Abstract
Stomatal movements require massive changes in guard cell osmotic content, and both stomatal opening and stomatal closure have been shown to be energy-requiring processes. A possible role for glycolysis in contributing to the energetic, reducing requirements, or signalling processes regulating stomatal movements has not been investigated previously. Glycolysis, oxidization of glucose to pyruvate, is a central metabolic pathway and yields a net gain of 2 ATP and 2 NADH. 2,3-biphosphoglycerate-independent phosphoglycerate mutase (iPGAM) is a key enzymatic activity in glycolysis and catalyses the reversible interconversion of 3-phosphoglycerate to 2-phosphoglycerate. To investigate functions of iPGAMs and glycolysis in stomatal function and plant growth, Arabidopsis insertional mutants in At1g09780 and At3g08590, both of which have been annotated as iPGAMs on the basis of sequence homology, were identified and characterized. While single mutants were indistinguishable from the wild type in all plant phenotypes assayed, double mutants had no detectable iPGAM activity and showed defects in blue light-, abscisic acid-, and low CO2-regulated stomatal movements. Vegetative plant growth was severely impaired in the double mutants and pollen was not produced. The data demonstrate that iPGAMs and glycolytic activity are critical for guard cell function and fertility in Arabidopsis.
Keywords: Abscisic acid, glycolysis, guard cell, phosphoglycerate mutase, stomata
Introduction
During stomatal movements, massive changes in guard cell solute content occur, and the bioenergetic requirements of this process have been estimated (Assmann and Zeiger, 1987). Studies using pharmacological inhibitors have demonstrated an energetic requirement for both stomatal opening and stomatal closure (Weyers et al., 1982; Schwartz and Zeiger, 1984), and have implicated both photophosphorylation and oxidative phosphorylation as sources of energy for stomatal movements, with the relative contribution from these two sources dependent on environmental conditions, particularly light quality and quantity.
Several decades ago, oxidative phosphorylation by isolated guard cell protoplasts (Shimazaki et al., 1983) and oxygen evolution and photophosphorylation by isolated guard cell protoplasts and chloroplasts (Shimazaki et al., 1982; Shimazaki and Zeiger, 1985; Wu and Assmann, 1993) were quantified. More recently, the first characterizations of guard cell proteomes have been published (Zhao et al., 2008; Zhu et al., 2009). In Arabidopsis, gene ontology (GO) analysis of the 1734 guard cell proteins identified by a combination of gel-free and gel-based methods (Zhao et al., 2008) indicated that a large proportion of these proteins are localized in the chloroplast (Fig. 1). Further TopGO analysis (Alexa et al., 2006) suggested that proteins localized in chloroplast thylakoid membranes and mitochondria are enriched in this proteome (Fig. 2A). In addition, all the enzymatic steps of glycolysis are represented by one or more proteins in the Arabidopsis guard cell proteome (Fig. 2B); in fact, from TopGO analysis, the biological process ‘glycolysis’ ranked third in the guard cell proteome but 14th in the leaf proteome (Zhao et al., 2008). It was therefore hypothesized that glycolysis might also contribute significantly to the energy and reducing equivalents required for stomatal movement, or provide a source of metabolites (Voll et al., 2009; Munoz-Bertomeu et al., 2011) that regulate this process.
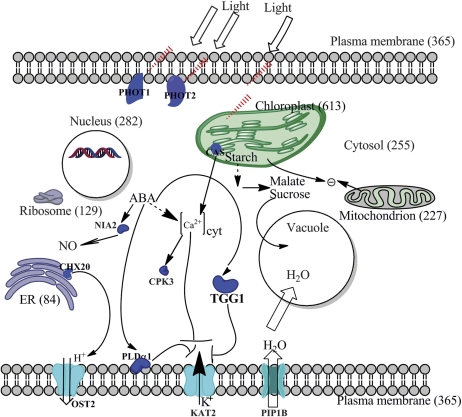
Fig. 1.
Proteins identified in guard cells are localized in all known subcellular localizations. Numbers in parentheses are the numbers of proteins in each cellular compartment identified in the Arabidopsis guard cell proteome of Zhao et al. (2008, 2010). PHOT1 (At3g45780, blue light photoreceptor), PHOT2 (At5g58140, blue light photoreceptor), CAS (At5g23060, calcium-sensing receptor), CHX20 (At3g53720, cation/H+ exchanger 20), NIA2 (At1g37130, nitrogen reductase 2), CPK3 (At4g23650, calcium-dependent protein kinase 3), OST2 (At2g18960, open stomata 2), KAT2 (At4g18290, potassium channel), and PLDα1 (At3g15730, phospholipase D alpha 1) are proteins identified in that study (Zhao et al., 2008, 2010) which are known to be involved in light/ABA signalling in guard cells according to previous studies. TGG1 (At5g26000, myrosinase) was identified to be the most abundant protein in guard cells by mass spectrometry methods (Zhao et al., 2008). OST2 is a H+ ATPase, KAT2 is a K+ channel, and PIP1B is an aquaporin. (This figure is available in colour at JXB online.)
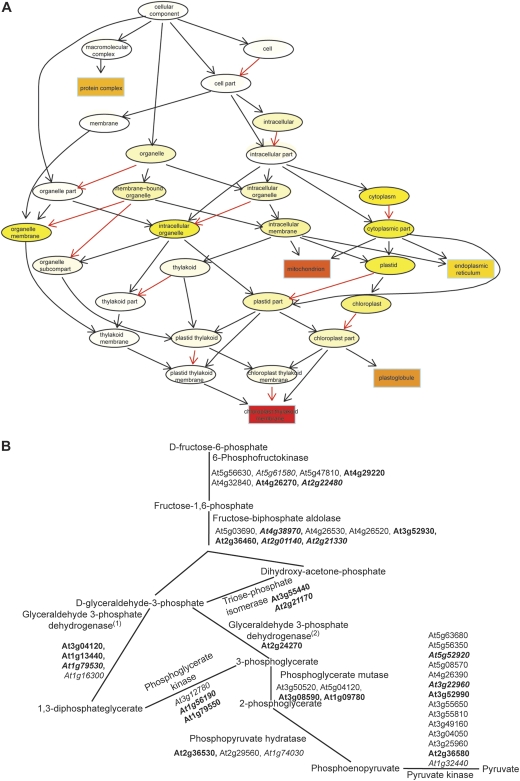
Fig. 2.
Proteins involved in energy provision are enriched in the identified guard cell proteome. (A) Proteins localized in chloroplast thylakoid membranes and mitochondria are enriched in the guard cell proteome. This figure contains the top five GO terms found using the TopGO package (Alexa et al., 2006) for ‘cellular component’. Dark red (P=1E-30), light yellow (P=1E-20), white (non-significant). A black arrow indicates ‘is a relationship’, a red arrow indicates ‘is part of a relationship’. (B) Identified guard cell proteins can cover all steps in glycolysis. Glycolysis proteins downloaded from KEGG (http:www.genome.jp) are depicted in B. Proteins in bold were detected in the guard cell proteome. Proteins in italics are predicted to have chloroplast transit peptides by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). (1) and (2) indicate phosphorylating and non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase, respectively.
Glycolysis hydrolyses glucose into two three-carbon sugars which are then further oxidized and converted into two molecules of pyruvate. In plants, glycolysis occurs in parallel in the cytosol and plastids (Plaxton, 1996). Phosphoglycerate mutase (PGAM) is an important enzyme in the pathway of glycolysis and catalyses the interconversion of the phosphate group between the C-3 carbon of 3-phosphoglycerate (3-PGA) and the C-2 carbon of 2-phosphoglycerate (2-PGA). PGAMs are divided into two evolutionarily unrelated groups based on whether they require 2,3-biphosphoglycerate as a cofactor: cofactor-dependent PGAMs (dPGAMs) and cofactor-independent PGAM (iPGAMs). The dPGAMs are commonly present in vertebrates, certain fungi, and bacteria, while the iPGAMs are found in higher plants, some invertebrates, fungi, and bacteria (Jedrzejas, 2000).
Three genes, At1g09780, At1g22170, and At3g08590, are annotated as PGAMs in TAIR, and four genes, At1g09780, At3g50520, At3g08590, and At5g04120, are annotated as PGAMs in KEGG. Five genes, At1g58280, At2g17280, At3g50520, At5g04120, and At5g64460, are reported by Mazarei et al. (2003) to contain a dPGAM/bPGAM (cofactor-dependent PGAM/bisphosphoglycerate mutase) ‘phosphoglyceromutase family’ element. The protein encoded by At2g17280 was shown to have the highest sequence similarity to dPGAMs from other organisms (Mazarei et al., 2003). Recently, At1g22170 was also annotated as a dPGAM by Stein et al. (2010) on the basis of similarity of sequence alignment and electrostatic potentials, and this enzyme was reported, among all the enzymes of glycolysis, to show the lowest variation in the electrostatic potential between species (Stein et al., 2010). Andriotis et al. (2010) reported that six genes, At1g09780, At1g22170, At1g78050, At3g08590, At3g30840, and At4g09520, encode putative PGAM proteins. Two of these, At1g22170 and At1g78050, were categorized by Andriotis and colleagues as dPGAMs, and At1g22170 was further confirmed to be localized to plastids by transient transformation. Mutants lacking the At1g22170 transcript have no significant phenotypes compared with control plants (Andriotis et al., 2010).
Only two genes, At1g09780 and At3g08590, are annotated as iPGAMs in both TAIR and KEGG, and the encoded proteins are reported to be highly similar to iPGAMs from other higher plants (Andriotis et al., 2010). iPGAM1 (At1g09780) and iPGAM2 (At3g08590) are 90% identical, and both of these proteins were identified in the guard cell proteomes (Zhao et al., 2008). The small size of the iPGAM gene family facilitates functional characterization by mutant analysis. Accordingly, in the present study a biochemical and phenotypic characterization of homozygous ipgam1, ipgam2, and ipgam1 ipgam2 double mutants was performed. All ipgam single mutants have comparable iPGAM enzyme activity and show similar phenotypes to wild-type Col. However, the double mutants have no detectable iPGAM enzyme activity and show severe impairment in stomatal movements and in vegetative, and reproductive growth, suggesting that these enzymes of primary metabolism (glycolysis) are key components of energy and/or metabolite provision for multiple pathways.
Materials and methods
Plant material and growth conditions
Arabidopsis seeds were first plated on 1/2 MS plates and then transplanted into soil as described in Zhao et al. (2008). Plants were grown in growth chambers under short day conditions (8/16 h light/dark cycles, 19 °C, 110 μmol m−2 s−1 of light with ∼75% relative humidity).
Mutant identification
Seeds of T-DNA insertion lines [SALK_003321 (ipgam1-1), SALK_029822 (ipgam1-2), SALK_016231 (ipgam2-1), and SALK_002280 (ipgam2-2)] were acquired from the Arabidopsis Biological Resource Center (ABRC). T-DNA insertion sites were confirmed by sequencing genomic PCR products, obtained using gene-specific primers plus T-DNA border-specific primers (Supplementary Table S1 available at JXB online).
iPGAM enzymatic assay
Healthy leaves from Col, single and double ipgam mutant plants were excised and frozen in liquid nitrogen. Total proteins were extracted and the iPGAM enzyme assay was performed according to Bourgis et al. (2005). Chemicals were from Sigma.
Stomatal aperture assays
For blue light treatment, excised leaves were placed in opening buffer and subjected to darkness for 2 h to close the stomata. Stomatal apertures were measured at this time to obtain a baseline, and the remaining leaves were transferred to blue light (10 μmol m−2 s−1) for an additional 3 h. For low CO2 treatments, 200 ml of opening solution (10 mM KCl, 7.5 mM IDA, 10 mM MES, pH 6.15 with KOH) in a 600 ml beaker was pre-bubbled overnight with CO2-free air generated by passing air over the soda lime cartridge of a LICOR 6400 photosynthesis system; this scrubs CO2 from the air. The flow rate of the CO2-free air was set at 500 μmol s−1 overnight and then reduced to 200 μmol s−1 upon addition of leaves to the buffer. Before low CO2 treatment, all leaves were put in opening solution without bubbling under darkness for 2 h. Then control leaves were left in the untreated opening solution and treatment leaves were transferred to opening solution bubbled with low CO2 air. Stomatal apertures were measured after an additional 2.5 h. Both control and treated leaves were put in darkness until measurement. For abscisic acid (ABA) inhibition of stomatal opening, the dishes containing excised leaves were placed in darkness for 2 h to promote stomatal closure. For ABA to promote stomatal closure, dishes were placed under light (200 μmol m−2 s−1) for 2 h to induce stomatal opening. A 5 μl aliquot of 50 mM ABA (A.G. Scientific) (50 μM final concentration) or 100% ethanol (solvent control for ABA) was then added to each well for treatment or control, respectively, and leaves were further left under light (200 μmol m−2 s−1) for a further 2 h for both treatment and solvent controls.
For all aperture experiments, two leaves were used for each treatment, except for blue light and low CO2 experiments, where one leaf was used so that measurement time was minimized. To avoid the possible low confidence caused by one leaf per replicate, five and eight replicates were performed for blue light and low CO2 experiments. Four replicates were performed for ABA regulation of stomatal aperture experiments. Epidermal peels were prepared and 10 epidermal images were photographed per leaf. At least 50 stomatal apertures were measured per leaf. All stomatal apertures were measured using Image J software.
Results
Forty-five out of the 78 genes involved in photosynthetic carbon reduction, 41 out of the 60 genes of the tricarboxylic acid (TCA) cycle, and 35 out of the 77 Arabidopsis genes involved in photophosphorylation as annotated in KEGG were identified by guard cell proteomic studies (Zhao et al., 2008; Supplementary Tables S1–S3 at JXB online). Only 47 out of the 162 genes involved in oxidative phosphorylation are identified in the guard cell proteome (Supplementary Table S4). Of the 68 glycolytic proteins, 34 (50%) were identified in the Arabidopsis guard cell proteome and together they cover all the steps of glycolysis (Fig. 2B). Glycolysis is catalysed by parallel pathways in the cytosol and plastids (Plaxton, 1996) and glycolytic enzymes from both compartments were identified: 11 of the 34 identified glycolytic proteins in the guard cell proteome are predicted to have chloroplast transit peptides by ChloroP (Emanuelsson et al., 1999) (Fig. 2B). Both iPGAMs were identified in the guard cell proteome: peptides unique to each isoform were identified from both gel-based and gel-free MudPIT methods (Fig. 3A). Protein coverage was ∼32.5% for At1g09780 and 38.2% for At3g08590. Both iPGAMs are predicted to be localized in the cytosol by GO annotation, and At1g09780 is also predicted to be localized in the chloroplast in the GO database even though no chloroplast transit peptide is detected by ChloroP.
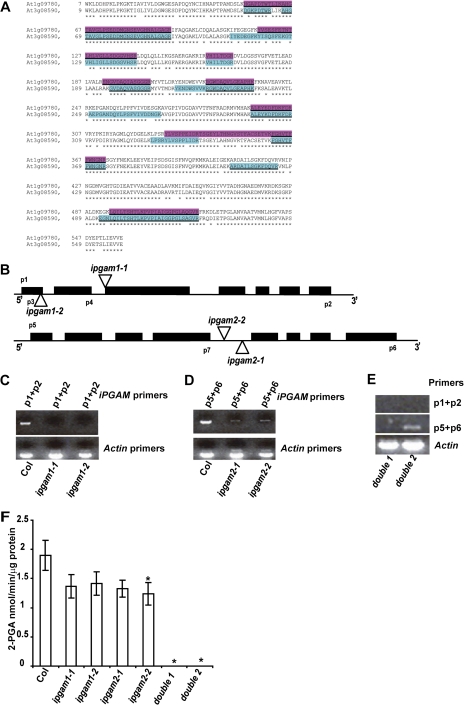
Fig. 3.
Double ipgam1 ipgam2 mutants have no detectable iPGAM enzyme activity. (A) iPGAM1 and iPGAM2 are highly similar to each other. Peptides highlighted were identified by mass spectrometry (MS) methods from At1g09780 and At3g08590, respectively. About 32.5% of amino acids from At1g09780 and 38.2% amino acids from At3g08590 were identified by the MS methods. All peptides identified multiple times or in both biological replicated are underlined. (B) ipgam1-1 and ipgam1-2 have a T-DNA insertion in the third or first exon of iPGAM1, respectively; ipgam2-1 and ipgam2-2 have T-DNA insertions in the fourth intron of iPGAM2. (C) ipgam1 mutants are knock-out mutants as indicated by RT-PCR assays using forward primer p1 and reverse primer p2. (D) ipgam2 mutants are knock-down mutants as indicated by RT-PCR assays using forward primer p5 and reverse primer p6. (E) The full-length transcript of iPGAM1 is absent in double 1 and double 2 mutants (double 1=ipgam1-1 ipgam2-2, double 2=ipgam1-2 ipgam2-1), while the full-length transcript of iPGAM2 is detected at low levels in the double 2 mutant. (F) Double mutants have no detectable iPGAM enzyme activity. Single mutants have somewhat lower iPGAM enzyme activity than Col. n=4, data are mean ±SE. P <0.05 (Student's t-test) was considered significant (*). (This figure is available in colour at JXB online.)
iPGAM enzyme activity is undetectable in ipgam double mutants
To study the roles of iPGAMs and their corresponding metabolic pathways in stomatal movements, ipgam1 and ipgam2 T-DNA insertional mutants were identified and characterized. Two independent T-DNA insertional alleles for each iPGAM gene were identified (Fig. 3B). RT-PCR assay showed that both ipgam1 alleles are null mutants, lacking full-length transcripts (Fig. 3C). The two ipgam2 alleles are knock-down mutants (Fig. 3D): full-length transcripts of iPGAM2 are present at reduced levels in both ipgam2 mutants as compared with Col (Fig. 3D). Two fully independent double mutants (ipgam1-1 ipgam2-2=double 1; ipgam1-2 ipgam2-1=double 2) were obtained by crossing (Fig. 3E).
Enzymatic assays of iPGAM activity (Westram et al., 2002; Bourgis et al., 2005) in all mutants using 5-week-old leaves showed that all the single ipgam mutants have reduced iPGAM enzyme activities as compared with the wild type (Col) but the rates are not significantly different among these single mutants except possibly for ipgam2-2 (P=0.04). Importantly, ipgam double mutants have no detectable iPGAM enzyme activity (Fig. 3F).
Blue light-, low CO2-, and ABA-regulated stomatal movements are defective in ipgam double mutants
Guard cell size and stomatal density weree first compared in Col and ipgam double mutants, and no significant differences were observed in these parameters (data not shown). Blue light induces stomatal opening (Shimazaki et al., 2007), and previous analysis showed that blue light-stimulated stomatal opening was diminished in Arabidopsis plants deficient in phosphoglucomutase, which catalyses the conversion of glucose 1-phosphate to glucose 6-phosphate upstream of glycolysis (Lasceve et al., 1997). There is also evidence from fava bean guard cells for light activation of phosphofructokinase (PFK), which generates fructose 1,6-phosphate, the substrate for glycolysis, or pyrophosphate:fructose 6-phosphate phosphotransferase (PFP; or pyrophosphate-dependent phosphofructokinase) (Hedrich et al., 1985). Accordingly, blue light promotion of stomatal opening was evaluated in the ipgam mutants. Low CO2 induction of stomatal opening under darkness was also studied since stomatal opening under darkness is thought to rely mainly on energy from oxidative phosphorylation (Schwartz and Zeiger, 1984). ABA regulation of stomatal movement is also energy requiring (Weyers et al., 1982; Lasceve et al., 1997) and was also studied.
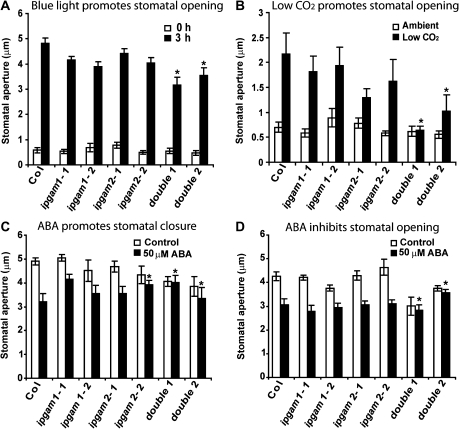
ipgam single mutants had responses similar to Col in most stomatal aperture assays, which could indicate functional redundancy of the two iPGAM proteins. Indeed, both independent double mutants were hyposensitive to blue light (double1 P=0.01, double2 P=0.008) or low CO2 promotion of stomatal opening (double1 P=0.001, double2 P=0.009) (Fig. 4A, B). ABA inhibition of stomatal opening (double1 P=0.002, double2 P=0.008) and ABA promotion of stomatal closure (double1 P=0.002, double2 P=0.006) were both abolished in the double mutants (Fig. 4C, D). The single ipgam2-2 mutant is hyposensitive to ABA promotion of stomatal closure (P=0.003), consistent with its relatively low iPGAM enzyme activities (Fig. 3F).
Fig. 4.
ipgam1 ipgam2 double mutants have multiple stomatal phenotypes. (A) Double mutants are hyposensitive to blue light (10 μmol m−2 s−1) promotion of stomatal opening. (B) Double mutants are hyposensitive to low CO2 promotion of stomatal opening under darkness. (C) Double mutants are hyposensitive to ABA (50 μM) promotion of stomatal closure. (D) Double mutants are hyposensitive to ABA (50 μM) inhibition of stomatal opening. For A–D, n=5, 8, 4, and 4, respectively. P <0.05 (Student's t-test) was considered significant (*).
Plant vegetative and reproductive growth are both defective in ipgam double mutants
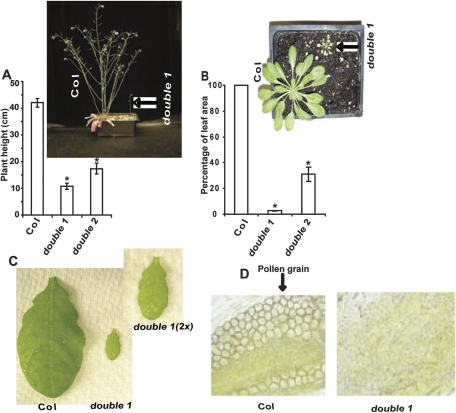
None of the single ipgam mutants showed any evident morphological phenotypes under either long (12 h light) or short day (8 h light) growth conditions (data not shown). However, both the vegetative and the reproductive growth of the double mutants are dramatically reduced (Fig. 5). The mature leaf areas of double 1 and double 2 are only 3% and 30%, respectively, of those from Col, and the height of the tallest bolt of ∼10-week-old double 1 and double 2 plants is only 26% and 41%, respectively, of that of the wild type (Fig. 5A, B). A pale reticulate leaf phenotype was also present in double mutants but not in Col control plants (Fig. 5C).
Fig. 5.
ipgam1 ipgam2 double mutants have vegetative and reproductive phenotypes. (A) Double mutant plants are dramatically shorter than Col plants. (B) Double mutant rosette leaf areas are dramatically reduced compared with Col plants. For A and B, n=10. Data are mean ±SE. (D) Leaves from double ipgam mutants exhibit a pale, slightly reticulate phenotype. (C) No visible pollen grains were found in double mutant anthers. Col and double 1 plants are shown. The same phenotype was present in double 2 (data not shown).
Seeds from double mutants were never obtained. Crosses were therefore attempted between the double mutants and Col using either the double mutants or Col as the pollen donor. Interestingly, seeds were only obtained when Col served as the pollen donor, indicating that the double mutants were female fertile and probably male sterile. Further inspection showed that no visible pollen grains were produced by the double mutants (Fig. 5D). These results led us to conclude that the ipgam double mutants are self-sterile due to failure to produce pollen grains.
Discussion
Plant iPGAMs have been isolated from wheat germ (Leadlay et al., 1977; Smith and Hass, 1985), castor bean (Huang and Dennis, 1995), maize (Grana et al., 1989), and lily (Wang et al., 1996), and share high similarity in amino acid sequence (Grana et al., 1995). However, the regulatory mechanisms of iPGAM in plants are still unclear. It was suggested that iPGAM in lily may have multiple functions, since iPGAM was ubiquitiously detected in all tissues via immunolocalization (Wang et al., 1996). On the basis of this information, transgenic potato plants with reduced iPGAM enzyme activity were generated via antisense technology. Phenotypic analysis showed that the growth of these transgenic potato plants was retarded due to a reduced photosynthetic rate (Westram et al., 2002). However, the mechanism by which photosynthesis is regulated by iPGAM enzyme activity still awaits investigation (Westram et al., 2002).
iPGAMs are important to guard cell function
Both stomatal opening and stomatal closure, including ABA-induced stomatal closure, require energy (Weyers et al., 1982; Schwartz and Zeiger, 1984; Assmann and Zeiger, 1987). The enrichment in the guard cell proteome of proteins localized in the chloroplast thylakoid membrane and in mitochondria (Fig. 2A) is indicative of important roles for both photophosphorylation and oxidative phosphorylation in guard cells (Schwartz and Zeiger, 1984; Vavasseur and Raghavendra, 2005). Previous pharmacological studies using the photosynthetic inhibitor, 3(3,4-dichlorophenyl)-l,l-dimethylurea (DCMU), and the respiratory poison, poassium cyanide (KCN), suggested that stomatal opening under moderate to high intensity photosynthetically active radiation (PAR) mainly relies on photophosphorylation as a source of ATP, while stomatal opening induced by the low intensity blue light-specific response (Kinoshita et al., 2001; Shimazaki et al., 2007) or by low CO2 concentrations under darkness mainly relies on oxidative phosphorylation (Sharkey and Raschke, 1981; Schwartz and Zeiger, 1984).
The in silico analyses (Figs 1, 2; Supplementary Tables S1–S4 at JXB online) suggested an important role for glycolysis in stomatal function. This prediction was assessed and validated by wet-bench analyses. The hyposensitive phenotypes of ipgam mutants to low CO2 induction of stomatal opening under darkness and blue light promotion of stomatal opening (Fig. 4A, B) provide the first genetic evidence that glycolytic enzymes are critical for guard cell function in response to environmental signals. ATP can be generated via glycolysis under darkness (Plaxton, 1996), and glycolysis provides substrates to the TCA cycle and mitochondrial respiration for further production of ATP and reducing power. Studies with the respiratory poison KCN have in particular implicated oxidative phosphorylation in energy provision during stomatal opening in darkness and in stomatal opening mediated specifically by the blue light photoreceptor system (Schwartz and Zeiger, 1984); ATP from photophosphorylation can also contribute to blue light induction of proton pumping under some conditions (Mawson, 1993). In addition, malate2–, an important osmoticum and counter-ion for K+, is produced in guard cells, plausibly through carboxylation and reduction of phosphoenolpyruvate (PEP) derived from glycolysis, under blue light (particularly in the presence of a red light background) (Ogawa et al., 1978). Stomatal aperture under 200 μmol m−2 s−1 white light, where the PAR response of stomata will dominate, is not significantly different between Col and ipgam double mutants (Fig. 4C, D, control). Therefore, a straightforward hypothesis to explain the present results is that glycolysis regulates stomatal movements via energy provision under darkness and via production of ATP, reducing equivalents, and malate2– under blue light.
A more complicated explanation of the guard cell phenotypes would involve an impact of loss of the intact glycolytic pathway on production of other metabolites that have a signalling or regulatory effect. The differential importance of such a metabolite to guard cell blue light, CO2, and ABA signalling could explain why these responses, if calculated on a percentage rather than an absolute aperture basis, are differentially sensitive to the double iPGAM mutation.
As an example of a signalling metabolite that can originate from glycolysis, serine can be synthesized from 3-PGA in plastids (Ho et al., 1999), and this amino acid has been proposed to provide a connection between primary metabolism and ABA signalling (Munoz-Bertomeu et al., 2011). gapcp mutants which lack plastidial glycolytic GAPC exhibit reduced serine content in roots and ABA hyposensitivity; serine supplementation can restore sensitivity to ABA in inhibition of seed germination and seedling growth in these mutants (Munoz-Bertomeu et al., 2011). gapcp mutants also show hyposensitivity to ABA induction of stomatal closure. In potato plants, 3-PGA overaccumulation was reported when iPGAM enzyme activity was reduced by antisense inhibition (Westram et al., 2002). Therefore, an alternative hypothesis for the ABA-hyposensitive phenotype of the Arabidopsis double ipgam mutants is that the resultant high concentrations of 3-PGA in guard cells alter serine homeostasis which further affects the sensitivity of ABA regulation of stomatal movements. However, if and how 3-PGA and/or serine specifically regulates stomatal apertures awaits further investigation.
iPGAMs are important for plant growth and reproduction
In total, seven genes have been annotated as dPGAMs in Arabidopsis on the basis of sequence similarity (At1g22170, At1g58280, At1g78050, At2g17280, At3g50520, At5g04120, and At5g64460) (Mazarei et al., 2003; Andriotis et al., 2010; Stein et al., 2010), and four genes have been annotated as iPGAMs (At1g09780, At3g08590, At3g30840, and At4g09520) (Andriotis et al., 2010). Considering the total number of PGAM genes in Arabidopsis, it was not assured that a double iPGAM knockout would exhibit any phenotypes. Indeed, Arabidopsis plants with null mutation of At1g22170 show no obvious phenotypes (Andriotis et al., 2010). In contrast, the severe growth and reproductive phenotypes of the ipgam double mutants that are observed (Fig. 5) suggest a major role in glycolysis for the two iPGAMs that were targeted. Given that there are 11 total PGAMs in Arabidopsis, however, it is reasonable to conclude that these double mutants also still retain some PGAM activity.
The double ipgam mutants show severely retarded growth and a pale reticulate leaf phenotype (Fig. 5C). In ipgam double mutants, conversion between 3-PGA and 2-PGA is impaired. iPGAM antisense potato plants retaining only 25% iPGAM enzyme activity contain ∼30% more 3-PGA and 40% less PEP, the direct downstream product of 2-PGA, than control plants, and also showed retarded growth (Westram et al., 2002). PEP is produced from 2-PGA in a reaction catalysed by enolase. eno-1 tobacco plants with antisense inhibition of enolase activity exhibit increased concentrations of 3-PGA and 2-PGA, decreased PEP (∼75% less), and a retarded growth phenotype accompanied by a reticulate leaf phenotype (Voll et al., 2009). Interestingly, eno-1 mutants have essentially unchanged pyruvate content and respiratory activity, but show strong reduction in aromatic amino acid content (Voll et al., 2009), consistent with the fact that PEP imported into plastids is the precursor for production of aromatic amino acids and other aromatic compounds via the shikimate pathway. Therefore, an alternative hypothesis is that the severe retardation of plant growth and the reticulate leaf phenotypes in ipgam double mutants are not due to reduced energy provision but rather to altered aromatic acid metabolism. Several reticulate leaf mutants have been characterized recently, including cue1 (Voll et al., 2003), ven3 and ven6 (Molla-Morales et al., 2011), and trp2 (Jing et al., 2009). Analysis of these mutants suggests that mesophyll development is restricted and amino acid biosynthesis is impaired. There is a decreased amount of aromatic amino acids in cue1, a defect of arginine biosynthesis in ven3 and ven6, and a defect in tryptophan biosynthesis in trp2. Whether the leaf phenotypes of ipgam double mutants are due to a disorder in amino acid metabolism awaits further investigation.
The retarded growth phenotype of ipgam double mutants alternatively might be caused by a reduced photosynthetic rate. The pale and reticulate leaf phenotype (Fig. 5C) is suggestive of chlorosis. Both transgenic eno-1 tobacco (Voll et al., 2009) and ipgam antisense potato (Westram et al., 2002) showed a reduced photosynthetic rate. Unfortunately, ipgam double mutants of Arabidopsis are extremely small, precluding assessment of photosynthesis via gas exchange technology.
Pollen development requires high energy provision (Lee and Warmke, 1979; Li et al., 2010). The failure of double ipgam mutants to produce pollen may indicate high energetic requirements for pollen production, consistent with the fact that many other male-sterile mutants are associated with disruptions of genes encoding mitochondrial proteins (Chase, 2007). In transgenic potato plants with reduced iPGAM activity, the content of PEP is decreased (Westram et al., 2002). PEP is the precursor of pyruvate, which can be imported into mitochondria for respiration and energy production. It has been shown in Arabidopsis that reduced pyruvate levels caused by impairment of the cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC-1), which converts glyceraldehyde-3-phosphate to 1,3-bis-phosphoglycerate upstream of 3-PGA, results in impaired oxidative phosphorylation (Rius et al., 2008). Recently, it was also reported that plants with deficient plastidic glyceraldehyde-3-phosphate dehydrogenase, which converts 1,3-PGA to 3-PGA, produce non-viable pollen grains with shrunken and collapsed shapes (Munoz-Bertomeu et al., 2010). These studies suggest that glycolytic enzymes are required for normal pollen development, possibly via energy provision.
In conclusion, it has been shown that transgenic elimination of iPGAM activity has dramatic phenotypic effects on stomatal movement, vegetative biomass production, and reproduction in Arabidopsis. The simplest explanation for these phenotypes is that they result from diminished production of ATP and reducing power from glycolysis and the downstream reactions of the TCA cycle and oxidative phosphorylation. However, given that transgenic reduction of iPGAM in potato also negatively impacts photosynthesis, and that transgenic impairment of the glycolytic enzyme, enolase, results in phenotypes associated with disruption of the shikimic acid pathway, the specific mechanisms underlying the ipgam phenotypes which are observed await characterization of the ipgam metabolome.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer information.
Table S2. Genes involved in photosynthesis in Arabidopsis.
Table S3. Genes involved in oxidative phosphorylation in Arabidopsis.
Table S4. Genes involved in the TCA cycle in Arabidopsis.
Table S5. Genes involved in the Calvin cycle in Arabidopsis.
Acknowledgments
The authors gratefully acknowledge Sarah Nilson for help with setting up of the low CO2 experiments. This study was funded by NSF grants MCB-0618402 and MCB-0817954 to SMA.
References
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Andriotis VM, Kruger NJ, Pike MJ, Smith AM. Plastidial glycolysis in developing Arabidopsis embryos. New Phytologist. 2010;185:649–662. doi: 10.1111/j.1469-8137.2009.03113.x. [DOI] [PubMed] [Google Scholar]
- Assmann SM, Zeiger E. Guard cell bioenergetics. In: Zeiger E, Farquhar G, Cowan I, editors. Stomatal function. Stanford, CA: Stanford Unversity Press; 1987. pp. 163–194. [Google Scholar]
- Bourgis F, Botha FC, Mani S, Hiten FN, Rigden DJ, Verbruggen N. Characterization and functional investigation of an Arabidopsis cDNA encoding a homologue to the d-PGMase superfamily. Journal of Experimental Botany. 2005;56:1129–1142. doi: 10.1093/jxb/eri105. [DOI] [PubMed] [Google Scholar]
- Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends in Genetics. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grana X, Perez de la Ossa P, Broceno C, Stocker M, Garriga J, Puigdomenech P, Climent F. 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase is conserved among different phylogenic kingdoms. Comparative Biochemistry and Physiology. Part B, Biochemistry and Molecular Biology. 1995;112:287–293. doi: 10.1016/0305-0491(95)00076-3. [DOI] [PubMed] [Google Scholar]
- Grana X, Urena J, Ludevid D, Carreras J, Climent F. Purification, characterization and immunological properties of 2,3-bisphosphoglycerate-independent phosphoglycerate mutase from maize (Zea mays) seeds. European Journal of Biochemistry. 1989;186:149–153. doi: 10.1111/j.1432-1033.1989.tb15189.x. [DOI] [PubMed] [Google Scholar]
- Hedrich R, Raschke K, Stitt M. A role for fructose 2,6-bisphosphate in regulating carbohydrate metabolism in guard cells. Plant Physiology. 1985;79:977–982. doi: 10.1104/pp.79.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito K. Plastidic pathway of serine biosynthesis. Molecular cloning and expression of 3-phosphoserine phosphatase from Arabidopsis thaliana. Journal of Biological Chemistry. 1999;274:11007–11012. doi: 10.1074/jbc.274.16.11007. [DOI] [PubMed] [Google Scholar]
- Huang Y, Dennis DT. Histidine residues 139, 363 and 500 are essential for catalytic activity of cofactor-independent phosphoglyceromutase from developing endosperm of the castor plant. European Journal of Biochemistry. 1995;229:395–402. doi: 10.1111/j.1432-1033.1995.tb20480.x. [DOI] [PubMed] [Google Scholar]
- Jedrzejas MJ. Structure, function, and evolution of phosphoglycerate mutases: comparison with fructose-2,6-bisphosphatase, acid phosphatase, and alkaline phosphatase. Progress in Biophysics and Molecular Biology. 2000;73:263–287. doi: 10.1016/s0079-6107(00)00007-9. [DOI] [PubMed] [Google Scholar]
- Jing Y, Cui D, Bao F, Hu Z, Qin Z, Hu Y. Tryptophan deficiency affects organ growth by retarding cell expansion in Arabidopsis. The Plant Journal. 2009;57:511–521. doi: 10.1111/j.1365-313X.2008.03706.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Lasceve G, Leymarie J, Vavasseur A. Alterations in light-induced stomatal opening in a starch-deficient mutant of Arabidopsis thaliana L deficient in chloroplast phosphoglucomutase activity. Plant, Cell and Environment. 1997;20:350–358. [Google Scholar]
- Leadlay PF, Breathnach R, Gatehouse JA, Johnson PE, Knowles JR. Phosphoglycerate mutase from wheat germ: studies with isotopically labeled 3-phospho-D-glycerates showing that the catalyzed reaction is intramolecular. Appendix: phosphoglycerate mutase from wheat germ: isolation, crystallization, and properties. Biochemistry. 1977;16:3045–3053. doi: 10.1021/bi00633a001. [DOI] [PubMed] [Google Scholar]
- Lee S-LJ, Warmke HE. Organelle size and number in fertile and T-cytoplasmic male-sterile corn. American Journal of Botany. 1979;66:141–148. [Google Scholar]
- Li WQ, Zhang XQ, Xia C, Deng Y, Ye D. MALE GAMETOPHYTE DEFECTIVE 1, encoding the FAd subunit of mitochondrial F1F0-ATP synthase, is essential for pollen formation in Arabidopsis thaliana. Plant and Cell Physiology. 2010;51:923–935. doi: 10.1093/pcp/pcq066. [DOI] [PubMed] [Google Scholar]
- Mawson BT. Regulation of blue-light-induced proton pumping by Vicia faba L. guard cell protoplasts: energetic contributions by chloroplastic and mitochondrial activities. Planta. 1993;191:293–301. [Google Scholar]
- Mazarei M, Lennon KA, Puthoff DP, Rodermel SR, Baum TJ. Expression of an Arabidopsis phosphoglycerate mutase homologue is localized to apical meristems, regulated by hormones, and induced by sedentary plant-parasitic nematodes. Plant Molecular Biology. 2003;53:513–530. doi: 10.1023/B:PLAN.0000019062.80459.80. [DOI] [PubMed] [Google Scholar]
- Molla-Morales A, Sarmiento-Manus R, Robles P, Quesada V, Perez-Perez JM, Gonzalez-Bayon R, Hannah MA, Willmitzer L, Ponce MR, Micol JL. Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. The Plant Journal. 2011;65:335–345. doi: 10.1111/j.1365-313X.2010.04425.x. [DOI] [PubMed] [Google Scholar]
- Munoz-Bertomeu J, Bermudez MA, Segura J, Ros R. Arabidopsis plants deficient in plastidial glyceraldehyde-3-phosphate dehydrogenase show alterations in abscisic acid (ABA) signal transduction: interaction between ABA and primary metabolism. Journal of Experimental Botany. 2011;62:1229–1239. doi: 10.1093/jxb/erq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Bertomeu J, Cascales-Minana B, Irles-Segura A, Mateu I, Nunes-Nesi A, Fernie AR, Segura J, Ros R. The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiology. 2010;152:1830–1841. doi: 10.1104/pp.109.150458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Ishikawa H, Shimada K, Shibata K. Synergistic action of red and blue light and action spectra for malate formation in guard cells of Vicia faba L. Planta. 1978;142:61–65. doi: 10.1007/BF00385121. [DOI] [PubMed] [Google Scholar]
- Plaxton WC. The organization and regulation of plant glycolysis. Annual Review of Plant Physiology and Plant Molecular Biololgy. 1996;47:185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF. Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiology. 2008;148:1655–1667. doi: 10.1104/pp.108.128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A, Zeiger E. Metabolic energy for stomatal opening. Roles of photophosphorylation and oxidative phosphorylation. Planta. 1984;161:129–136. doi: 10.1007/BF00395472. [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Raschke K. Effect of light quality on stomatal opening in leaves of Xanthium strumarium L. Plant Physiology. 1981;68:1170–1174. doi: 10.1104/pp.68.5.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki K, Doi M, Assmann SM, Kinoshita T. Light regulation of stomatal movement. Annual Review of Plant Biology. 2007;58:219–247. doi: 10.1146/annurev.arplant.57.032905.105434. [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Gotow K, Kondo N. Photosynthetic properties of guard-cell protoplasts from Vicia faba L. Plant and Cell Physiology. 1982;23:871–879. [Google Scholar]
- Shimazaki K, Gotow K, Sakaki T, Kondo N. High respiratory activity of guard-cell protoplasts from Vicia faba L. Plant and Cell Physiology. 1983;24:1049–1056. [Google Scholar]
- Shimazaki K, Zeiger E. Cyclic and noncyclic photophosphorylation in isolated guard cell chloroplasts from Vicia faba L. Plant Physiology. 1985;78:211–214. doi: 10.1104/pp.78.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, Hass LF. Wheat germ phosphoglycerate mutase: purification, polymorphism, and inhibition. Biochemical and Biophysical Research Communations. 1985;131:743–749. doi: 10.1016/0006-291x(85)91301-4. [DOI] [PubMed] [Google Scholar]
- Stein M, Gabdoulline RR, Wade RC. Cross-species analysis of the glycolytic pathway by comparison of molecular interaction fields. Molecular Biosystems. 2010;6:152–164. doi: 10.1039/b912398a. [DOI] [PubMed] [Google Scholar]
- Vavasseur A, Raghavendra AS. Guard cell metabolism and CO2 sensing. New Phytologist. 2005;165:665–682. doi: 10.1111/j.1469-8137.2004.01276.x. [DOI] [PubMed] [Google Scholar]
- Voll L, Hausler RE, Hecker R, Weber A, Weissenbock G, Fiene G, Waffenschmidt S, Flugge UI. The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. The Plant Journal. 2003;36:301–317. doi: 10.1046/j.1365-313x.2003.01889.x. [DOI] [PubMed] [Google Scholar]
- Voll LM, Hajirezaei MR, Czogalla-Peter C, Lein W, Stitt M, Sonnewald U, Bornke F. Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytologist. 2009;184:607–618. doi: 10.1111/j.1469-8137.2009.02998.x. [DOI] [PubMed] [Google Scholar]
- Wang JL, Walling LL, Jauh GY, Gu YQ, Lord EM. Lily cofactor-independent phosphoglycerate mutase: purification, partial sequencing, and immunolocalization. Planta. 1996;200:343–352. doi: 10.1007/BF00200302. [DOI] [PubMed] [Google Scholar]
- Westram A, Lloyd JR, Roessner U, Riesmeier JW, Kossmann J. Increases of 3-phosphoglyceric acid in potato plants through antisense reduction of cytoplasmic phosphoglycerate mutase impairs photosynthesis and growth, but does not increase starch contents. Plant, Cell and Environment. 2002;25:1133–1143. [Google Scholar]
- Weyers JDB, Paterson NW, Fitzsimons PJ, Dudley JM. Metabolic inhibitors block ABA-induced stomatal closure. Journal of Experimental Botany. 1982;33:1270–1278. [Google Scholar]
- Wu WH, Assmann SM. Photosynthesis by guard-cell chloroplasts of Vicia faba L—effects of factors associated with stomatal movement. Plant and Cell Physiology. 1993;34:1015–1022. [Google Scholar]
- Zhao Z, Stanley BA, Zhang W, Assmann SM. ABA-regulated G protein signaling in Arabidopsis guard cells: a proteomic perspective. Journal of Proteome Research. 2010;9:1637–1647. doi: 10.1021/pr901011h. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang W, Stanley BA, Assmann SM. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. The Plant Cell. 2008;20:3210–3226. doi: 10.1105/tpc.108.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Dai S, McClung S, Yan X, Chen S. Functional differentiation of Brassica napus guard cells and mesophyll cells revealed by comparative proteomics. Molecular and Cellular Proteomics. 2009;8:752–766. doi: 10.1074/mcp.M800343-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.