555), encoded a 606 amino acid polypeptide, and shared high identity with other CACTA superfamily transposable elements [41% identical to TNP2 in A. majus Tam1 (Nacken et al., 1991), 33% identical to TNP2 in Glycine max Tgm9 (Xu et al., 2010), and 32% identical to En-1 in Zea mays (Pereira et al., 1986)]. To test whether the insertion sequence has the ability to excise from the DvIVS gene, 600 bp fragments containing the insertion site in MJW were amplified and cloned into plasmid vectors. A total of 23 clones were obtained with the DvIVS gene fragments with small rearrangements that were presumed to be footprint sequences of the insertion sequence (Table 3, Fig. 5B). From these results, the insertion sequence was regarded as an active transposable element and named transposable element of D. variabilis 1 (Tdv1). The DvIVS fragments could be obtained from MJOr but not from MJY. In addition to the DvIVS gene fragment, two different gene fragments were obtained. Both of them were shorter than the DvIVS fragments, and showed 97% identity to the DvIVS gene. This suggested that the dahlia lines used here have at least three types of DvIVS genes.
Table 2.
Comparison of TIRs with the reported transposable elements of the CACTA superfamily
| Name | TIR sequences | Length (bp) | Species | Reference |
| Tdv1 | CACTACAA | 8 | Dahlia variabilis | This study |
| En1 | CACTACAAGAAAA | 13 | Zea mays | Pereira et al. (1986) |
| Tam1 | CACTACAACAAAA | 13 | Antirrhinum majus | Nacken et al. (1991) |
| Cac1 | CACTACAA | 8 | Arabidopsis thaliana | Miura et al. (2001) |
| Tpn1 | CACTACAAGAAAAATGCACATAGCAAC | 27 | Pharbitis nil (Ipomoea nil) | Inagaki et al. (1994) |
| Tgm1 | CACTATTAGAAAA | 13 | Glycine max | Vodkin et al. (1983) |
| Cs1 | CACTATGTGAAAAAAGCTTA | 20 | Sorghum bicolor | Chopra et al. (1999) |
| Pis1 | CACTACGCCAAA | 12 | Pisum sativum | Shirsat (1988) |
| Psl | CACTACAAAAAA | 12 | Petunia hybrida | Snowden and Napoli (1998) |
| Tdc1 | CACTACAAGAAAACGCGAGA | 20 | Daucus carota | Ozeki et al. (1997) |
| Tnr3 | CACTAGAAGGGAT | 13 | Oryza sativa | Motohashi et al. (1996) |
This table is based on Tian (2006).
Table 3.
Amplified fragment with IVS-G2241F and IVS-G2869R
| Type | Length (bp) | Identity to the DvIVS genome (%) | MJOr | MJY | MJW |
| 1 | 628 | 100 | 10 | 0 | 23a |
| 2 | 625 | 97 | 6 | 16 | 24 |
| 3 | 615 | 97 | 1 | 3 | 16 |
Footprint sequences were not considered in type 1 of MJW.
To test whether all three DvIVS genes are expressed in the flowers, DvIVS cDNA fragments from MJOr were amplified, cloned into a plasmid vector, and the cDNA sequence was analysed. Although 20 clones of DvIVS cDNA were sequenced, all of them were derived from the DvIVS gene (data not shown). The result suggested that the anthocyanin pigmentation in the flowers depend on the DvIVS gene and that the two other DvIVS genes are not expressed in the flowers.
To characterize the DvIVS transcripts in MJY, RT-PCR was performed using three primer sets. Successful amplification was achieved only by the primer set amplifying the region upstream of Tdv1 insertion sites in both MJOr and MJY; however, other primer sets did not successfully amplify regions in MJY (Fig. 5C). This suggested that Tdv1 insertion into the fourth intron of DvIVS resulted in truncated DvIVS mRNA in MJY. On analysing DvIVS mRNA expression in MJY by 3′ RACE, truncated DvIVS mRNA containing the fourth exon and a short part of the fourth intron (nucleotides 125–250 downstream of the fourth exon/fourth intron junction) was detected (data not shown). Thus, Tdv1 insertion into the DvIVS gene of MJY resulted in generation of short-sized DvIVS transcripts lacking the bHLH domain, thereby leading to an inability for transcriptional regulation of anthocyanin synthesis genes.
Discussion
DvIVS is a bHLH transcription factor associated with anthocyanin synthesis in dahlia
The following transcription regulators of anthocyanin synthesis pathway enzymes are well known: bHLH, R2R3-MYB, and WDR (Koes et al., 2005). To date, many studies have been conducted to determine flavonoid synthesis pathway transcription factors. An1 and JAF13, An2, and An11 have been isolated as bHLH, R2R3-MYB, and WDR transcription factors, respectively, regulating anthocyanin synthesis in petunia (Quattrocchio et al., 1993, 1998; De Vetten et al., 1997; Spelt et al., 2000, 2002). Similarly, bHLH, R2R3-MYB, and WDR transcription factors control anthocyanin synthesis in the three Ipomoea species (Park et al., 2004, 2007; Morita et al., 2006).
bHLH proteins are found in both animals and plants, and constitute a superfamily of transcription factors. Arabidopsis has at least 147 bHLH genes, and these are divided into 21 subfamilies (Toledo-Ortiz et al., 2003). The bHLH genes associated with regulation of anthocyanin synthesis were first found in Z. mays (Ludwig et al., 1989), and have subsequently been found in many other species. Recent studies showed that AtTT8 (Nesi et al., 2000), AtGL3 (Payne et al., 2000), and AtEGL3 (Zhang et al., 2003) regulate anthocyanin and proanthocyanidin synthesis in A. thaliana. These bHLH genes regulating anthocyanin synthesis are usually divided into two subgroups on the basis of their phylogenetic tree (Spelt et al., 2000): one subgroup includes AtTT8, An1, and InIVS, and the other includes AtGL3, AtEGL3, JAF13, and InDEL. Usually, the bHLH domain in the former subgroup is not separated by an intron, while that in the latter group is separated by an intron. For example, the bHLH domains of An1 (Spelt et al., 2000), IpIVS (Park et al., 2007), AtTT8 (Nesi et al., 2000), and GtbHLH1 in Gentiana triflora (Nakatsuka et al., 2008) are encoded within a single exon, while those of JAF13 and ZmIn1 are encoded within two exons. DvIVS shows high identity with An1 (50%), InIVS (48%), and AtTT8 (48%) in deduced amino acid sequences and is classified into the same subgroup with these bHLHs in the phylogenetic tree (Fig. 4B). DvIVS has a bHLH domain encoded within a single exon (Fig. 5A), suggesting that the structure of the DvIVS gene is similar to that of the An1 gene. These data suggest that DvIVS is an orthologue of AtTT8, An1, and InIVS, and that DvIVS regulates transcription of DvCHS1, DvF3H, DvDFR, and DvANS, but not of DvCHS2 and DvCHI. Real-time RT-PCR showed that the expression patterns of DvIVS and DvCHS1 were highly coordinated, while that of DvCHS2 was not, supporting the suggestion (Fig. 3C). A slight decrease in the expression levels of DvCHS2 was observed in MJY; this may have been caused by the excess production of substrate resulting from an inability to synthesize anthocyanin. In many cases, although the particular set of genes may differ between species and tissues within a species, a subset of the anthocyanin pathway genes is affected by these transcription factors (Gonzalez et al., 2008). Transcription regulation in the three Ipomoea species (Park et al., 2004, 2007; Morita et al., 2006) is similar to that in dahlia.
The cDNA fragment derived from the 5′ end of DvIVS mRNA containing the first to third exons was detected in MJY; however, the cDNA fragment from the 3′ end of DvIVS mRNA was not observed. This truncated mRNA did not contain a bHLH domain in the eighth exon, suggesting that the bHLH domain of DvIVS is important for anthocyanin synthesis in dahlia.
Flower colour variegation is caused by Tdv1
Flower colour variegation is often caused by recurrent excision of transposable elements in a pigment biosynthesis gene. For example, Tpn1, a CACTA superfamily transposable element in DFR in I. nil (Inagaki et al., 1994); Tip100, a hAT superfamily transposable element in CHS-D in I. purpurea (Habu et al., 1998); and Tgm9, a CACTA superfamily transposable element in DFR2 in G. max (Xu et al., 2010) resulted in variegated flower phenotypes. Therefore, an association of transposable elements with floret colour was expected in MJW. It was hypothesized that MJOr was a complete revertant mutant line and MJY was a loss-of-function mutant line, where the transposable element could not transpose from a gene associated with anthocyanin synthesis.
A 5.4 kb insertion sequence, named Tdv1, was found in the fourth intron of the DvIVS gene in MJY. Tdv1 shares a number of CACTA superfamily characteristic features. CACTA superfamily transposable elements are DNA transposons directly transposed from DNA to DNA (Feschotte et al., 2002), and almost all CACTA transposable elements harbour ‘CACTA’ in the outermost region of TIR (Tian, 2006). Tdv1 also has a CACTA superfamily characteristic TIR (CACTACAA) (Table 2) and a 3 bp (TGG) TSD was observed in the MJY genome (Fig. 5B). The 606 amino acid ORF in Tdv1 shared high identity with other CACTA superfamily transposable elements [41% identical to TNP2 in A. majus Tam1 (Nacken et al., 1991), 33% identical to TNP2 in G. max Tgm9 (Xu et al., 2010), and 32% identical to En-1 in Z. mays (Pereira et al., 1986)]. Further analysis of this translated amino acid sequence by NCBI’s CDD (Marchler-Bauer et al., 2011) showed high identity with Transposase_21 (pfam02992). The footprint sequences formed by excision of the transposable element were also found in the MJW genome (Table 3, Fig. 5C), indicating that Tdv1 is an active CACTA superfamily transposable element. Because no PCR fragments with footprint sequences of Tdv1 in MJY could be amplified, transposition of Tdv1 is completely suppressed in MJY.
Anthocyanin and butein synthesis in dahlia
Flavones and butein accumulated in the ray florets of both MJOr and MJY. Thus, to synthesize these compounds, at least one CHS other than DvCHS1 must function in MJY. Indeed, another CHS, DvCHS2, coding for a 398 amino acid protein, was identified whereas DvCHS1 coded for a 389 amino acid protein. The nucleotides from the start to the stop codon and the amino acid sequence in DvCHS1 were 69% and 82% identical, respectively, to those in DvCHS2. A phylogenic tree was constructed using the deduced amino acid sequences associated with flavonoid synthesis, including DvCHS1 and DvCHS2 (Supplementary Fig. S1 at JXB online). DvCHS1 showed high identity with GhCHS4 (88%) and DvCHS2 showed high identity with GhCHS1 (89%) and GhCHS3 (84%). GhCHS1 and GhCHS3 are associated with anthocyanin and flavonol synthesis in the corolla of gerbera (Helariutta et al., 1995), and GhCHS4 is expressed in ray florets and carpels and is a typical CHS that synthesizes naringenin chalcone (Laitinen et al., 2008). CHS generally functions as a homodimer (Ferrer et al., 1999); therefore, amino acid residues associated with the formation of active sites are commonly conserved in plants. Cys164, His303, and Asn336, as CoA-binding sites, Val98, Thr132, Ser133, Met137, Gly163, Thr194, Val196, Thr197, Gly211, Gly216, Ile254, Gly256, Ser338, and Pro375 as active site residues, and Phe215 and Phe265 as gatekeepers are conserved in CHS2 of Medicago sativa (Ma et al., 2009). DvCHS1 and DvCHS2 possess all these conserved residues. These observations indicate that DvCHS1 and DvCHS2 encode functional CHS proteins. Therefore, although whether DvCHS1 and DvCHS2 have functional selectivity is still unclear, presumably DvCHS2 but not DvCHS1 contributes to synthesis of flavones or butein in MJY, and regulation of anthocyanin and flavone/butein synthesis is different in dahlia.
Redundant copies regulated by DvIVS
Dahlia is an autoallooctoploid (Lawrence, 1929, 1931b; Lawrence and Scott-Honcrieff, 1935; Gatt et al., 1998). DvCHS1 (Supplementary Table S7 at JXB online), DvCHI (Supplementary Table S8), DvF3H (Supplementary Table S9), and DvDFR (Supplementary Table S10) mRNA sequences expressed in MJOr and MJY ray florets wer sequenced and the number of different sequences expressed in both lines was determined on the basis of SNPs. Sequences of DvCHI, DvF3H, and DvDFR cDNA were almost the same in MJOr and MJY. However, a drastic difference was detected in the expression sequence of DvCHS1. At least three different major transcripts were detected in MJOr, but only one transcript was detected in MJY. This difference was confirmed by DGGE analysis (Supplementary Fig. S2 at JXB online). The genomic region of DvCHS1 of MJOr, MJW, and MJY was sequenced (Supplementary Tables S11, S12). Three common sequences were detected among these three lines. Thus, mRNA differences were derived not from genomic differences, but presumably from differences in DvIVS transcriptional regulation. Although the expression level of DvCHS1 was much lower in MJY (Fig. 3C), one type of DvCHS1 mRNA expressed in MJY was also expressed in MJOr, suggesting that DvCHS1 expression is not completely regulated by DvIVS. DvCHS1 can be divided into two groups: the regulation of the first is totally dependent on DvIVS, while the other can also be activated independently of DvIVS. The common sequence belongs to the latter group and the others belong to the former group. This imperfect regulation might be derived from genetic redundancy. Nevertheless, further study is needed in this regard. The manner in which plants with higher polyploidy regulate redundant genes is very interesting.
Finally, a putative model of anthocyanin and butein synthesis in MJOr and MJY was constructed (Fig. 6). In MJOr, DvIVS functions, followed by activation of DvCHS1, DvF3H, DvDFR, and DvANS and synthesis of anthocyanidin, flavone, and butein. In MJY, because of Tdv1 insertion, only partial DvIVS mRNA is expressed and transcriptional regulation cannot function. Although DvCHS1, DvF3H, DvDFR, and DvANS are not activated, DvCHS2 and DvCHI are not affected by DvIVS, resulting in accumulation of flavone and butein. Therefore, MJOr produces orange ray florets on account of the accumulation of anthocyanin and butein, while MJY produces yellow ray florets on account of the accumulation of butein only.
Fig. 6.
The putative model of anthocyanin and butein synthesis in MJOr and MJY. For simplicity, the 3′ hydroxylation pathway is abbreviated. (A) The model for MJOr. DvIVS can function, then DvCHS1, DvF3H, DvDFR, and DvANS are activated, and anthocyanidin, flavone, and butein are synthesized. (B) The model for MJY. Because of Tdv1 insertion, only truncated DvIVS mRNA is expressed and transcriptional regulation cannot function. Although DvCHS1, DvF3H, DvDFR, and DvANS are not activated, DvCHS2 and DvCHI are not affected by DvIVS, resulting in accumulation of flavone and butein.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic tree for CHS associated with flavonoid synthesis.
Figure S2. DGGE analysis of DvCHS1 cDNA from MJOr and MJY.
Table S1. Degenerate primers used for isolating the cDNA fragment.
Table S2. Primers used for RACE.
Table S3. Primers used for semi-quantitative RT-PCR.
Table S4. Primers used for real-time RT-PCR.
Table S5. Primers used for analysing the DvIVS genomic region and Tdv1 region.
Table S6. Primers used for confirming the sequence.
Table S7. SNPs of DvCHS1 cDNA in MJOr and MJY.
Table S8. SNPs of DvCHI cDNA in MJOr and MJY.
Table S9. SNPs of DvF3H cDNA in MJOr and MJY.
Table S10. SNPs of DvDFR cDNA in MJOr and MJY.
Table S11. SNPs in the exon of the DvCHS1 genomic region in MJOr, MJY, and MJW.
Table S12. SNPs in the intron of the DvCHS1 genomic region in MJOr, MJY, and MJW.
Acknowledgments
We thank Yukihiro Fukuda for supplying ‘Michael J’, and Dr Yoshikazu Tanaka (Suntory, Japan) for supplying the dahlia cDNA library.
References
- Bate-Smith EC, Swain T. The isolation of 2, 4, 4′-trihydroxychalcone from yellow varieties of Dahlia variabilis. Journal of the Chemical Society. 1953:2185–2187. [Google Scholar]
- Bate-Smith EC, Swain T, Nördstrom CG. Chemistry and inheritance of flower colour in the Dahlia. Nature. 1955;176:1016–1018. [Google Scholar]
- Bomati EK, Austin MB, Bowman ME, Dixon RA, Noel JP. Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. Journal of Biological Chemistry. 2005;280:30496–30503. doi: 10.1074/jbc.M502239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broertjes C, Ballego JM. Mutation breeding of Dahlia variabilis. Euphytica. 1967;16:171–176. [Google Scholar]
- Chopra S, Brendel V, Zhang J, Axtell JD, Peterson T. Molecular characterization of a mutable pigmentation phenotype and isolation of the first active transposable element from Sorghum bicolor. Proceedings of the National Academy of Sciences, USA. 1999;96:15330–15335. doi: 10.1073/pnas.96.26.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Robbins T, Nijjar C, Ralston E, Courtney-Gutterson N, Dooner HK. Tagging and cloning of a petunia flower color gene with the maize transposable element Activator. The Plant Cell. 1993;5:371–378. doi: 10.1105/tpc.5.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vetten N, Quattrocchio F, Mol J, Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes and Development. 1997;11:1422–1434. doi: 10.1101/gad.11.11.1422. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV, Furtek DB, Nelson OE. Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac) Proceedings of the National Academy of Sciences, USA. 1984;81:3825–3829. doi: 10.1073/pnas.81.12.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nature Structural Biology. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nature Reviews Genetics. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Fischer D, Stich K, Britsch L, Grisebach H. Purification and characterization of (+) dihydroflavonol (3-hydroxyflavanone) 4-reductase from flowers of Dahlia variabilis. Archives of Biochemistry and Biophysics. 1988;264:40–47. doi: 10.1016/0003-9861(88)90567-x. [DOI] [PubMed] [Google Scholar]
- Gatt M, Ding H, Hammett K, Murray B. Polyploidy and evolution in wild and cultivated Dahlia species. Annals of Botany. 1998;81:647–656. [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Habu Y, Hisatomi Y, Iida S. Molecular characterization of the mutable flaked allele for flower variegation in the common morning glory. The Plant Journal. 1998;16:371–376. doi: 10.1046/j.1365-313x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- Harborne JB, Greenham J, Eagles J. Malonylated chalcone glycosides in Dahlia. Phytochemistry. 1990;29:2899–2900. [Google Scholar]
- Helariutta Y, Elomaa P, Kotilainen M, Griesbach RJ, Schroder J, Teeri TH. Chalcone synthase-like genes active during corolla development are differentially expressed and encode enzymes with different catalytic properties in Gerbera hybrida (Asteraceae) Plant Molecular Biology. 1995;28:47–60. doi: 10.1007/BF00042037. [DOI] [PubMed] [Google Scholar]
- Inagaki Y, Hisatomi Y, Suzuki T, Kasahara K, Iida S. Isolation of a Suppressor-mutator/Enhancer-like transposable element, Tpn1, from Japanese morning glory bearing variegated flowers. The Plant Cell. 1994;6:375–383. doi: 10.1105/tpc.6.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Laitinen RAE, Ainasoja M, Broholm SK, Teeri TH, Elomaa P. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. Journal of Experimental Botany. 2008;59:3691–3703. doi: 10.1093/jxb/ern216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence WJC. The genetics and cytology of Dahlia species. Journal of Genetics. 1929;21:125–159. [Google Scholar]
- Lawrence WJC. The genetics and cytology of Dahlia variabilis. Journal of Genetics. 1931a;24:257–306. [Google Scholar]
- Lawrence WJC. The secondary association of chromosomes. Cytologia. 1931b;2:352–384. [Google Scholar]
- Lawrence WJC, Scott-Honcrieff R. The genetics and chemistry of flower colour in Dahlia: a new theory of specific pigmentation. Journal of Genetics. 1935;30:155–226. [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proceedings of the National Academy of Sciences, USA. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LQ, Pang XB, Shen HY, Pu GB, Wang HH, Lei CY, Wang H, Li GF, Liu BY, Ye HC. A novel type III polyketide synthase encoded by a three-intron gene from Polygonum cuspidatum. Planta. 2009;229:457–469. doi: 10.1007/s00425-008-0845-7. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Research. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Carpenter R, Sommer H, Saedler H, Coen ES. Molecular analysis of instability in flower pigmentation of Antirrhinum majus, following isolation of the pallida locus by transposon tagging. EMBO Journal. 1985;4:1625–1630. doi: 10.1002/j.1460-2075.1985.tb03829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClaren M. Dahlia: history and species. In: McClaren B, editor. Encyclopedia of dahlias. Portland, OR: Timber Press; 2009. pp. 161–166. [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant and Cell Physiology. 2006;47:457–470. doi: 10.1093/pcp/pcj012. [DOI] [PubMed] [Google Scholar]
- Motohashi R, Ohtsubo E, Ohtsubo H. Identification of Tnr3, a Suppressor-Mutator/Enhancer-like transposable element from rice. Molecular and General Genetics. 1996;250:148–152. doi: 10.1007/BF02174173. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacken WKF, Piotrowiak R, Saedler H, Sommer H. The transposable element Tam1 from Antirrhinum majus shows structural homology to the maize transposon En/Spm and has no sequence specificity of insertion. Molecular and General Genetics. 1991;228:201–208. doi: 10.1007/BF00282466. [DOI] [PubMed] [Google Scholar]
- Nakatsuka T, Haruta KS, Pitaksutheepong C, Abe Y, Kakizaki Y, Yamamoto K, Shimada N, Yamamura S, Nishihara M. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant and Cell Physiology. 2008;49:1818–1829. doi: 10.1093/pcp/pcn163. [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix–loop–helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. The Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström CG, Swain T. The flavonoid glycosides of Dahlia variabilis. Part I. General introduction. Cyanidin, apigenin, and luteolin glycosides from the variety ‘Dandy’. Journal of the Chemical Society. 1953:2764–2773. [Google Scholar]
- Nordström CG, Swain T. The flavonoid glycosides of Dahlia variabilis. II. Glycosides of yellow varieties ‘Pius IX’ and ‘Coton’. Archives of Biochemistry and Biophysics. 1956;60:329–344. doi: 10.1016/0003-9861(56)90435-0. [DOI] [PubMed] [Google Scholar]
- Nordström CG, Swain T. The flavonoid glycosides of Dahlia variabilis. III. Glycosides from white varieties. Archives of Biochemistry and Biophysics. 1958;73:220–223. doi: 10.1016/0003-9861(58)90257-1. [DOI] [PubMed] [Google Scholar]
- Ogata J, Sakamoto T, Yamaguchi MA, Kawanobu S, Yoshitama K. Isolation and characterization of anthocyanin 5-O-glucosyltransferase from flowers of Dahlia variabilis. Journal of Plant Physiology. 2001;158:709–714. [Google Scholar]
- Ozeki Y, Davies E, Takeda J. Somatic variation during long term subculturing of plant cells caused by insertion of a transposable element in a phenylalanine ammonia-lyase (PAL) gene. Molecular and General Genetics. 1997;254:407–416. doi: 10.1007/s004380050433. [DOI] [PubMed] [Google Scholar]
- Park KI, Choi JD, Hoshino A, Morita Y, Iida S. An intragenic tandem duplication in a transcriptional regulatory gene for anthocyanin biosynthesis confers pale-colored flowers and seeds with fine spots in Ipomoea tricolor. The Plant Journal. 2004;38:840–849. doi: 10.1111/j.1365-313X.2004.02098.x. [DOI] [PubMed] [Google Scholar]
- Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S. A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. The Plant Journal. 2007;49:641–654. doi: 10.1111/j.1365-313X.2006.02988.x. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. Molecular analysis of the En/Spm transposable element system of Zea mays. EMBO Journal. 1986;5:835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. The Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Van der Woude K, Mol JNM, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. The Plant Journal. 1998;13:475–488. doi: 10.1046/j.1365-313x.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Saito N, Ishizuka K, Osawa Y. Paper-chromatographic identification of flavonoids from a scarlet-flowering dahlia and crystallization of pelargonidin and butein. Botanical Magazine Tokyo. 1970;83:229–232. [Google Scholar]
- Schlangen K, Miosic S, Halbwirth H. Allelic variants from Dahlia variabilis encode flavonoid 3′-hydroxylases with functional differences in chalcone 3-hydroxylase activity. Archives of Biochemistry and Biophysics. 2010a;494:40–45. doi: 10.1016/j.abb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Schlangen K, Miosic S, Thill J, Halbwirth H. Cloning, functional expression, and characterization of a chalcone 3-hydroxylase from Cosmos sulphureus. Journal of Experimental Botany. 2010b;61:3451–3459. doi: 10.1093/jxb/erq169. [DOI] [PubMed] [Google Scholar]
- Schlangen K, Miosic S, Topuz F, Muster G, Marosits T, Seitz C, Halbwirth H. Chalcone 3-hydroxylation is not a general property of flavonoid 3′-hydroxylase. Plant Science. 2009;177:97–102. [Google Scholar]
- Shirsat AH. A transposon-like structure in the 5′ flanking sequence of a legumin gene from Pisum sativum. Molecular and General Genetics. 1988;212:129–133. doi: 10.1007/BF00322455. [DOI] [PubMed] [Google Scholar]
- Singh JP, Arora RS, Dohare SR, Sengupta K. A spontaneous mutant for flower colour and shape in a white flowering dahlia. Euphytica. 1970;19:261–262. [Google Scholar]
- Snowden KC, Napoli CA. Psl: a novel Spm-like transposable element from Petunia hybrida. The Plant Journal. 1998;14:43–54. doi: 10.1046/j.1365-313x.1998.00098.x. [DOI] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JNM, Koes R. anthocyanin1 of petunia encodes a basic helix–loop–helix protein that directly activates transcription of structural anthocyanin genes. The Plant Cell. 2000;12:1619–1631. doi: 10.1105/tpc.12.9.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol J, Koes R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. The Plant Cell. 2002;14:2121–2135. doi: 10.1105/tpc.003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Nakayama T, Yonekura-Sakakibara K, Fukui Y, Nakamura N, Yamaguchi MA, Tanaka Y, Kusumi T, Nishino T. cDNA cloning, heterologous expressions, and functional characterization of malonyl-coenzyme A: anthocyanidin 3-O-glucoside-6′-O-malonyltransferase from dahlia flowers. Plant Physiology. 2002;130:2142–2151. doi: 10.1104/pp.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. The Plant Journal. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- Tian PF. Progress in plant CACTA elements. Acta Genetica Sinica. 2006;33:765–774. doi: 10.1016/S0379-4172(06)60109-1. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix–loop–helix transcription factor family. The Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin LO, Rhodes PR, Goldberg RB. Ca lectin gene insertion has the structural features of a transposable element. Cell. 1983;34:1023–1031. doi: 10.1016/0092-8674(83)90560-3. [DOI] [PubMed] [Google Scholar]
- Wimmer G, Halbwirth H, Wurst F, Forkmann G, Stich K. Enzymatic hydroxylation of 6'-deoxychalcones with protein preparations from petals of Dahlia variabilis. Phytochemistry. 1998;47:1013–1016. [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Brar HK, Grosic S, Palmer RG, Bhattacharyya MK. Excision of an active CACTA-like transposable element from DFR2 causes variegated flowers in soybean [Glycine max (L.) Merr.] Genetics. 2010;184:53–63. doi: 10.1534/genetics.109.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi MA, Oshida N, Nakayama M, Koshioka M, Yamaguchi Y, Ino I. Anthocyanidin 3-glucoside malonyltransferase from Dahlia variabilis. Phytochemistry. 1999;52:15–18. [Google Scholar]
- Yamasaki H, Uefuji H, Sakihama Y. Bleaching of the red anthocyanin induced by superoxide radical. Archives of Biochemistry and Biophysics. 1996;332:183–186. doi: 10.1006/abbi.1996.0331. [DOI] [PubMed] [Google Scholar]
- Zabala G, Vodkin L. A putative autonomous 20.5 kb-CACTA transposon insertion in an F3'H allele identifies a new CACTA transposon subfamily in Glycine max. BMC Plant Biology. 2008;8:124. doi: 10.1186/1471-2229-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.