Abstract
The cell wall is important for pollen tube growth, but little is known about the molecular mechanism that controls cell wall deposition in pollen tubes. Here, the functional characterization of the pollen-expressed Arabidopsis cellulose synthase-like D genes CSLD1 and CSLD4 that are required for pollen tube growth is reported. Both CSLD1 and CSLD4 are highly expressed in mature pollen grains and pollen tubes. The CSLD1 and CSLD4 proteins are located in the Golgi apparatus and transported to the plasma membrane of the tip region of growing pollen tubes, where cellulose is actively synthesized. Mutations in CSLD1 and CSLD4 caused a significant reduction in cellulose deposition in the pollen tube wall and a remarkable disorganization of the pollen tube wall layers, which disrupted the genetic transmission of the male gametophyte. In csld1 and csld4 single mutants and in the csld1 csld4 double mutant, all the mutant pollen tubes exhibited similar phenotypes: the pollen tubes grew extremely abnormally both in vitro and in vivo, which indicates that CSLD1 and CSLD4 are not functionally redundant. Taken together, these results suggest that CSLD1 and CSLD4 play important roles in pollen tube growth, probably through participation in cellulose synthesis of the pollen tube wall.
Keywords: Arabidopsis, cell wall, cellulose, CSLD1, CSLD4, pollen tube
Introduction
In flowering plants, the pollen tube generated from the vegetative cell of the pollen grain is essential for delivery of male gametes to the female gametophyte in sexual reproduction (Hülskamp et al., 1995; Johnson and Preuss, 2002; Lord and Russell, 2002). Studies have shown that biosynthesis of the pollen tube wall is involved in the regulation of pollen tube growth (Edlund et al., 2004; Geitmann and Steer, 2006). Although it is important for pollen tube growth, the molecular mechanisms that control pollen tube wall biosynthesis remain largely unknown.
The pollen tube undergoes polar growth, namely the pollen tube elongates in one direction. Therefore, cell wall synthesis mainly occurs in the tip region of growing pollen tubes. The newly formed cell wall at the pollen tube tip is mainly composed of pectins, such as highly methylesterified homogalacturonan and rhamnogalacturonan I (Lancelle and Hepler, 1992; Edlund et al., 2004; Geitmann and Steer, 2006; Dardelle et al., 2010). This pectic layer continues along the entire length of the pollen tube and the homogalacturonans in this layer are gradually demethylesterified by pectin methylesterase (Lennon and Lord, 2000; Jiang et al., 2005; Dardelle et al., 2010). Behind the tip, a secondary layer is formed beneath the outer pectic fibrous layer (Ferguson et al., 1998; Li et al., 1999a). The most abundant component in this secondary wall layer is callose. Cellulose is also present in this layer, but at a low level (Dong et al., 2005; Nishikawa et al., 2005; Geitmann and Steer, 2006). For example, the pollen tube wall of Nicotiana alata contains only 5–10% cellulose (Schlüpmann et al., 1994; Li et al., 1999a), whereas the stem cell wall of Arabidopsis thaliana contains 30% cellulose (Taylor et al., 2003). Electron microscopy studies have shown that cellulose microfibrils in the pollen tube are thinner and shorter than those in somatic cells. In addition, linkage assays indicate that cellulose from pollen tubes is a heteropolymer containing β-1,4 linkages and a few β-1,3 linkages, rather than a homogeneous β-1,4-linked glucan (Steer and Steer, 1989; Geitmann and Steer, 2006). These findings imply that the cellulosic material of pollen tubes differs from that of somatic cells and that it more closely resembles cellulose from lower eukaryotes than that of the somatic cells of higher plants. Moreover, treatment with cellulase or specific inhibitors of cellulose biosynthesis severely represses pollen tube growth (Anderson et al., 2002; Aouar et al., 2010). Furthermore, the distribution of cellulose synthase (CESA) in pollen tubes depends on actin filaments and endomembrane dynamics, but does not depend on microtubules (Cai et al., 2011), whereas movement of CESA in somatic cells is highly regulated by cortical microtubules (Crowell et al., 2009). These results show that there probably exists a special mechanism for cellulose deposition in pollen tubes. So far, although many genes have been predicted to be involved in cellulose synthesis in somatic cells (Mutwil et al., 2008), the genes responsible for cellulose synthesis in pollen tubes remain largely unknown.
In plant somatic cells, cellulose is synthesized by CESA at the plasma membrane (Kimura et al., 1999; Richmond and Somerville, 2000; Taylor, 2008). CESA genes belong to a superfamily that also includes nine cellulose synthase-like (CSL) families (CSLA/B/C/D/E/F/G/H/J; Yin et al., 2009). Apart from CESA genes, several CSL genes have also been shown to encode glycan synthases for hemicellulosic polysaccharides. For example, CSLA genes are involved in the formation of mannan (Dhugga et al., 2004; Liepman et al., 2005). CSLC genes encode enzymes that catalyse the elongation of the backbone of xyloglucan (Cocuron et al., 2007). CSLF and CSLH genes are responsible for β-(1–3,1–4)-D-glucan synthesis (Burton et al., 2006; Doblin et al., 2009). Although the products of these CSL genes have been identified, the functions of the other CSL genes including the CSLD genes remain unknown. To date, several mutants with lesions in CSLD genes have been described. Most of these mutations affect the polar growth of root hairs, pollen tubes, or xylem (Favery et al., 2001; Wang et al., 2001; Bernal et al., 2007, 2008; Kim et al., 2007; Li et al., 2009). For example, mutations in Arabidopsis CSLD1 and CSLD4 drastically impair pollen germination and pollen tube growth (Bernal et al., 2008). However, how these two genes affect pollen tube growth and whether they are involved in cell wall deposition still remain unclear.
In this study, a detailed functional characterization of CSLD1 and CLSD4 was performed. The results show that mutations in CSLD1 and CLSD4 dramatically impair the deposition of cellulose in the pollen tube wall and lead to severe defects in pollen tube growth. In addition, it was found that CSLD1 and CSLD4 proteins display a polar localization at the plasma membrane at the tip of the growing pollen tube, which is highly similar to the characteristics of the CESAs functioning in somatic cells. These new findings suggest that CSLD1 and CLSD4 could play important roles in cellulose synthesis for pollen tubes.
Materials and methods
Plant materials and mutant isolation
Mutant and wild-type seeds were surface sterilized and pre-germinated on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) plates with or without 50 mg ml−1 of kanamycin (Sigma) at 22 °C under a photoperiod of 16 h light/8 h dark. The plants were grown in soil at 22 °C under the same light cycle as for pre-germination. The generation of transposon dissociation (Ds) insertion mutants, csld4-2 and csld4-3, was performed as described by Sundaresan et al. (1995). The csld1-1 seeds (SALK_043260; Bernal et al., 2008) were obtained from the Arabidopsis Biological Resource Center (ABRC, www.arabidopsis.org), and the T-DNA insertion site was determined by PCR using the primer pair LBa1/D1-S1 (The sequences of all the primers used in this study are listed in Supplementary Table S2 available at JXB online). DNA preparation and Southern blotting were performed as described by Yang et al. (1999).
Phenotypic characterization
In vitro pollen tube growth assays were performed as described by Li et al. (1999b) with a small modification. Pollen grains from ∼6 anthers were plated onto the surface of agar plates (3 mm×3 mm). The pollen grains spread on the agar plates were cultured immediately at 22 °C and 100% relative humidity for ∼6 h and then visualized by light microscopy (Leitz DM2500; Leica, Wetzlar, Germany). For in vivo pollen growth assays, mature pistils of the male-sterile mutant male sterility 1 (ms1; Zinkl et al., 1999) were pollinated with a limited number (3–5) of pollen grains from the mutants and wild-type plants. The pollen tubes in the pistils were stained with aniline blue and viewed with a fluorescence microscope (Leitz DM2500) as described by Jiang et al. (2005) and Tan et al. (2010). 4',6-Diamidino-2-phenylindole (DAPI) staining was performed as described by McCormick (2004). Alexander staining was performed as described by Alexander (1969). β-Glucuronidase (GUS) staining was performed as described by Jia et al. (2009). For Calcofluor and Pontamine Fast Scarlet 4B (S4B; Sigma) staining (Hoch et al., 2005; Anderson et al., 2009), quartets were cultured in vitro at 16 °C for 12 h and then fixed and stained with Calcofluor and S4B as described by Anderson et al. (2009) and Dardelle et al. (2010). Specifically, 0.01% (w/v) S4B was dissolved in liquid pollen germination medium (without agar) instead of liquid half-strength MS medium. FM4-64 staining was performed as described by Szumlanski et al. (2009). Fluorescence recovery after photobleaching (FRAP) and brefeldin A (BFA) treatment were performed as described by Lee et al. (2008). Morphological observation of pollen grains and pollen tubes by scanning electronic microscopy (SEM; Hitachi, S-3400N) and transmission electronic microscopy (TEM; JEM1230) was carried out as described by Hülskamp et al. (1995).
Molecular cloning
Isolation of the flanking sequences adjacent to the Ds element by thermal asymmetric interlaced PCR (TAIL-PCR) (Liu et al., 1995) was performed as described previously (Yang et al., 2003). The insertion site was confirmed by PCR using the primer pair D4-Ds/Ds5-1.
Full-length CSLD1 and CSLD4 genomic DNA fragments were amplified by PCR using the primer pairs D4-F-AF/D4-FAR, D4-F-BF/D4-FBR, and D1-FAF/D1-FAR, D1-F-BF/D1-FBR, respectively. The resulting DNA fragments were cloned into a pMD-18 T-vector for sequencing. For complementation experiments, the full-length CSLD1 and CSLD4 genomic DNA fragments were subcloned into a pCAMBIA1300 vector (CAMBIA, http://www.cambia.org) and introduced into csld4-2, csld4-3, and csld1-1 heterozygous plants using the Agrobacterium-mediated infiltration method (Clough and Bent, 1998). For CSLD4, the transformant plants were selected on MS plates containing 25 mg l−1 of hygromycin B (Roche) and 50 mg l−1 of kanamycin, and confirmed by PCR using the primer pair 1300HindIII/D4-P1. For CSLD1, the transformant plants were selected by hygromycin B and confirmed by PCR using primer pair 1300HindIII/D1-P1.
The 2 kb CSLD4 promoter fragment was amplified by PCR with the primer pair D4-PF/D4-PR and subcloned upstream of the GUS reporter gene in the pCAMBIA1300 vector. CSLD1 and CSLD4 cDNAs were cloned using RT-PCR with the gene-specific primer pairs D1-CAF/D1-CAR, D1-CBF/D1-CBR, D4-CAF/D4-CAR, and D4-CBF/D4-CBR. These cDNA fragments were then subcloned to create an N-terminal fusion gene with the green fluorescent protein (GFP) coding sequence downstream of the CSLD4 promoter in pCAMBIA1300.
Confocal microscopy
A Zeiss LSM510 META laser-scanning microscope (Carl Zeiss, http://www.zeiss.com) was used in the experiments. GFP signals were excited at 488 nm and emission was collected at 505–530 nm. ROOT AND POLLEN ARFGAP (Song et al., 2006) (RPA)–DsRed2 was excited at 543 nm, and emission was collected at 585–615 nm.
Spinning disk microscopy
Images were taken using an OLYMPUS IX81 microscope (OLYMPUS) equipped with a Yokogawa CSU-X1 spinning disk and an Andor iXon DV887ESV-BV camera (Plateforme d'Imagerie Dynamique, Institut Pasteur, Paris, France). A 405, 488, or 561 nm laser was used for excitation, and emission was collected using band-pass 405/25, 488/25, and 561/25 filters for Calcofluor, GFP, and DsRed2, respectively. Time series were acquired at a frame rate of 1–20 frames s−1.
Results
Isolation and genetic analysis of csld1 and csld4 mutants
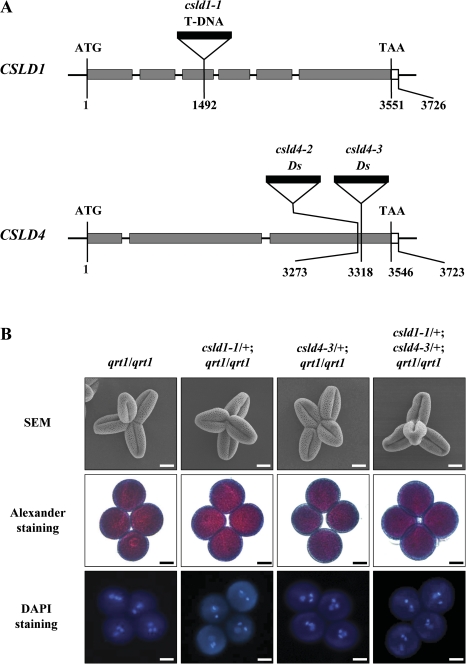
The two novel csld4 alleles used in this study, csld4-2 and csld4-3, were isolated in a screen for male gametophyte defective (mgp) mutants in Arabidopsis ecotype Landsberg erecta (Ler). Isolation of the genomic flanking sequences adjacent to the 5' end of the Ds element in these mutants revealed that both Ds insertions were located in the third exon of gene At4g38190, which encodes cellulose synthase-like protein CSLD4. The Ds elements were inserted 3273 bp and 3318 bp downstream of the ATG start codon in csld4-2 and csld4-3, respectively (Fig. 1A). The csld4-2 and csld4-3 mutants were found to exhibit identical phenotypes. Therefore, csld4-3 was used in most of the further characterization of the gene. The T-DNA in csld1-1 (SALK_043260) is located in the third exon of CSLD1 (At2g33100), 1492 bp downstream of the start codon (Fig. 1A).
Fig. 1.
Isolation of CSLD1 and CSLD4 knock-out mutants. (A) Schematic diagrams of the CSLD1 and CSLD4 gene structures, showing the T-DNA and Ds insertion sites. The grey and white boxes indicate translated and untranslated regions, respectively. (B) Scanning electron microscopy (SEM) analysis, Alexander staining, and DAPI staining of quartets from qrt1/qrt1, csld1-1/+; qrt1/qrt1, csld4-3/+; qrt1/qrt1, and csld4-3/+; csld1-1/+; qrt1/qrt1, respectively. For SEM analysis, pollen grains were coated with gold particles. Bars: 10 μm.
Genetic analysis of csld4 mutant lines was performed using a kanamycin selection marker carried by the Ds element and of csld1 mutant lines by PCR-aided genotyping (Table 1). The progeny from self-pollinated heterozygous csld1 (csld1-1/+) and heterozygous csld4 (csld4-2/+ and csld4-3/+) mutant plants all exhibited segregation ratios of ∼1 with insertion, to 1 without insertion (Table 1). These results indicate that the csld1 and csld4 mutants were probably defective in gametophyte function. Reciprocal crosses between the mutants and wild-type plants were then performed. When csld1-1/+, csld4-2/+, or csld4-3/+ plants were used as recipients in crosses with wild-type pollen, ∼50% of the resulting progeny contained the T-DNA or Ds insertions (Table 1). When wild-type plants were used as recipients in crosses with the mutant plants, none of the resulting offspring was identified as having the insertions (Table 1). Therefore, the csld1-1, csld4-2, and csld4-3 mutations completely suppressed the genetic transmission of male gametophytes and had no discernible influence on female gametophyte function. Furthermore, no homozygous mutant plants could be obtained in these mutant lines; therefore, heterozygous mutant plants were used for further phenotypic characterization of the male gametophytes.
Table 1.
Genetic analysis of csld4 and csld1 mutants
| Crosses (female×male) | With insertiona | Without insertionb | Ratio | TEFc | TEM |
| csld4-2/+×csld4-2/+ | 368 | 361 | 1.02 | NA | NA |
| csld4-3/+×csld4-3/+ | 516 | 529 | 0.98 | NA | NA |
| csld4-2/+×Ler | 1293 | 1314 | 0.98 | 100% | NA |
| csld4-3/+×Ler | 1085 | 1052 | 1.03 | 100% | NA |
| Ler×csld4-2/+ | 0 | 1793 | 0.00 | NA | 0 |
| Ler×csld4-3/+ | 0 | 1652 | 0.00 | NA | 0 |
| csld1-1/+×csld1-1/+ | 62 | 61 | 1.02 | NA | NA |
| csld1-1/+×Col | 84 | 102 | 0.82 | 82% | NA |
| Col×csld1-1/+ | 0 | 180 | 0.00 | NA | 0% |
| Col×csld1-1/+; gD1/gD1d | 55 | 40 | 1.38 | 100% | NA |
| Col×csld1-1/+; GFP-D1/GFP-D1 | 23 | 24 | 0.96 | 100% | NA |
| Col×csld1-1/+; GFP-D1/– | 11e | 21 | 0.52 | 52% | NA |
Kanamycin-resistant progeny in a csld4-2 or csld4-3 mutant background or progeny positive for the PCR analysis in the csld1-1 mutant background.
Kanamycin-sensitive progeny in a csld4-2 or csld4-3 mutant background or progeny negative for the PCR analysis in the csld1-1 mutant background.
TE, transmission efficiency; TE=(progeny with insertion/progeny without insertion)×100%; TEF and TEM, female and male transmission efficiency, respectively; NA, not applicable.
gD1 represents the CSLD1 genomic DNA construct. D1 represents CSLD1.
All of these lines are associated with the GFP-CSLD1 transgenic insertion.
The csld1 and csld4 mutations severely repress pollen tube growth both in vitro and in vivo
For characterization of mutant phenotypes, csld4-3 and csld1-1 were introgressed into the quartet1 (qrt1) mutant background (Preuss et al., 1994). In a qrt1 homozygous mutant background, the four microspores from a microsporocyte fail to separate after meiosis, but their functions are virtually unaffected (Preuss et al., 1994; Rhee and Somerville, 1998). Half of the pollen grains produced by the heterozygous mutant plants are mutant and half are wild type. Therefore, a quartet produced by the csld heterozygous mutant plants contains two mutant pollen grains (csld) and two wild-type pollen grains (CSLD). Pollen morphology, nuclear division, and pollen viability of the mutant plants were examined by SEM, DAPI staining, and Alexander staining, respectively. No obvious defects in the four pollen grains in the mature quartets (n >30) from the csld1/+; qrt1/qrt1 and csld4/+; qrt1/qrt1 plants were found compared with those from qrt1/qrt1 plants (Fig. 1B), indicating that the csld1 and csld4 mutations do not affect pollen grain formation.
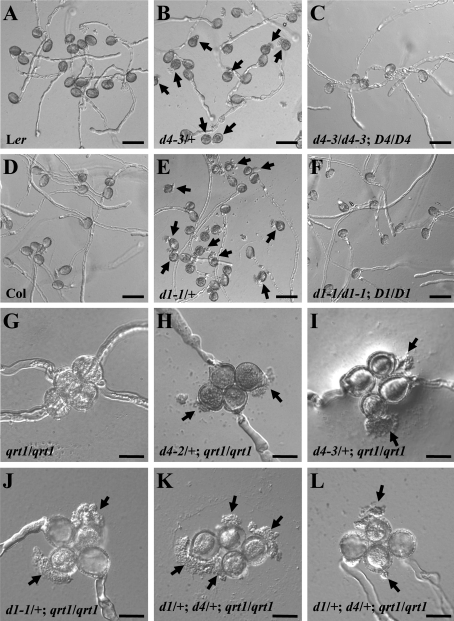
Germination and growth of the mutant pollen tubes were also examined (Table 2). Under in vitro conditions, ∼87.6% and 83.6% of wild-type Col and Ler pollen grains, respectively, produced normal pollen tubes (Fig. 2A, D). In contrast, only 45.4% of pollen grains from csld1-1/+ plants (Col background) and 43.9% of pollen grains from csld4-3/+ (Ler background) plants produced normal pollen tubes. A total of 47.1% and 45.5% of the pollen grains from csld1-1/+ and csld4-3/+ plants, respectively, ruptured just after germination (Fig. 2B, E), while only 6.9% of Col and 8.4% of the Ler pollen grains ruptured after germination. The percentages of ungerminated pollen grains in the mutants were similar to those in wild-type plants (Table 2). Germination assays were also performed in the qrt1 background (Table 3). In the control, ∼20.2% and 6.2% of qrt1/qrt1 quartets had three or four pollen grains that produced normal pollen tubes, respectively (Fig. 2G). In contrast, of the four pollen grains of each quartet from csld1-1/+; qrt1/qrt1 and csld4-3/+; qrt1/qrt1 mutant plants, a maximum of two produced normal pollen tubes; the rest ruptured just after germination or did not germinate (Fig. 2H–J).
Table 2.
In vitro germination of pollen grains from mutants, complemented mutants, and wild-type plants
| Genotype | Germinated normally (%) | Ruptured (%) | Failed to germinate (%) |
| Col (+/+) | 87.6±3.3 | 6.9±1.2 | 5.5±2.2 |
| csld1-1/+ | 45.4±5.7 | 47.1±4.0 | 7.5±2.2 |
| d1-1/d1-1; gD1/gD1 | 76.8±2.2 | 14.8±0.9 | 8.4±1.3 |
| Ler (+/+) | 83.6±3.4 | 8.4±2.5 | 8.0±0.9 |
| csld4-3/+ | 43.9±1.4 | 45.5±0.7 | 10.6±0.9 |
| d4-3/d4-3; gD4/gD4 | 78.5±1.7 | 15.2±1.5 | 6.3±1.1 |
Pollen grains were cultured in vitro at 22 °C for 6 h. All results represent the average of three biological replicates with the standard deviation. For each independent replicate, at least 300 pollen grains were analysed.
Fig. 2.
In vitro germination of pollen grains from wild-type, mutant, and complemented mutant plants. Pollen grains were cultured in vitro at 22 °C for 6 h. (A–F) In vitro germination of pollen grains from Ler (A), csld4-3/+ (B), complemented csld4-3 (C), Col (D), csld1-1/+ (E), and complemented csld1-1 (F) plants. Arrows indicate abnormal pollen tubes. (G–L) In vitro germination of quartets from qrt1/qrt1 (G), csld4-2/+; qrt1/qrt1 (H), csld4-3/+; qrt1/qrt1 (I), csld1-1/+; qrt1/qrt1 (J), and csld4-3/+; csld1-1/+; qrt1/qrt1 (K, L) plants. Arrows indicate ruptured pollen tubes. D1, D4, d1, and d4 represent CSLD1, CSLD1, csld1, and csld4, respectively. Bars: (A–F) 50 μm; (G–L) 20 μm.
Table 3.
In vitro germination of quartets from wild-type and mutants plants
| Genotype | No. of pollen grains germinated normally in a quartet |
||||
| 0 (%) | 1 (%) | 2 (%) | 3 (%) | 4 (%) | |
| qrt1/qrt1 | 8.1±1.8 | 30.6±9.3 | 34.9±1.2 | 20.2±8.1 | 6.2±2.4 |
| csld1-1/+; qrt1/qrt1 | 15.0±4.5 | 54.8±6.2 | 30.2±3.0 | 0.0 | 0.0 |
| csld4-3/+; qrt1/qrt1 | 13.0±2.6 | 43.0±4.1 | 44.0±5.2 | 0.0 | 0.0 |
| csld1-1/+; csld4-3/+; qrt1/qrt1 | 46.1±9.1 | 45.5±8.0 | 8.4±1.1 | 0.0 | 0.0 |
Quartets were cultured in vitro at 22 °C for 6 h. All results represent the average of three biological replicates with the standard deviation. For each independent replicate, at least 200 quartets were analysed.
Pollen germination was next investigated in vivo (see the Materials and methods). Most of the wild-type pollen grains produced normal pollen tubes (96% for Col, n=47; 95% for Ler, n=108; Fig. 3A, B). In contrast, some of the pollen grains from csld1/+ (25%, n=146) or csld4/+ (27%, n=82) mutant plants produced abnormal pollen tubes after adhesion to the stigma. Some of the abnormal pollen tubes stopped growing in papillar cells (Fig. 3G–J), and some stopped growing in stigmas with expanded shapes (Fig. 3C–F). In summary, the csld1 and csld4 mutant pollen tubes were unstable and severely defective both in vitro and in vivo.
Fig. 3.
In vivo germination of pollen grains from mutant and wild-type plants. In vivo germination of pollen grains from Ler (A, B), csld4/+ (C, D, G, H), and csld1-1/+ (E, F, I, J) plants. Compared with wild-type pollen tubes, mutant pollen tubes were arrested at the surfaces of stigma cells (G–J) or expanded in the stigma (C–F). The white arrows indicate aberrant pollen tubes. The white arrowheads indicate normal pollen tubes. Bars: 20 μm.
Phenotypes of csld1 and csld4 are rescued by wild-type CSLD1 and CSLD4 genes, respectively
To confirm that the phenotypes of csld4 and csld1 were attributable to defects in CSLD4 and CSLD1, respectively, complementation experiments were performed with the wild-type genomic DNAs and cDNAs of CSLD4 and CSLD1. First, CSLD4 and CSLD1 genomic DNA fragments, including the predicted promoters, transcribed regions, and 3′ end non-transcribed regions, were cloned (gCSLD4 and gCSLD1; see the Materials and methods) and introduced into csld4 and csld1 heterozygous plants. For csld4, 51 (in csld4-2) and 36 (in csld4-3) independent transformants were obtained in a screen; the progeny from 34 (in csld4-2) and 22 (in csld4-3) self-pollinated T1 plants segregated in a ratio of ∼2 kanR to 1 kanS (Supplementary Data File S1 at JXB online). For csld1, 12 independent transformants were obtained, and the plants heterozygous for csld1-1 and homozygous for the transgenic CSLD1 gene (csld1-1/+; gCSLD1/gCSLD1) in the T2 generation were selected. These plants were used as the males in a cross with wild-type Col plants. Approximately 50% of the resulting F1 seedlings carried the csld1-1 T-DNA insertion (Table 1). Plants homozygous for both insertions and transgenic genes (csld4/csld4; gCSLD4/gCSLD4 and csld1-1/ csld1-1; gCSLD1/gCSLD1) were obtained in the T3 generation. The in vitro germination of pollen grains from the transgenic csld4 homozygous plants was rescued from 43.9% to 78.5% (Fig. 2C, F; Table 2). In addition, expression of the N-terminal fusion of CSLD4 and CSLD1 with the GFP-encoding sequence driven by the CSLD4 promoter (pCSLD4:GFP-CSLD4 and pCSLD4:GFP-CSLD1) could also complement the phenotypes of csld4 and clsd1, respectively (Tables 1, 4; Supplementary Data File S1). These results demonstrate that the phenotypes of csld4 and csld1 were caused by mutations in CSLD4 and CSLD1, respectively.
Table 4.
Genetic analysis of the csld1/+; csld4/+; qrt1/qrt1 triple mutant
| Crosses (female×male) | D1/D1; D4/D4 | d1/D1; D4/D4 | D1/D1; d4/D4 | d1/D1; d4/D4 | No. of F1 seedlings |
| WT×d1/D1; d4/D4 | 100 | 0 | 0 | 0 | 100 |
| d1/D1; d4/D4×WT | 12 | 12 | 11 | 12 | 47 |
| WT×d1/D1; d4/D4; GFP-D4/– | 22 | 0 | 9a | 0 | 31 |
| d1/D1; d4/D4; GFP-D4/–×WT | 12 | 5b | 5 | 9c | 31 |
Progeny were analysed by PCR in all of the crosses.
All these lines are associated with the GFP-CSLD4 insertion.
Four of the five lines are associated with the GFP-CSLD4 insertion.
Four of the nine lines are associated with the GFP-CSLD4 insertion.
The csld1 csld4 double mutant shows no more severe defects
To investigate whether CSLD1 and CSLD4 serve redundant functions, a csld1 csld4 double mutant was generated. Because neither mutation can be transmitted through the male gametophyte, csld1/csld1; GFP-CSLD1/GFP-CSLD1 transgenic plants were used as the males in a cross with csld4-3/+. F1 plants with the csld1/+; csld4/+; GFP-CSLD1/– genotype were selected and self-pollinated. In the F2 generation, the double-heterozygous mutant plants with the csld1/+; csld4/+ genotype that did not carry the transgenic CSLD1 gene were selected (Supplementary Fig. S2 at JXB online). The qrt1 mutation was also introgressed into the double mutant to generate csld1/+; csld4/+; qrt1/qrt1 triple mutant plants. Because pollen grains in each quartet adhere to each other after meiosis, such plants produced three types of quartets: [2(csld1 CSLD4)+2(CSLD1 csld4)], [2(CSLD1 CSLD4)+2 (csld1 csld4)], and [(csld1 CSLD4)+(CSLD1 csld4)+(CSLD1 CSLD4)+(csld1 csld4)] (Supplementary Fig. S2). The (csld1 csld4) pollen grains represent the csld1 csld4 double mutant pollen grains.
First, genetic analysis on csld1/+; csld4/+; qrt1/qrt1 plants showed that neither csld1 nor csld4 could be transmitted through the male gametophyte, but that their transmission through the female gametophyte was not affected (Table 4). The construct pCSLD4:GFP-CSLD4 was also introduced into the triple mutant. The results showed that expression of GFP-CSLD4 in pollen could rescue the male transmission defects of csld4 but not csld1 pollen grains (Table 4).
Phenotypic analysis was then performed on quartets from csld1/+; csld4/+; qrt1/qrt1 plants with regard to morphology, cytoplasmic density, or nuclear constitution, and no defects were observed (n >30; Fig. 1B). Pollen germination and tube growth of this triple mutant were further investigated in vitro. Theoretically, quartets with the genotype [2(csld1 CSLD4)+2(CSLD1 csld4)] should not have produced any normal pollen tubes, quartets with the genotype [(csld1 CSLD4)+(CSLD1 csld4)+(CSLD1 CSLD4)+(csld1 csld4)] should have produced up to one normal pollen tube, and quartets with the genotype [2(CSLD1 CSLD4)+2(csld1 csld4)] should have produced up to two normal pollen tubes. It was observed that 45.5% (n=817) of the quartets from the triple mutant plants did not produce any normal pollen tubes (Fig. 2K, Table 3). Only 46.1% and 8.4% of the quartets (n=817) produced one or two normal pollen tubes (Fig. 2L; Table 3), respectively. The two pollen grains producing normal tubes in a quartet should be CSLD1 CSLD4, and the other two should be csld1 csld4; most of the latter (63.0%, n=216) ruptured after germination, like the csld1 or csld4 single mutant pollen grains (Supplementary Table S1 at JXB online). These results indicate that the phenotype of the the csld1 csld4 double pollen grains was no more severe than that of the csld1 and csld4 single mutant pollen grains.
The structures of csld1 and csld4 pollen tube cell walls are obviously disorganized
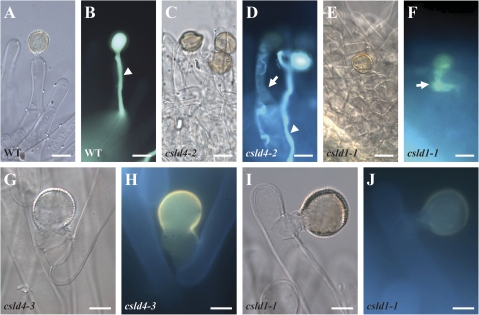
To understand why the csld1 and csld4 mutant pollen grains ruptured easily after germination, the morphology of the mutant pollen grains was examined using TEM. No obvious difference in surface appearance or cell wall structure was observed between mutant and wild-type pollen grains before germination (Supplementary Fig. S1 at JXB online). TEM images of the pollen grains germinated in vitro showed that newly emerging wild-type pollen tubes (n=18 for Col, n=19 for Ler) had a characteristic cell wall consisting of an outer fibrous pectin layer and an inner non-fibrous callose layer (Fig. 4A–D), whereas about half of the pollen grains from csld1/+ and csld4/+ plants (five out of 11 for csld1-1/+, four out of 10 for csld4-3/+) produced pollen tubes that exhibited thickened and highly irregular cell walls (Fig. 4E–L). In particular, some of the csld1 (Fig. 4I, J) and csld4 (Fig. 4K, L) pollen grains produced pollen tubes without obvious non-fibrous materials in the wall (two out of 11 for csld1-1/+, two out of 10 for csld4-3/+) and blocks of fibrillar inclusions could be observed embedded inside the inner wall (Fig. 4F, H, J, L), which were probably caused by uneven deposition of cell wall polymers in the csld1 and csld4 pollen tube walls.
Fig. 4.
Electronic microscopic observation of mutant and wild-type pollen tubes. Pollen grains were cultured in vitro at 16 °C for 3 h and then used for ultrathin sections. All of the ultrathin sections were stained with osmium tetroxide. Images show an overview of the newly emerging pollen tubes of Col (A), csld1 (E, I), Ler (C), and csld4 (G, K), and high magnification of the cell wall of Col (B), csld1 (F, J), Ler (D), and csld4 (H, L) pollen tubes. Wild-type Col (A, B) and Ler (C, D) pollen tubes had a characteristic cell wall consisting of an outer fibrous pectin layer and an inner non-fibrous callose layer. Mutant pollen grains from csld1/+ (E–H) and csld4/+ (I–L) plants produced pollen tubes that exhibited thickened and highly irregular cell walls. Some of the csld1 (I, J) and csld4 (K, L) pollen grains produced pollen tubes without obvious non-fibrous materials in the tube wall. Arrows indicate blocks of fibrillar inclusions. ow, outer wall; iw, inner wall; w, wall; cyt, cytoplasm; pm, plasma membrane. Bars: (A, E, I, C, G and K) 2 μm; (B, F, J, D, H and L) 1 μm.
Both CSLD1 and CSLD4 are highly expressed in mature pollen grains and pollen tubes
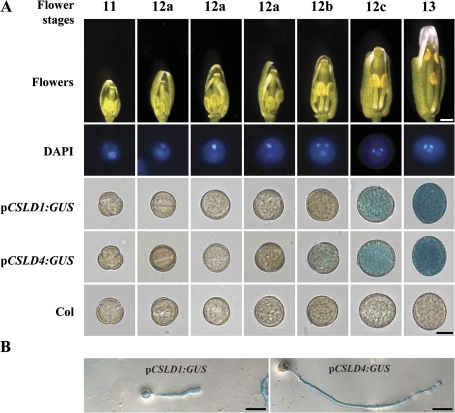
To study the detailed expression patterns of CSLD1 and CSLD4, transgenic lines transformed with the GUS gene driven by the CSLD1 (CS70765) or CSLD4 (CS70768) promoter from the ABRC were generated. GUS activity was detected in mature pollen and pollen tubes and was not detected in most other tissues/organs in these transgenic lines (Fig. 5; Supplementary Fig. S4 at JXB online). In pCSLD1:GUS transgenic lines, GUS activity was not detected in pollen until flower stage 12c and pCSLD4:GUS activity was not detected until flower stage 12b (Fig. 5). These data show that both CSLD1 and CSLD4 are specifically expressed in mature pollen and pollen tubes.
Fig. 5.
Expression of CSLD1 and CSLD4 in pollen and pollen tubes. (A) Different stages of flowers (Smyth et al., 1990) from pCSLD1:GUS and pCSLD4:GUS transgenic lines and wild-type plants were examined for GUS activity. Typical pollen grains with GUS staining are shown. (B) pCSLD1:GUS and pCSLD4:GUS pollen tubes showed GUS activity. Pollen grains were cultured in vitro at 22 °C for 6 h and then used for GUS staining. Bars: top of (A) 0.5 mm; bottom of (A) 10 μm; (B) 50 μm.
CSLD1 and CSLD4 are transported to the plasma membrane in the tip region of the pollen tube
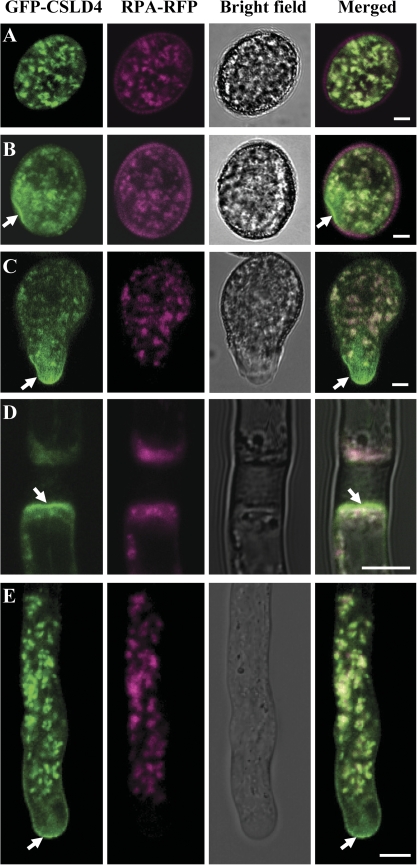
As demonstrated above, pCSLD4:GFP-CSLD1 and pCSLD4:GFP-CSLD4 could complement the phenotypes of csld1 and csld4, respectively, indicating that the N-terminal GFP fusion does not affect the normal function of CSLD1 and CSLD4. This suggests that the GFP fusion proteins probably present the correct localization of CSLD1 and CSLD4 in vivo. Accordingly, transgenic Arabidopsis lines expressing GFP–CSLD1 and GFP–CSLD4 were subjected to further analysis using confocal laser scanning microscopy (CLSM). The results showed that GFP–CSLD1 and GFP–CSLD4 are similarly located in pollen and pollen tubes. Before germination, GFP–CSLD1/4 (representing GFP–CSLD1 and GFP–CSLD4) exhibited punctate fluorescence signals in pollen grain cytoplasm, probably in the Golgi apparatus (Fig. 6A; Supplementary Fig. S5A at JXB online). To confirm this result, the GFP–CSLD1/4 plants were crossed with transgenic lines expressing the Golgi marker protein RPA (Song et al., 2006; Deng et al., 2010) with a C-terminal-fused red fluorescent tag, DsRed2, under the control of the pollen-specific LAT52 promoter (RPA–DsRed2). In mature pollen grains, as shown in Fig. 6A and Supplementary Fig. S5A, the green fluorescence signals of GFP–CSLD1/4 showed the same localization pattern as the red fluorescence signals of RPA–DsRed2, indicating that CSLD1/4 products were indeed present in the Golgi apparatus before germination.
Fig. 6.
Subcellular localization of CSLD4 in vivo. GFP–CSLD4 (green) co-localized with RPA–DsRed2 (magenta) in pollen grains before germination (A). GFP–CSLD4 signals were also observed at the cell periphery at the future pollen tube exit site (B, arrow) and at the plasma membrane of the freshly emerged pollen tube tip (C, arrow) and of growing pollen tubes (E, arrow). GFP–CSLD4 signals were also found at the plasma membrane adjacent to pollen tube plugs (arrow). Bars: 5 μm.
When the pollen grains began to germinate, the GFP–CSLD1/4 signals were found not only in the Golgi apparatus, but also in small vesicles, the cell periphery at the germinating point before emergence of the pollen tube (Fig. 6B; Supplementary Fig. S5B), and at the tip region of newly emerged (Fig. 6C; Supplementary Fig. S5C) and elongating (Fig. 6E; Supplementary Fig. S5E) pollen tubes where RPA–DsRed2 signals were absent. GFP–CSLD1/4 signals were also found at the periphery of the pollen tube plug (Fig. 6D; Supplementary Fig. S5D). To confirm that the GFP–CSLD1/4 signals on the cell periphery were located at the plasma membrane, the pollen tubes expressing GFP–CSLD4 were stained with the membrane-specific dye FM4-64 and it was found that the stain signals overlapped with the GFP–CSLD4 signals (Supplementary Fig. S6A at JXB online). Pollen tube plasmolysis also showed that GFP–CSLD1/D4 signals were localized to the plasma membrane (Supplementary Fig. S6B, C).
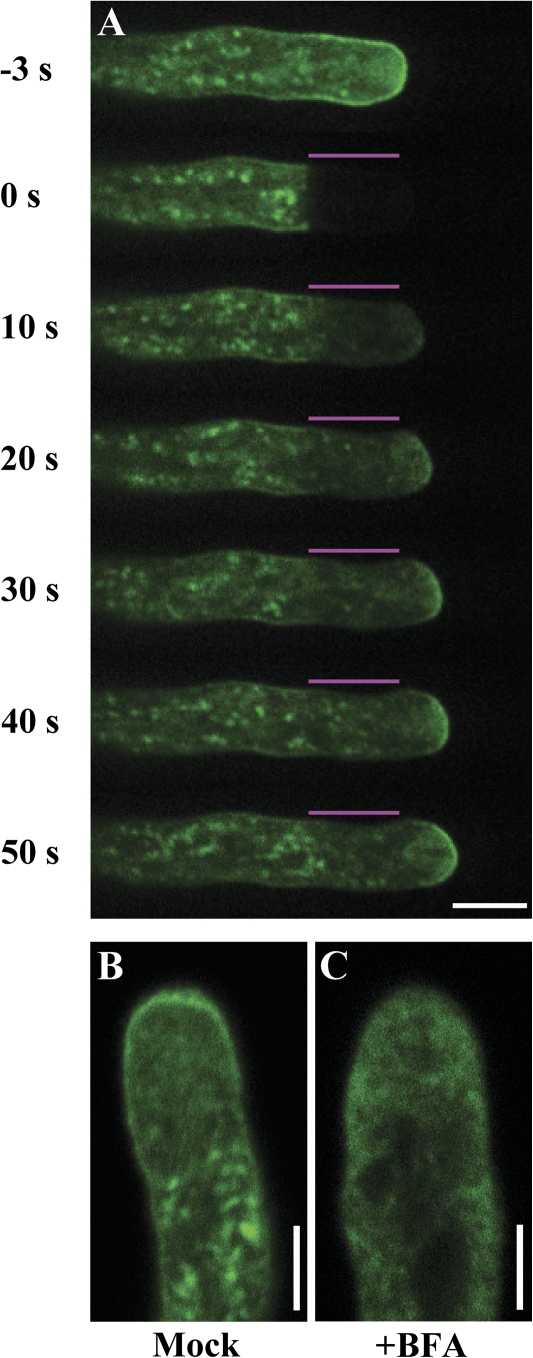
The above results show that GFP–CSLD1/4 proteins are likely to be associated with dynamic small vesicles exhibiting a characteristic ‘V’-shaped pattern within the apical clear zone (Fig. 6E; Supplementary Fig. S5E and Videos S1, S2 at JXB online). The presence of vesicles arranged in this pattern is consistent with possible exocytosis and endocytosis at the apical region (Moscatelli and Idilli, 2009). To confirm whether GFP–CSLD1/4 proteins were inserted into the plasma membrane via exocytosis, FRAP was performed. The visible GFP–CSLD4 signals in the apical region of the cells, including those associated with the plasma membrane, were reduced to almost background levels after photobleaching. Recovery of the GFP–CSLD4 signals was first observed in the centre of the apical dome and then gradually appeared in the lateral region of the plasma membrane (10 independent experiments; Fig. 7A; Supplementary Video S3 at JXB online). The fluorescence fully recovered in the apical region of the plasma membrane ∼30 s after photobleaching. In addition, plasma membrane localization of GFP-CSLD4 in the apical region was completely disrupted by BFA treatment (Fig. 7B, C). These results suggest that targeting of GFP–CSLD4 to the plasma membrane depends on exocytosis in the central region of the plasma membrane of the pollen tube tip. Based on these results, it is proposed that both CSLD1 and CSLD4 are preferentially located in the Golgi apparatus before germination and that during pollen germination and pollen tube growth they are transported to the plasma membrane at the tube tip via exocytosis.
Fig. 7.
FRAP analysis of GFP–CSLD4 in growing pollen tubes. Pollen grains were cultured in vitro at 22 °C for 6 h. (A) Spinning disk microscopy was used to perform FRAP in the tip region of growing pollen tubes expressing GFP–CSLD4. The fluorescence recovered at the plasma membrane of the pollen tube tip after photobleaching (10 independent experiments). Typically, a series of time-lapse images were taken every 10 s for 1 min. The numbers to the left of each image indicate elapsed time after photobleaching. The bleached area is marked by lines above the pollen tube. (B, C) Images showing pollen tubes expressing GFP–CSLD4 after mock treatment with 0.1% methanol (B), or treatment with 5 μg ml−1 BFA for 1 h (C). BFA completely disrupted the plasma membrane localization of GFP–CSLD4. Images were taken from the midplane. Bars: (A) 10 μm; (B, C) 5 μm.
csld1 and csld4 pollen tube walls exhibit cellulose deficiencies
To understand the roles of CSLD1 and CSLD4 in pollen tube growth, the effects of csld1 and csld4 mutations on the distribution of cell wall components in pollen tubes were investigated. Quartets from qrt1/qrt1, csld1/+; qrt1/qrt1, csld4/+; qrt1/qrt1, and csld1/+; csld4/+; qrt1/qrt1 plants were cultured in vitro at 16 °C. As shown in Supplementary Fig. S3 at JXB online, qrt1/qrt1 quartets produced up to four normal pollen tubes. Under the same conditions, up to two pollen grains in the csld1 and csld4 mutant quartets produced normal pollen tubes (representing wild-type); the other two pollen grains produced short and irregular pollen tubes (representing the mutant). These abnormal mutant pollen tubes were used for further analyses.
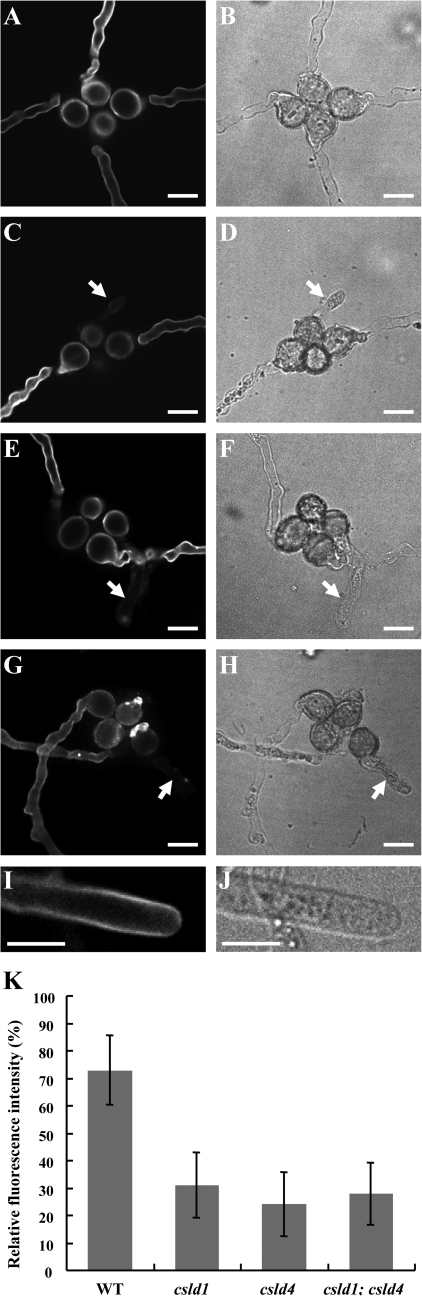
To investigate the distribution of β-glucans, such as callose and cellulose, in the mutant pollen tubes, cytochemical staining of the mutant pollen tubes with Calcofluor, aniline blue, and S4B was performed. Calcofluor stains both callose and cellulose (Sauter et al., 1993; Dardelle et al., 2010). Aniline blue specifically stains callose, and S4B specifically stains cellulose (Hoch et al., 2005; Anderson et al., 2009). The Calcofluor-stained pollen tubes were observed using spinning disk microscopy. Most of the abnormal mutant pollen tubes showed almost no fluorescent signal (Fig. 8C, E, G). Only a few abnormal mutant pollen tubes (three of 26 for csld1-1, three of 24 for csld4-3, and two of 18 for the csld1 csld4 double mutant) displayed punctate signals (Supplementary Fig. S7, D, H at JXB online) in comparison with the homogeneous signals of the control pollen tubes (Supplementary Fig. S7B). These signals may have been caused by the uneven deposition of cell wall materials that could be stained by Calcofluor. Thereafter, images were taken in the same focal plane and the fluorescence intensity along the pollen tube wall was measured with ImageJ (Abramoff et al., 2004; http://rsbweb.nih.gov/ij/). Among pollen tubes of the same quartet, the pollen tube with the weakest fluorescence was compared with the tube showing the strongest fluorescence (Fig. 8K). The results showed that mutant pollen tube walls had much lower fluorescence intensity than did the control pollen tubes.
Fig. 8.
Calcofluor staining of mutant and wild-type pollen tubes. Quartets were cultured in vitro at 16 °C for 12 h and then used for S4B staining. (A–H) Spinning disk confocal and bright field images of wild-type qrt1/qrt1 (A, B), csld4-3/+; qrt1/qrt1 (C, D), csld1-1/+; qrt1/qrt1 (E, F), and csld1-1/+; csld4-3/+; qrt1/qrt1 (G, H) pollen tubes stained with Calcofluor. Arrows indicate abnormal mutant pollen tubes. (I) and (J) Calcofluor staining of the tip region of a normal pollen tube, which cannot be shown in the same images above. (K) The fluorescence intensity of mutant pollen tubes was much weaker than that of control tubes. Images were taken at the same focal plane and the fluorescence intensity along the pollen tube wall was quantified by the grey value measured with ImageJ (Abramoff et al., 2004; http://rsbweb.nih.gov/ij/). The fluorescence intensity of the brightest pollen tube was set to 100% and the weakest was measured (n=20 for wild-type qrt1/qrt1, n=26 for csld1-1, n=24 for csld4-3, and n=18 for the csld1 csld4 double mutant). Bars: 20 μm.
To examine the callose deposition in pollen tube walls, wild-type and mutant pollen tubes were stained with aniline blue. In wild-type pollen tubes, callose was mainly distributed in the tube shank region and was absent from the apical region of the elongated pollen tubes (Supplementary Fig. S8A, B at JXB online). The distribution of callose staining along csld4 mutant pollen tubes was highly irregular (Supplementary Fig. S8C–J). In particular, 15 out of 21 csld4 pollen tubes observed exhibited that the intensity of callose staining in the mutant pollen tube walls was weaker than that in the normal pollen tubes (Supplementary Fig. S8C–F), and six out of 21 csld4 pollen tubes observed showed that callose was unevenly accumulated (Supplementary Fig. S8G–J). These results indicated that csld4 mutation affected the deposition of callose in pollen tube walls.
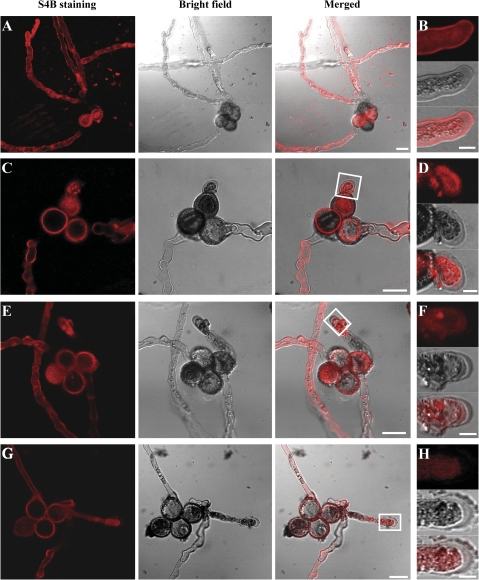
Accordingly, S4B staining was used to investigate the content and distribution of cellulose in wild-type and mutant pollen tube walls. Although S4B staining of the cytoplasm of both mutant and wild-type pollen tubes was intense, there was a marked decrease in the intensity of S4B fluorescence in the cell wall of csld1 (Fig. 9E, F), csld4 (Fig. 9C, D), and csld1 csld4 double mutant (Fig. 9G, H) pollen tubes compared with that of wild-type pollen tubes (Fig. 9A, B), especially in the pollen tube tip (five independent observations). These results suggest that the cellulose content was significantly reduced in the mutant pollen tube walls.
Fig. 9.
S4B staining of mutant and wild-type pollen tubes. Confocal sections of wild-type qrt1/qrt1 (A, B), csld4-3/+; qrt1/qrt1 (C, D), csld1-1/+; qrt1/qrt1 (E, F), and csld1-1/+; csld4-3/+; qrt1/qrt1 (G, H) pollen tubes stained with S4B. The S4B fluorescence intensity in the cell wall of csld4 (C, D), csld1 (E, F), and csld1 csld4 double mutant (G, H) pollen tubes was much weaker than that of wild-type pollen tubes (A, B). It is noteworthy that the S4B staining is weak at some parts of the pollen tube that are out of the focal planes shown here (especially the pollen tube base near the pollen grain). (B) High magnification image of a wild-type pollen tube showing S4B staining associated with the tube wall and cytoplasm. D, F, and H correspond to the white boxes in C, E, and G, respectively, showing that fluorescence was difficult to detect in the mutant pollen tube wall (five independent observations). Bars: (A, C, E, G) 20 μm; (B, D, F, H) 50 μm.
Discussion
In this study, a detailed characterization of several mutant alleles of the Arabidopsis CSLD1 and CSLD4 genes was performed. The results show that these mutations disrupt pollen tube growth and reduce the cellulose content of the pollen tube walls. It was also found that the CSLD1 and CSLD4 proteins are transported to the plasma membrane of the pollen tube tip region during pollen germination and tube growth. Taken together, these results suggest that CSLD1 and CSLD4 play important roles in pollen tube growth, possibly by participating in pollen tube cellulose synthesis.
A regular framework of cellulose microfibrils is required for the deposition of other cell wall polymers such as callose and pectins. Disruption of this framework can result in thickening and irregularities in the cell wall (Persson et al., 2007). Studies have shown that reduction or lack of cellulose in pollen tubes leads to severe defects in tube growth (Anderson et al., 2002; Aouar et al., 2010). Here it is shown that growth of csld1 or csld4 mutant pollen tubes is severely defective; moreover, the deposition of cellulose in the mutant pollen tubes is significantly reduced and the deposition of other cell wall polymers such as callose is irregular. This strongly implies that CSLD1 and CSLD4 may be involved in cellulose synthesis during pollen tube growth. This hypothesis is also supported by the following three observations.
First, both CSLD1 and CSLD4 encode putative CSLD proteins that, among the CSLs, share the highest sequence similarity to CESAs. Together with CSLFs, CSLDs and CESAs are closely phylogenetically related to each other and form a special category within the cellulose synthase superfamily (Yin et al., 2009). CESAs are β-1,4-d-glucan (cellulose) synthases and CSLFs are β-(1–3,1–4)-d-glucan (MLG) synthases (Yin et al., 2009). Accordingly, CSLDs have a high potential to be β-linked glucan synthases functioning as β-1,4-d-glucan synthases or β-(1–3,1–4)-d-glucan synthases.
Secondly, CSLD1 and CSLD4 are expressed abundantly in mature pollen and pollen tubes. To date, most CSLD genes have been found to be highly expressed in tip growing cells, while CESA genes are expressed at much lower levels in these cells. For example, tobacco pollen tubes express CSLD genes but do not express detectable CESA genes (Doblin et al., 2001). In Arabidopsis, published microarray analyses (http://www.arabidopsis.org) and the results of the present study show that expression of CSLD1 and CSLD4 cannot be detected until the tricellular stage and that their expression increases significantly during pollen maturation and pollen tube growth. In contrast, in mature pollen and pollen tubes, the expression levels of all CESA genes are much lower than those of CSLD1 and CSLD4 (Honys and Twell, 2003, 2004; Qin et al., 2009; this work, see Supplementary Data File S2 at JXB online). In the moss Physcomitrella patens, CSLD family members are most highly represented among P. patens CESA superfamily expressed sequence tags (ESTs), especially in cultures containing only protonemata that are undergoing tip growth (Roberts and Bushoven, 2007). These results indicate that CSLD genes are predominantly expressed in tip growing cells, in which CESA genes are present at low levels. Moreover, the CESA and CSL genes are reported to have distinct spatiotemporal expression patterns and may have distinct functions (Hamann et al., 2004). Therefore, CSLDs may function as special cellulose synthases for tip growing cells.
Thirdly, the location of CSLD1 and CSLD4 within cells is consistent with their having a role in cellulose deposition in pollen tubes. Previous studies used transient expression of CSLD N-terminal fluorescently tagged fusion proteins in tobacco leaves (Bernal et al., 2007, 2008; Li et al., 2009) or proteomic analysis of Arabidopsis cellular compartments (Dunkley et al., 2006) have shown that CSLDs are located in the Golgi apparatus. In this study, the recombinant pCSLD4:GFP-CSLD1/4 constructs could successfully complement the mutant phenotype, indicating that GFP–CSLD1/4 fusion proteins were correctly targeted to the appropriate intracellular locations. The results showed that CSLD1/4 move from the Golgi apparatus to the plasma membrane by vesicle transport. The CSLD protein has also been identified in the plasma membrane proteome in rice (Natera et al., 2008). The plasma membrane localization of CSLD1/4 is consistent with the known location of cellulose biosynthesis within the cell (Taylor, 2008) and with the phenotype of the mutant pollen tubes which exhibit defective pollen tube growth and reduction of cellulose deposition in the pollen tube wall (this study). The fusion proteins were also detected at the plasma membrane adjacent to the pollen tube plugs, where cellulose is also deposited (Ferguson et al., 1998).
In addition, the localization pattern observed for CLSD1/4 in pollen tubes is similar to that of CESAs in somatic cells. Previous electron microscopic and proteomic analysis studies have shown that CESAs are located in both the plasma membrane and the Golgi apparatus (Haigler and Brown, 1986; Kimura et al., 1999; Dunkley et al., 2006; Natera et al., 2008). Recent observations also show that fluorescent-tagged CESA3 and CESA6 move from the Golgi apparatus to the plasma membrane via vesicle exocytosis (Paredez et al., 2006; Crowell et al., 2009). Thus, the localization and movement of CESAs are consistent with their role as cellulose synthases in somatic cells. At the present time, it is not known if CESAs participate in cellulose biosynthesis of pollen tubes or not. To address the question, N-terminal GFP fusions of Arabidopsis CESA3 and CESA8 were expressed under the control of the CSLD4 promoter in pollen tubes. The results showed that expression of CESA3 and CESA8 in pollen grains and pollen tubes could not complement the csld1 and csld4 mutants, implying that the CESAs could not replace the functions of CLSD1 and CLSD4 in pollen tubes. In addition, GFP–CESA3 and GFP–CESA8 expressed in pollen tubes are located in the Golgi apparatus, but not in the vesicles or plasma membrane of the pollen tube tip (Supplementary Fig. S10 at JXB online). Based on immunofluorescence microscopy data, it was recently reported that CESA proteins localize to the plasma membrane at the tip of Nicotiana alata pollen tubes (Cai et al., 2011). However, the CESA antibody used in this study was generated against a peptide sequence that has high similarity (with identities of 85%) to NaCSLD1 (AAK49455.1) identified in N. alata pollen tubes (Cai et al., 2011). Beside this, CESA genes are not expressed detectably in N. alata pollen tubes, while NaCSLD1 is highly expressed (Doblin et al., 2001). These CESA localization data can therefore not rule out the possibility that CSLD proteins are located in the plasma membrane of the pollen tube tip and CESAs are not. Taken together, the present data imply that CSLDs may be involved in the synthesis of cellulose at the tip region of the pollen tube and that CESAs may not be involved in this process. Thus, CSLD1 and CSLD4 may be specifically involved in biosynthesis of cellulose in growing pollen tubes. Nevertheless, the actual mechanisms that control cellulose synthesis in pollen tubes and somatic cells remain unknown. Further study is required to address this question.
CESAs from land plants and some algae assemble into hexagonal arrangements known as ‘rosettes’ or terminal complexes (TCs) (Tsekos, 1999; Roberts and Roberts, 2007; Taylor, 2008). This raises the interesting question of whether CSLD1 and CSLD4 also function in a complex. Indeed, TCs have been described in many tip growing cells including pollen tubes (Wada and Staehelin, 1981; Emons, 1985; Reiss et al., 1985; Tsekos and Reiss, 1994). In most tip growing cells, the density of TCs is high in the tip region and decreases toward the base of the cell, a distribution that is consistent with the polar localization of CSLD1 and CSLD4 in pollen tubes observed in this study. In addition, although CSLD1 and CSLD4 have similar expression patterns, csld1 and csld4 single mutants, as well as the csld1 csld4 double mutant, have similar phenotypes even at lower temperature. The phenotype of the csld1 mutant could not be complemented by the pCSLD4:GFP-CSLD4 construct that could complement the phenotype of the csld4 mutants (Table 4). These results suggest that CSLD1 and CSLD4 are not functionally redundant and that they probably function as a complex in cellulose synthesis. The N-terminal portions of CESAs contain Zn-binding domains that are proposed to serve in CESA–CESA interactions (Kurek et al., 2002). CSLD4 and most of the other CSLDs have similar Zn-binding domains near the N-terminus, but CSLD1 does not have the domain (Supplementary Fig. S9 at JXB online) (Doblin et al., 2001). Moreover, thus far, attempts to address this question have failed to achieve any conclusive results. Whether CSLD1 and CSLD4 assemble into TCs remains unknown, representing an interesting question to be addressed further.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. TEM observation of mutant pollen grains before germination.
Figure S2. Generation of csld1 csld4 double mutants.
Figure S3. In vitro germination of quartets from mutant and wild-type plants at different temperatures.
Figure S4. Expression patterns of CSLD1 and CSLD4.
Figure S5. Subcellular localization of CSLD1 in Arabidopsis pollen grains and pollen tubes.
Figure S6. GFP–CSLD1 and GFP–CSLD4 are located at the plasma membrane of the pollen tube tip.
Figure S7. Calcofluor staining of wild-type and mutant pollen tubes.
Figure S8. Callose distribution in wild-type and mutant pollen tubes.
Figure S9. Homology of the predicted amino acid sequences of the CSLD and CESA proteins.
Figure S10. Subcellular localization of CESA3 and CESA8 in Arabidopsis pollen tubes.
Table S1. Primers used for cloning and PCR assays.
Table S2. In vitro germination of quartets from mutants and wild-type plants.
Videos S1. and S2. GFP–CSLD1 (Video S1) and GFP–CSLD4 (Video S2) move in pollen tubes.
Video S3. FRAP analysis of GFP–CSLD4 in the pollen tube tip of growing pollen tubes.
Data File S1. Genetic analysis of CSLD4 transgenic csld4 mutant lines.
Data File S2. Expression levels of CESA and CSLD genes during pollen development.
Supplementary Material
Acknowledgments
We thank Dr V. Sundaresan, Dr Wei-Cai Yang, and Ms Li-Fen Xie for their kind help with the mutant screens. We also thank Dr Youqun Wang for assistance with TEM analysis, and undergraduate students Yi Wang and Jilong Wang for their assistance with genetic analysis and molecular cloning. This work was supported by research grants from the Natural Science Foundation of China (grant nos 30770133, J0730639); the Ministry of Sciences and Technology (grant nos 2007CB947603, 2007CB108700); and the Ministry of Education, PR China (grant no. B06003).
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Alexander MP. Differential staining of aborted and non-aborted pollen. Stain Technology. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C. Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiology. 2009;152:787–796. doi: 10.1104/pp.109.150128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Barnes WS, Bedinger P. 2,6-Dichlorobenzonitrile, a cellulose biosynthesis inhibitor, affects morphology and structural integrity of petunia and lily pollen tubes. Journal of Plant Physiology. 2002;159:61–67. [Google Scholar]
- Aouar L, Chebli Y, Geitmann A. Morphogenesis of complex plant cell shapes: the mechanical role of crystalline cellulose in growing pollen tubes. Sexual Plant Reproduction. 2010;23:15–27. doi: 10.1007/s00497-009-0110-7. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Jensen JK, Harholt J, et al. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. The Plant Journal. 2007;52:791–802. doi: 10.1111/j.1365-313X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sorensen I, Blancaflor EB, Scheller HV, Willats WG. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiology. 2008;148:1238–1253. doi: 10.1104/pp.108.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Cai G, Faleri C, Del Casino C, Emons AMC, Cresti M. Distribution of callose synthase, cellulose synthase, and sucrose synthase in tobacco pollen tube is controlled in dissimilar ways by actin filaments and microtubules. Plant Physiology. 2011;155:1169–1190. doi: 10.1104/pp.110.171371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proceedings of the National Academy of Sciences, USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Hofte H, Vernhettes S. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. The Plant Cell. 2009;21:1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardelle F, Lehner A, Ramdani Y, Bardor M, Lerouge P, Driouich A, Mollet JC. Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiology. 2010;153:1563–1576. doi: 10.1104/pp.110.158881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wang W, Li WQ, Xia C, Liao HZ, Zhang XQ, Ye D. MALE GAMETOPHYTE DEFECTIVE 2, encoding a sialyltransferase-like protein, is required for normal pollen germination and pollen tube growth in Arabidopsis. Journal of Integrative Plant Biology. 2010;52:829–843. doi: 10.1111/j.1744-7909.2010.00963.x. [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, et al. Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM. Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiology. 2001;125:2040–2052. doi: 10.1104/pp.125.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2009;106:5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DP. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. The Plant Journal. 2005;42:315–328. doi: 10.1111/j.1365-313X.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, et al. Mapping the Arabidopsis organelle proteome. Proceedings of the National Academy of Sciences, USA. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D. Pollen and stigma structure and function: the role of diversity in pollination. The Plant Cell. 2004;16 Suppl:S84–S97. doi: 10.1105/tpc.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emons AMC. Plasma-membrane rosettes in root hairs of Equisetum hyemale. Planta. 1985;163:350–359. doi: 10.1007/BF00395143. [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes and Development. 2001;15:79–89. doi: 10.1101/gad.188801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C, Teeri TT, Siika-aho M, Read SM, Bacic A. Location of cellulose and callose in pollen tubes and grains of Nicotiana tabacum. Planta. 1998;206:452–460. [Google Scholar]
- Geitmann A, Steer M. The architecture and properties of the pollen tube cell wall. In: Malhó R, editor. The pollen tube. Plant Cell Monographs. Vol. 3. Berlin: Springer Verlag; 2006. pp. 177–200. [Google Scholar]
- Haigler CH, Brown RM. Transport of rosettes from the golgi apparatus to the plasma membrane in isolated mesophyll cells of Zinnia elegans during differentiation to tracheary elements in suspension culture. Protoplasma. 1986;134:111–120. [Google Scholar]
- Hamann T, Osborne E, Youngs HL, Misson J, Nussaume L, Somerville C. Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose. 2004;11:279–286. [Google Scholar]
- Hoch HC, Galvani CD, Szarowski DH, Turner JN. Two new fluorescent dyes applicable for visualization of fungal cell walls. Mycologia. 2005;97:580–588. doi: 10.3852/mycologia.97.3.580. [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiology. 2003;132:640–652. doi: 10.1104/pp.103.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Schneitz K, Pruitt RE. Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. The Plant Cell. 1995;7:57–64. doi: 10.1105/tpc.7.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia DJ, Cao X, Wang W, Tan XY, Zhang XQ, Chen LQ, Ye D. GNOM-LIKE 2, encoding an adenosine diphosphate-ribosylation factor-guanine nucleotide exchange factor protein homologous to GNOM and GNL1, is essential for pollen germination in Arabidopsis. Journal of Integrative Plant Biology. 2009;51:762–773. doi: 10.1111/j.1744-7909.2009.00858.x. [DOI] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. The Plant Cell. 2005;17:584–596. doi: 10.1105/tpc.104.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Preuss D. Plotting a course: multiple signals guide pollen tubes to their targets. Developmental Cell. 2002;2:273–281. doi: 10.1016/s1534-5807(02)00130-2. [DOI] [PubMed] [Google Scholar]
- Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han CD. OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiology. 2007;143:1220–1230. doi: 10.1104/pp.106.091546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM., Jr. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. The Plant Cell. 1999;11:2075–2086. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proceedings of the National Academy of Sciences, USA. 2002;99:11109–11114. doi: 10.1073/pnas.162077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancelle SA, Hepler PK. Ultrastructure of freeze-substituted pollen tubes of Lilium longiflorum. Protoplasma. 1992;167:215–230. [Google Scholar]
- Lee YJ, Szumlanski A, Nielsen E, Yang Z. Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. Journal of Cell Biology. 2008;181:1155–1168. doi: 10.1083/jcb.200801086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon KA, Lord EM. In vivo pollen tube cell of Arabidopsis thaliana I. Tube cell cytoplasm and wall. Protoplasma. 2000;214:45–56. [Google Scholar]
- Li H, Bacic A, Read SM. Role of a callose synthase zymogen in regulating wall deposition in pollen tubes of Nicotiana alata Link et Otto. Planta. 1999a;208:528–538. [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. The Plant Cell. 1999b;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. The Plant Journal. 2009;60:1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x. [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proceedings of the National Academy of Sciences, USA. 2005;102:2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Lord EM, Russell SD. The mechanisms of pollination and fertilization in plants. Annual Review of Cell and Developmental Biology. 2002;18:81–105. doi: 10.1146/annurev.cellbio.18.012502.083438. [DOI] [PubMed] [Google Scholar]
- McCormick S. Control of male gametophyte development. The Plant Cell. 2004;16 Suppl:S142–S153. doi: 10.1105/tpc.016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscatelli A, Idilli AI. Pollen tube growth: a delicate equilibrium between secretory and endocytic pathways. Journal of Integrative Plant Biology. 2009;51:727–739. doi: 10.1111/j.1744-7909.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Mutwil M, Debolt S, Persson S. Cellulose synthesis: a complex complex. Current Opinion in Plant Biology. 2008;11:252–257. doi: 10.1016/j.pbi.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Natera SH, Ford KL, Cassin AM, Patterson JH, Newbigin EJ, Bacic A. Analysis of the Oryza sativa plasma membrane proteome using combined protein and peptide fractionation approaches in conjunction with mass spectrometry. Journal of Proteome Research. 2008;7:1159–1187. doi: 10.1021/pr070255c. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biology. 2005;5:22. doi: 10.1186/1471-2229-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2007;104:15566–15571. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264:1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genetics. 2009;5:e1000621. doi: 10.1371/journal.pgen.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss HD, Herth W, Schnepf E. Plasma-membrane ‘rosettes’ are present in the lily pollen tube. Naturwissenschaften. 1985;72:276. doi: 10.1007/BF00448693. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Somerville CR. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. The Plant Journal. 1998;15:79–88. doi: 10.1046/j.1365-313x.1998.00183.x. [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiology. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Bushoven JT. The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Molecular Biology. 2007;63:207–219. doi: 10.1007/s11103-006-9083-1. [DOI] [PubMed] [Google Scholar]
- Roberts AW, Roberts E. Evolution of the cellulose synthase (CesA) gene family: insights from green algae and seedless plants. In: Brown RM, Saxena IM, editors. Cellulose: molecular and structural biology. Dordrecht: Springer; 2007. pp. 17–34. [Google Scholar]
- Sauter M, Seagull RW, Kende H. Internodal elongation and orientation of cellulose microfibrils and microtubules in deepwater rice. Planta. 1993;190:354–362. [Google Scholar]
- Schlüpmann H, Bacic A, Read SM. Uridine diphosphate glucose metabolism and callose synthesis in cultured pollen tubes of Nicotiana alata Link et Otto. Plant Physiology. 1994;105:659–670. doi: 10.1104/pp.105.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabídopsis. The Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Yang CY, Liu J, Yang WC. RPA, a class II ARFGAP protein, activates ARF1 and U5 and plays a role in root hair development in Arabidopsis. Plant Physiology. 2006;141:966–976. doi: 10.1104/pp.106.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer MW, Steer JM. Pollen tube tip growth. New Phytologist. 1989;111:323–358. doi: 10.1111/j.1469-8137.1989.tb00697.x. [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes and Development. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Szumlanski AL, Nielsen E. The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. The Plant Cell. 2009;21:526–544. doi: 10.1105/tpc.108.060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan XY, Liu XL, Wang W, Jia DJ, Chen LQ, Zhang XQ, Ye D. Mutations in the Arabidopsis nuclear-encoded mitochondrial phage-type RNA polymerase gene RPOTm led to defects in pollen tube growth, female gametogenesis and embryogenesis. Plant and Cell Physiology. 2010;51:635–649. doi: 10.1093/pcp/pcq029. [DOI] [PubMed] [Google Scholar]
- Taylor NG. Cellulose biosynthesis and deposition in higher plants. New Phytologist. 2008;178:239–252. doi: 10.1111/j.1469-8137.2008.02385.x. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences, USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsekos I. The sites of cellulose synthesis in algae: diversity and evolution of cellulose-synthesizing enzyme complexes. Journal of Phycology. 1999;35:635–655. [Google Scholar]
- Tsekos I, Reiss HD. Tip cell growth and the frequency and distribution of cellulose microfibril-synthesizing complexes in the plasma membrane of apical shoot cells of the red alga Porphyra yezoensis. Journal of Phycology. 1994;30:300–310. [Google Scholar]
- Wada M, Staehelin LA. Freeze-fracture observations on the plasma membrane, the cell wall and the cuticle of growing protonemata of Adiantum capillus-veneris L. Planta. 1981;151:462–468. doi: 10.1007/BF00386540. [DOI] [PubMed] [Google Scholar]
- Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M. AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiology. 2001;126:575–586. doi: 10.1104/pp.126.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D. Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. The Plant Cell. 2003;15:2792–2804. doi: 10.1105/tpc.016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes and Development. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Huang J, Xu Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biology. 2009;9:99. doi: 10.1186/1471-2229-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. Pollen–stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development. 1999;126:5431–5440. doi: 10.1242/dev.126.23.5431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.