Abstract
The mitogen-activated protein kinase (MAPK) cascade plays pivotal roles in diverse signalling pathways related to plant development and stress responses. In this study, the cloning and functional characterization of a group-I MAPK gene, PtrMAPK, in Poncirus trifoliata (L.) Raf are reported. PtrMAPK contains 11 highly conserved kinase domains and a phosphorylation motif (TEY), and is localized in the nucleus of transformed onion epidermal cells. The PtrMAPK transcript level was increased by dehydration and cold, but was unaffected by salt. Transgenic overexpression of PtrMAPK in tobacco confers dehydration and drought tolerance. The transgenic plants exhibited better water status, less reactive oxygen species (ROS) generation, and higher levels of antioxidant enzyme activity and metabolites than the wild type. Interestingly, the stress tolerance capacity of the transgenic plants was compromised by inhibitors of antioxidant enzymes. In addition, overexpression of PtrMAPK enhanced the expression of ROS-related and stress-responsive genes under normal or drought conditions. Taken together, these data demonstrate that PtrMAPK acts as a positive regulator in dehydration/drought stress responses by either regulating ROS homeostasis through activation of the cellular antioxidant systems or modulating transcriptional levels of a variety of stress-associated genes.
Keywords: Abiotic stress tolerance, antioxidant system, mitogen-activated protein kinase, Poncirus trifoliata (L.) Raf., reactive oxygen species, stress-responsive gene
Introduction
As sessile organisms, plants are frequently challenged by various harsh environmental cues, among which drought has been shown to be the most devastating one that adversely affects plant growth, development, and crop productivity. On the other hand, during the long process of evolution plants have evolved a set of versatile acclimation and adaptation mechanisms that provide resistance to environmental stresses, ranging from the perception of the stress signal to activation of a series of metabolic, physiological, and biochemical alterations (Umezawa et al., 2006). This process is regulated by an array of highly complex and intricate signalling networks enabling plants to fight against the abiotic stresses. It is now well accepted that protein phosphorylation plays a crucial role in mediating the signal transduction involved in abiotic stress response. Although several families of proteins may orchestrate protein phosphorylation, the mitogen-activated protein kinase (MAPK) cascade acts as part of the major transduction pathway that transfers the extracellular stimuli into an intracellular response (Tena et al., 2001; Ortiz-Masia et al., 2008).
The MAPK cascade is composed of three hierarchically organized kinase modules, MAPK, MAPKK (MAPK kinase), and MAPKKK (MAPKK kinase). They are functionally linked and operate as an important network for amplifying, integrating, and channelling a broad spectrum of signals from the upstream receptors to the downstream cellular effectors, leading to an adaptive stress response at cellular and organismal levels (T Zhang et al., 2006; Nadarajah and Sidek, 2010). MAPK, the terminal module of the MAPK cascade, is activated by its upstream dual specific MAPKK via phosphorylation of conserved threonine (T) and tyrosine (Y) residues in the catalytic subdomain. MAPKK itself is activated via phosphorylation of two serine/threonine residues in a conserved S/T-X3–5-S/T motif by an upstream MAPKKK (Stulemeijer et al., 2007; Zaïdi et al., 2010). After activation, the MAPK module is translocated into the nucleus or cytoplasm to initiate the cellular responses through phosphorylation of downstream proteins (Pedley and Martin, 2005; Fiil et al., 2009; Nadarajah and Sidek, 2010). In this sense, MAPK, the last component of the MAPK cascade, is the major port of signal transduction from upstream to the target.
MAPKs are ubiquitous proteins in eukaryotes and exist in the form of a gene family. For example, the Arabidopsis thaliana genome contains a total of 20 MAPK genes, and 17 MAPK genes have been identified in the rice genome (Rohila and Yang, 2007; Nadarajah and Sidek, 2010), indicating the complexity of the MAPK cascade in the plant kingdom. MAPKs have been demonstrated to take part in a myriad of cellular processes, including growth, differentiation, defence, and cell death (Nakagami et al., 2005; Kosetsu et al., 2010; Nadarajah and Sidek, 2010; Suarez Rodriguez et al., 2010). Since the first plant MAPK-encoding gene was cloned in the 1990s, MAPK genes have been isolated from several plant species to date (Nadarajah and Sidek, 2010; Zaïdi et al., 2010, and references therein). Among these, some MAPK genes involved in drought signal transduction have been identified, such as AtMPK4 and AtMPK6 in Arabidopsis (Ichimura et al., 2000; Nadarajah and Sidek, 2010), and OsMAPK5 and OsMAPK2 in rice (Xiong and Yang, 2003; Rolila and Yang, 2007). Unravelling of these signalling factors offers a valuable approach for engineering drought tolerance.
It has to be pointed out that although MAPK genes have been cloned from diverse plants, current studies give priority to cDNA cloning, analysis of expression, or kinase activity under various conditions, whereas the functions of the isolated MAPK genes have been less well characterized. On the other hand, it is also noticeable that knowledge of the MAPK cascade of fruit crops under abiotic stresses is scarce as compared with other plants, such as Arabidopsis, rice, tobacco, and tomato. It has been suggested that despite the evolutionary conservation of the MAPK signalling pathway in eukaryotes, there might exist certain differences in the composition and function of a specific component in this cascade (Morris, 2001; Zaïdi et al., 2010). Trifoliate orange (Poncirus trifoliata L. Raf) is a widely used rootstock in citrus-producing regions. Nevertheless, susceptibility to drought poses constraints on its use in regions with limited water supply and the occurrence of periodic drought. Since trifoliate orange is polyembryonic by nature, slow progress has been made in the improvement of drought tolerance via traditional cross-hybridization. Accumulating evidence suggests that genetic engineering provides a new tool for improving stress tolerance (Umezawa et al., 2006). As a first step towards creation of trifoliate orange transgenic plants with enhanced stress tolerance, efforts have been made to isolate and functionally characterize a MAPK gene in this plant.
Materials and methods
Plant materials and stress treatments
Uniform and healthy shoots were collected from 8-month-old trifoliate orange seedlings and subjected to stress treatment (dehydration, salt, and cold). For dehydration treatment, the shoots were put onto dry filter papers (90×90 mm) and allowed to dehydrate for 0, 1, 3, and 6 h in an ambient environment. Salt stress was produced by incubating the shoots in 200 mM NaCl solution for 0, 1, 5, 24, 48, and 72 h. For cold stress, the shoots were placed in a growth chamber set at 4 °C for 0, 1, 6, 48, and 72 h. Leaves were independently harvested at the designated time point, immediately frozen in liquid nitrogen, and stored at –80 °C until further use.
Cloning and bioinformatics analysis of PtrMAPK
The sequence of AtMPK3 (At3g45640) was used as a bait for a homology search against the citrus expressed sequence tag (EST) database, HarvEST (http://harvest.ucr.edu). Seven ESTs were obtained, and merged into an 831 bp sequence. Sequence analysis by Open Reading Frame (ORF) Finder showed that the 5'-end was missing. Thus, 5′-RACE (rapid amplification of cDNA ends) was used to amplify the 5′-end sequence. For this purpose, total RNA was extracted from the leaves sampled from the shoots dehydrated for 6 h using TRIZOL reagent (TaKaRa, Dalian, China). A 1 μg aliquot of the total RNA was used to synthesize RACE-Ready 5′-RACE cDNA with coding sequence (CDS) primers provided by the SMART RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) following the manufacturer’s instructions. The cDNA was then used for the 5′-RACE PCR with a gene-specific primer (GSP, 5′-CCACACATCTATTGCAGCAGTGTAGTCAG-3′) designed based on the merged sequence. The PCR product was purified, subcloned into the pMD18-T vector (TaKaRa), and sequenced (UnitedGene, Shanghai, China). The putative 5′-end sequence and the merged sequence were overlapped with DNAStar to generate a cDNA contig. In order to validate the sequence accuracy, reverse transcription-PCR (RT-PCR) was carried out with a pair of primers (GSP1, Table 1), designed according to the contig, covering 72 bp upstream and 139 bp downstream of the deduced ORF. The RT-PCR reaction, in a total volume of 25 μl, consisted of 100 ng cDNA, 1× reaction buffer, 2.5 mM MgCl2, 0.25 mM dNTP, 1 U of Taq DNA polymerase (Fermentas) and 0.5 μM of each primer. The amplification consisted of 35 cycles of 30 s at 94 °C, 50 s at 60 °C, 90 s at 72 °C, and a 10 min extension at 72 °C. The resulting single PCR band of the expected size (∼1339 bp) was subcloned and sequenced. After confirmation of the accuracy of the full-length sequence (PtrMAPK), a homology search was carried out in the NCBI (National Center for Biotechnology Information) using protein BLAST. The retrieved sequences were aligned with PtrMAPK using ClustalW (http://align.genome.jp/), and a phylogenetic tree was constructed by the Neighbor–Joining (NJ) method using the MEGA 4 program.
Table 1.
Primer sequences used for cloning, subcellular localization, vector construction, transgenic confirmation, and expression analysis
| Genes | Primers | Sequences (5′–3′) |
|
| Forward | Reverse | ||
| PtrMAPK | GSP1 | GAGACTCGATAAAAACACAACCAC | AGGTAACCGAAAGATTGTCGGCCACC |
| Actin | Actin | ATTGTAAGCAACTGGGATGATA | AGAGGTGCCTCAGTGAGAAG |
| PtrMAPK | GSP2 | CGTTTGCTCGGTGTTGAATA | CTCTTCGTAACGGTGGAGGA |
| PtrMAPK | GSP3 | ATACCATGGATGGCTGACGTGGCGCAGGTCAA (NcoI site is underlined) | GCCACTAGTTCCTGGATTGAGTGCTAATGCCTCC (SpeI site is underlined) |
| PtrMAPK | GSP4 | ATAGGATCCATGGCTGACGTGGCGCAGGTCAA (BamHI site is underlined) | GCGCTCGAGTTATCCTGGATTGAGTGCTAATGCC (XhoI site is underlined) |
| PtrMAPK | GSP5 | CGGTCGACGGAGAGAGAGTAAAATGGCTGACG (SalI site is underlined) | AAGGTACCGAAAGATTGTCGGCCACCTGCAG (KpnI site is underlined) |
| PtrMAPK | GSP6 | CGTTTGCTCGGTGTTGAATA | CTCTTCGTAACGGTGGAGGA |
| NPTII | NPTII | AGACAATCGGCTGCTCTGAT | TCATTTCGAACCCCAGAGTC |
| NtAPX | CAAATGTAAGAGGAAACTCAGAGGA | CAGCCTTGAGCCTCATGGTACCG | |
| NtCAT | AGGTACCGCTCATTCACACC | AAGCAAGCTTTTGACCCAGA | |
| NtSOD | AGCTACATGACGCCATTTCC | CCCTGTAAAGCAGCACCTTC | |
| NtADC1 | CTTGCTGATTACCGCAATTTATC | TAGGATCAGCAGCCCCCATAGCC | |
| NtSAMDC | CATTCACATTACCCCGGAAG | AGCAACATCAGCATGCAAAG | |
| NtGST | CCCCTAGTTTGCTCCCTTCT | TTCTTAGCTGCCTCCTGCTC | |
| NtPOX2 | CTTGGAACACGACGTTCCTT | TCGCTATCGCCATTCTTTCT | |
| NtDREB3 | GCCGGAATACACAGGAGAAG | CCAATTTGGGAACACTGAGG | |
| NtNCED1 | AAGAATGGCTCCGCAAGTTA | GCCTAGCAATTCCAGAGTGG | |
| NtERD10C | AACGTGGAGGCTACAGATCG | GTTCCTCTTGGGCATGAGTT | |
| NtERD10D | GAGGACACGGCTGTACCAGT | GCGCCACTTCCTCTGTCTT | |
| NtLEA5 | TTGAATCTGGGGTTTTGGTT | GGAAGCATTGACGAGCTAGG | |
| NtUbiquitin | TCCAGGACAAGGAGGGTAT | CATCAACAACAGGCAACCTAG | |
Subcellular localization of PtrMAPK
The complete ORF of PtrMAPK was amplified by PCR using primer GSP3 containing an NcoI or SpeI restriction site. The PCR product, after confirmation by sequencing, was digested with NcoI and SpeI and cloned into the pCAMBIA1302 vector digested using the same restriction enzymes to create a fusion construct (pCAMBIA1302-PtrMAPK-GFP). Both the fusion construct and the control vector (pCAMBIA1302-GFP) were mobilized into Agrobacterium tumefaciens strain EHA105 by heat shock. Transformation of onion epidermal cells was done based on the method of HY Liu et al. (2009). After culture on MS (Murashige and Skoog, 1962) medium for 2 d at 28 °C in darkness, the transformed cells were visualized using a universal fluorescence microscope (Olympus BX61, Tokyo, Japan) equipped with a geen fluorescent protein (GFP)-optimized ND filter set. The images were collected as JPEG digital files by an Olympus DP70 CCD camera (Cool-SNAP-Pro; Media Cybernetics, USA) and processed with the dedicated software IPP (Image-Pro Plus, version 5.1, Media cybernetics).
Protein kinase activity assay
The full-length PtrMAPK ORF was amplified by PCR from the pMD18-T vector harbouring PtrMAPK with GSP4 containing a BamHI or XhoI restriction site, and cloned in-frame with glutathione S-transferase (GST) into the pGEX-6P1 expression vector, generating a fusion construct of pGST-PtrMAPK, which was then introduced into the Escherichia coli strain DE3. The bacterial cells were incubated overnight at 28 °C in LB medium supplemented or not with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside). The pGST-PtrMAPK protein was purified on glutathione–Sepharose 4B beads (52-2303-00 AK, USA) based on the manufacturer’s instructions. Kinase activity assay was carried out as described by Ning et al. (2010). Each reaction (20 μl final volume) contained 50 mM Tris-HCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM dithiothreitol (DTT), 0.2 mM ATP, 2 μCi of [γ-32P]ATP, and 2 μg of the purified protein, supplemented or not with 2 μg myelin basic protein (MBP). The mixture was incubated for 30 min at room temperature, followed by addition of 5 μl of sample buffer to stop the reaction. After boiling for 5 min at 100 °C, the samples were separated on a 15% SDS–polyacrylamide gel, followed by detection of the 32P-labelled bands using Kodak X-ray film.
Plant transformation and generation of transgenic plants
To construct a vector for plant transformation, the full-length cDNA of PtrMAPK was amplified by PCR with GSP5 including SalI and KpnI restriction sites on their respective 5′-ends (Table 1). The PCR-generating fragment was digested with SalI and KpnI and inserted in the sense orientation into the SalI/KpnI sites of a pBI121 binary vector to replace the β-glucuronidase (GUS) gene, under control of the Cauliflower mosaic virus 35S (CaMV 35S) promoter. After sequence confirmation the construct was introduced into A. tumefaciens strain EHA105. To produce transgenic tobacco plants, Agrobacterium-mediated transformation of leaf discs was carried out according to Horsh et al. (1985). The presence of the transgene in the kanamycin-resistant seedlings was verified by PCR using GSP5, as described by Huang et al. (2010). Overexpression of PtrMAPK in two of the selected putative transgenic plants was examined by semi-quantitative RT-PCR using primer GSP6, while the Ubiquitin gene was used as an internal control (Table 1). T2 seeds of the overexpressing lines were harvested according to HY Liu et al. (2009) for the stress tolerance assay.
Stress tolerance assays of the wild-type (WT) and transgenic plants
The WT and transgenic lines were subjected to dehydration (in vitro seedlings or leaves) and drought (potted plants) in order to examine their stress tolerance capacity. For dehydration stress, two sets of experiments were designed. First, 30-day-old in vitro seedlings of the three lines removed from germination medium (MS+30 g l−1 sucrose+0.75% agar, pH 5.7) were allowed to dry for up to 90 min on the laboratory bench in an ambient environment. The fresh weight (FW) of the seedlings was measured every 10 min using a scale, and the percentage FW loss was determined relative to the initial weight. The leaves were sampled at the completion of dehydration and used for the determination of ion leakage (IL), malondialdehyde (MDA) content, and histochemical staining of O2− and H2O2. The dehydration was repeated twice with three replicates composed of three seedlings for each line. In the second set of experiment, 60-day-old in vitro seedlings of the transgenic lines were incubated for 2 d in MS medium supplemented with either 1 mM diethyldithiocarbamate [DDC; a superoxide dismutase (SOD) inhibitor] or 5 mM NaN3 [a peroxidase (POD) inhibitor]. Then, leaves of the WT and transgenic lines treated or not with the enzyme inhibitors were detached and dehydrated for 50 min as mentioned above. At the end of dehydration the leaves were sampled for determination of IL and O2− or H2O2 staining.
For drought treatment, the seedlings were first transplanted into plastic containers filled with a mixture of soil and sand (1:1) where they were regularly watered for 2 months until the drought treatment. At the onset of and after 7 d of withholding water, the leaves were collected from some plants for analysis of antioxidant enzyme activity, cell death, relative water content (RWC), metabolite levels, and expression of stress-responsive genes, whereas those plants without sample collection were exposed to continuous drought for up to 21 d. At the end of the drought treatment, the leaves were harvested for assay of IL, O2− and H2O2 staining, and total chlorophyll content. In order to assess the survival rate under drought conditions, 30-day-old plants grown in the plastic containers were not watered for 20 d, followed by 3 d of re-watering. Then the phenotypes were examined, the plants were photographed, and the survival rate (number of surviving plants/total number of experimented plants) was calculated. Drought treatment was repeated three times with three replicates in both the WT and transgenic lines composed of at least 17 plants for each.
Analysis of IL, MDA, chlorophyll content, O2−, and H2O2 accumulation, antioxidant enzyme activity, and metabolite levels
IL, MDA, and total chlorophyll content were measured as described in previous studies (Huang et al., 2010; Wang et al., 2011). In situ accumulation of O2− and H2O2 was examined based on histochemical staining by nitroblue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB), respectively. The activity of three antioxidant enzymes, catalase (CAT; EC 1.11.1.6), POD (EC 1.11.1.7), and SOD (EC 1.15.1.1), was spectrophotometrically measured. Details of these assays were the same as in previous reports (Huang et al., 2010; Shi et al., 2010; Wang et al., 2011). The contents of ascorbic acid and glutathione (GSH) were measured using two detection kits (Nanjing Jiancheng Bioengineering Institute, China). Proline content was assessed as described by Zhao et al. (2009) with slight modification.
Detection of cell death and measurement of RWC
Cell death was analysed by trypan blue staining based on the method of Pogány et al. (2004). The stock solution of trypan blue was prepared by mixing 10 g of phenol, 10 ml of glycerol, 10 ml of lactic acid, 10 ml of distilled water, and 0.02 g of trypan blue (Sigma). The stock solution was diluted with ethanol (96%, 1:2, v/v) to obtain a working solution. The tobacco leaves were soaked in the working solution, boiled for 1 min in a water bath, and then incubated in the working solution for 1 d. The leaves were transferred to saturated chloral hydrate solution (50 g of chloral hydrate dissolved in 20 ml of distilled water), followed by observation and photography. RWC was assessed using a Moisture Balance (Mettler, HG63, Switzerland) according to the manufacturer’s instructions.
Expression analysis by quantitative real-time PCR (qRT-PCR)
The transcript level of PtrMAPK in the trifoliate orange shoots under various stresses and expression patterns of ROS-related and stress-responsive genes in tobacco (WT and transgenic lines) before and after drought treatment were analysed by qRT-PCR using the SYBR Green dye method. Total RNA was isolated from the samples and used for cDNA synthesis with the same procedures as detailed above. Each reaction buffer (10 μl) was composed of 50 ng of cDNA samples, 5 μl of 2× SYBR Green MasterMix Reagent (Applied Biosystems), and 0.2 μM of gene-specific primers (Table 1). Actin or Ubiquitin was used as an internal control to normalize the relative expression level of the analysed genes in trifoliate orange or tobacco, respectively. The thermal cycles used were as follows: 95 °C for 10 min, and 40 cycles of 95 °C for 15 s, 58 °C for 1 min. Each sample was amplified in four independent replicates. Relative gene expression was calculated according to the delta-delta Ct method of the system.
Statistical analysis
The data, shown as mean ±SD, were analysed using SAS software (version 8.0, SAS Institution, NC, USA), and analysis of variance (ANOVA) was used to compare the statistical difference based on Fisher’s LSD test, at a significance level of P <0.05, P <0.01, and P <0.001.
Results
Cloning of a full-length cDNA of the PtrMAPK gene and sequence analysis
The search in the HarvEST database using the AtMPK3 sequence as bait yielded seven partial MAPK homologous ESTs that were merged into a sequence of 831 bp. As the 5′-end sequence was lacking, 5′-RACE was carried out so as to obtain a partial sequence that could be overlapped with the merged sequence to obtain a full-length sequence. 5′-RACE PCR produced a single fragment of 750 bp. Overlapping of the 5′-end sequence with the merged sequence gave rise to a 1339 bp sequence. Sequence analysis indicated that it contained a complete ORF of 1128 bp, flanked by a 72 bp 5′-untranslated region (UTR) and a 139 bp 3′-UTR. In order to verify the overlapped sequence, PCR was carried out with primers spanning upstream and downstream of the ORF. The resulting sequence and the overlapped sequence matched each other perfectly, suggesting the accuracy of the full-length sequence. The gene, referred to as PtrMAPK, encodes a polypeptide of 375 amino acids with a calculated molecular mass of 43 kDa and a pI of 5.51.
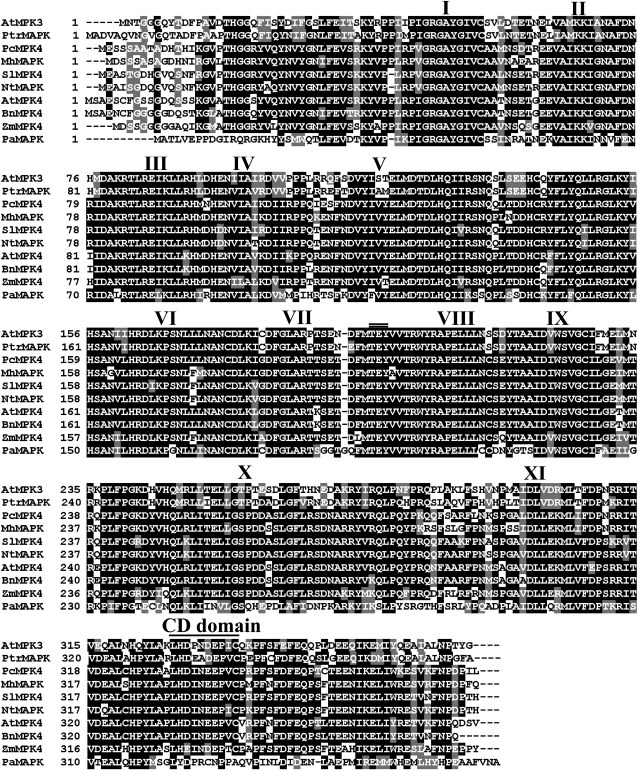
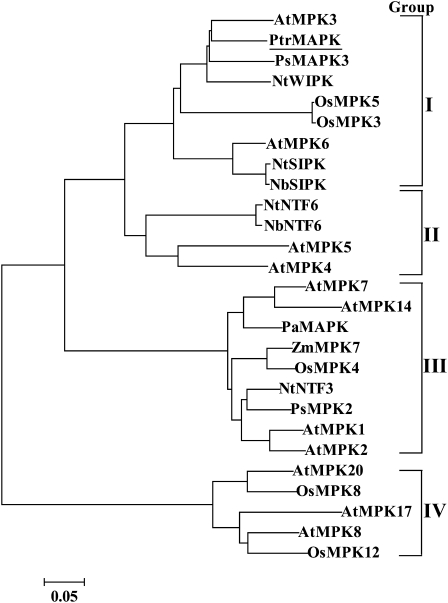
Multiple alignments indicated that PtrMAPK shares a significant degree of sequence identity with nine other MAPK proteins. The alignment showed that PtrMAPK has a highly conserved sequence, including the 11 subdomains that are important features of serine/threonine protein kinase and a putative phosphorylation site, TEV (201–203), located close to the kinase domain VIII (Fig. 1). At the C-terminus there is a common docking (CD) domain (LHDLNDEPVC, 332–341) that functions as the binding site of MAPKKs. A phylogenetic tree constructed based on the PtrMAPK and 26 MAPKs from other plant species reveals that they could be grouped into four major groups, and PtrMAPK belongs to the first group (Fig. 2).
Fig. 1.
Multiple alignments of the predicted protein sequence of PtrMAPK and MAPKs from other plants, including Arabidopsis thaliana (AtMPK3, At3g45640; AtMPK4, NP_192046.1), Brassica napus (BnMPK4, ABB69023), Malus hupehensis (MhMAPK, ABR10070), Nicotiana tabacum (NtMAPK, BAE46985), Petroselinum crispum (PcMPK4, AAN65180), Prunus armeniaca (PaMAPK, AF134730), Solanum lycopersicum (SlMPK4, ADH43227), and Zea mays (ZmMPK4, ACG31917). Identical and similar amino acid residues among the aligned sequences are indicated by black and grey shading, respectively. The 11 major conserved subdomains of a protein kinase are marked by Roman numerals. The dual phosphorylation motif (TEY) is shown by a double line, while a common docking (CD) motif at the C-terminus is indicated by a single line.
Fig. 2.
A phylogenetic tree created based on PtrMAPK and its homologous sequence from different plant species. Information about the proteins included in the tree (GenBank accession numbers) is given in Supplementary Table S1 available at JXB online.
Expression patterns of PtrMAPK under various stress conditions
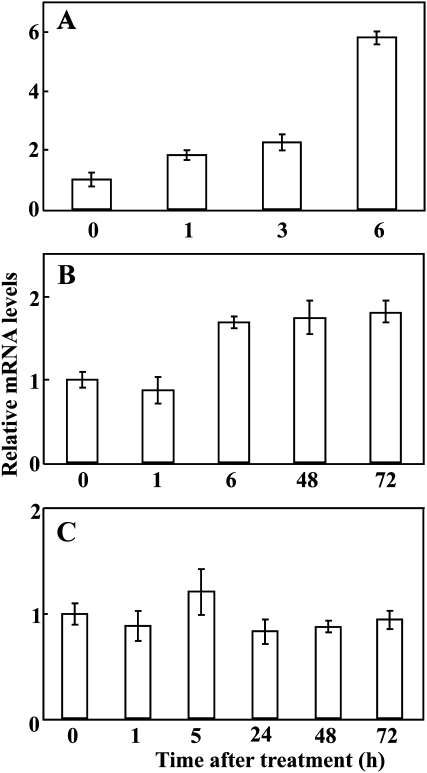
To investigate the response of PtrMAPK to abiotic stresses, the steady-state mRNA levels of this gene in response to dehydration, salinity, and cold were monitored. The transcript level of PtrMAPK was up-regulated rapidly within 1 h of exposure to dehydration, which was increased progressively until it was induced by nearly 6-fold at the end of dehydration (Fig. 3A). Low temperature treatment led to a slight induction of PtrMAPK abundance between 6 h and 72 h (Fig. 3B), whereas salt treatment did not noticeably alter the expression level of the gene (Fig. 3C).
Fig. 3.
Expression patterns of PtrMAPK in trifoliate orange under dehydration (A), low temperature (B), and salt (C), as analysed by qRT-PCR. For each stress, the expression level at time point 0 (the beginning of the relevant treatment) was defined as 1.0, and the expression level at other time points was normalized accordingly. Error bars show standard deviations for four independent replicates.
PtrMAPK was localized in the nucleus
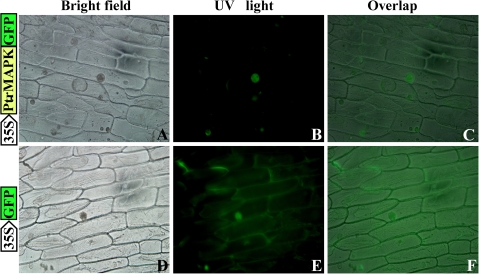
In order to identify the subcellular localization of PtrMAPK, an in-frame fusion protein (PtrMAPK–GFP) was generated by subcloning the PtrMAPK ORF without the stop codon into the upstream of the GFP gene in pCAMBIA1302, under the control of the CaMV 35S promoter. The subcellular localization of PtrMAPK was examined by monitoring the GFP fluorescence in onion epidermis cells. In the cells transformed with the PtrMAPK–GFP fusion construct, green fluorescence was exclusively observed in the nucleus (Fig. 4A–C), whereas fluorescence was detected in both the nucleus and the cytoplasm of the cells transformed with the control vector (Fig. 4D–F). These results indicate that PtrMAPK might be located in the nucleus.
Fig. 4.
Cellular localization of PtrMAPK in onion epidermal cells. PtrMAPK was fused to the N-terminus of green fluorescence protein (PtrMAPK–GFP), which was transformed into onion epidermal cells through Agrobacterium-mediated infection, using GFP as a control. The expression of PtrMAPK–GFP or GFP alone was examined under a universal fluorescence microscope. Bright-field images (A and D), fluorescence images (B and E), and the overlapped images (C and F) of representative cells expressing PtrMAPK–GFP fusion protein (upper panels) or GFP (lower panels) are shown.
PtrMAPK has kinase activity
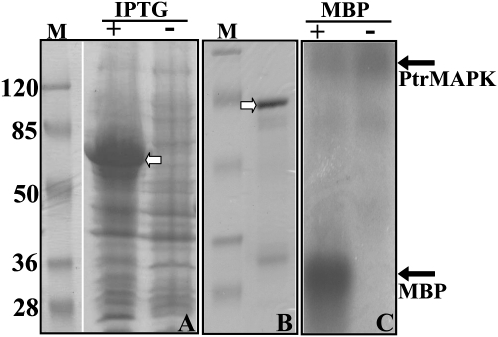
As PtrMAPK contains 11 kinase domains, a question is thus raised as to whether or not PtrMAPK possesses kinase activity. In order to answer this question, a kinase activity assay was performed using MBP as a substrate; MBP has been used extensively to examine kinase activity (Eichberg and Iyer, 1996). SDS–PAGE analysis showed that the extracted protein from E. coli cells transformed with the empty vector (pGEX-6P1) accumulated a 28 kDa protein after induction by IPTG (data not shown). However, in the E. coli cells transformed with pGST-PtrMAPK, a larger protein (71 kDa) was detected after IPTG induction (Fig. 5A), suggesting that eukaryotic expression of PtrMAPK could give rise to the predicted protein. Purification of the fusion protein on glutathione–Sepharose 4B beads gave rise to an intense band of the expected size on the polyacrylamide gel (Fig. 5B). Kinase assay of the purified protein showed that a clearly visible band of 17 kDa was detected in the presence of MBP, whereas it was absent in the reaction without MBP (Fig. 5C). These results indicate that MBP has been successfully phosphorylated, suggesting that PtrMAPK displays kinase activity.
Fig. 5.
Analysis of kinase activity in PtrMAPK. (A) SDS–PAGE separation of protein extracted from E. coli transformed with pGEX-PtrMAPK induced (+) or not (–) with 1 mM IPTG. (B) SDS–PAGE separation of the purified protein derived from the IPTG-induced extract in A. The predicted protein in A and B is shown by an open arrow. M, a molecular weight marker (kDa). (C) Detection of protein phosphorylation. The purified PtrMAPK protein was incubated in a buffer to which MBP for kinase reaction was added (+) or not (–), followed by detection of phosphorylation with SDS–PAGE and autoradiography. The positions of the proteins are shown by the filled arrows.
Transformation and regeneration of transgenic plants overexpressing PtrMAPK
As PtrMAPK was induced by dehydration in a stronger manner, transgenic tobacco plants overexpressing PtrMAPK were generated to examine the role of PtrMAPK in dehydration and drought stress tolerance. In total, seven putative transgenic lines were confirmed by PCR using primers specific to PtrMAPK and NPTII (data not shown). RT-PCR analysis showed that PtrMAPK mRNA was detected in the transgenic plants, but not in the WT (data not shown). Two of the lines (OE-2 and OE-19), of which OE-19 had a higher PtrMAPK expression level, were used for the stress tolerance test.
Assessment of stress tolerance in the transgenic lines
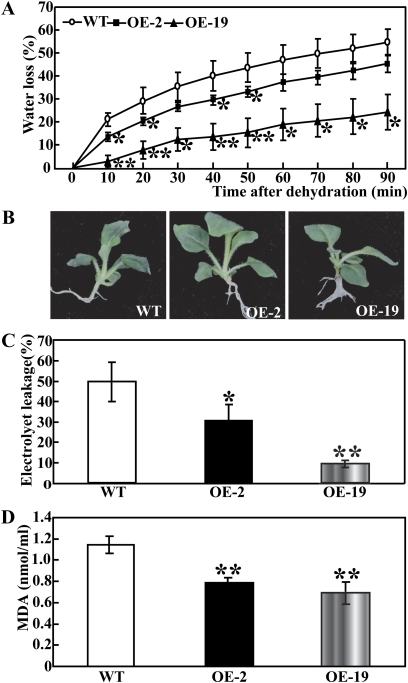
To investigate whether transgenic overexpression of PtrMAPK was correlated with stress tolerance, the WT and transgenic lines were subjected to water stress by means of either short-term dehydration or long-term drought. When the 30-day-old in vitro seedlings were dehydrated in an ambient environment, a steady decrease in FW was observed in both the WT and the two transgenic lines. However, at any time point within 90 min of dehydration, the two transgenic lines lost remarkably less water compared with the WT (Fig. 6A). At the end of dehydration, phenotype differences were prominent between the WT and transgenic lines, as the former lost its turgor to a greater extent (Fig. 6B). IL, an important indicator of membrane injury, was significantly higher in the WT (49.7%) than in OE-2 (30.8%) or OE-19 (9.4%), indicating that the WT suffered from severer membrane damage (Fig. 6C). In addition, the MDA level exhibited a profile similar to the IL, significantly lower in the transgenic lines relative to the WT (Fig. 6D). These findings demonstrate that the transgenic lines were more resistant to dehydration.
Fig. 6.
Overexpression of PtrMAPK enhances dehydration tolerance in transgenic tobacco. (A) Relative water loss rate in the wild type (WT) and transgenic lines (OE-2, OE-19) under dehydration for 90 min in an ambient environment, as measured by reduction of fresh weight every 10 min. * (P <0.05) and ** (P <0.01) indicate that the water loss rate in the two transgenic lines is significantly lower than that of the WT at the same time point. (B) A representative photograph showing the dehydrated seedlings. (C and D) Ion leakage (C) and MDA level (D) in the WT, OE-2, and OE-19 after 90 min dehydration. Asterisks indicate significant difference between the WT and the two transgenic lines (*P <0.05; **P <0.01). (This figure is available in colour at JXB online.)
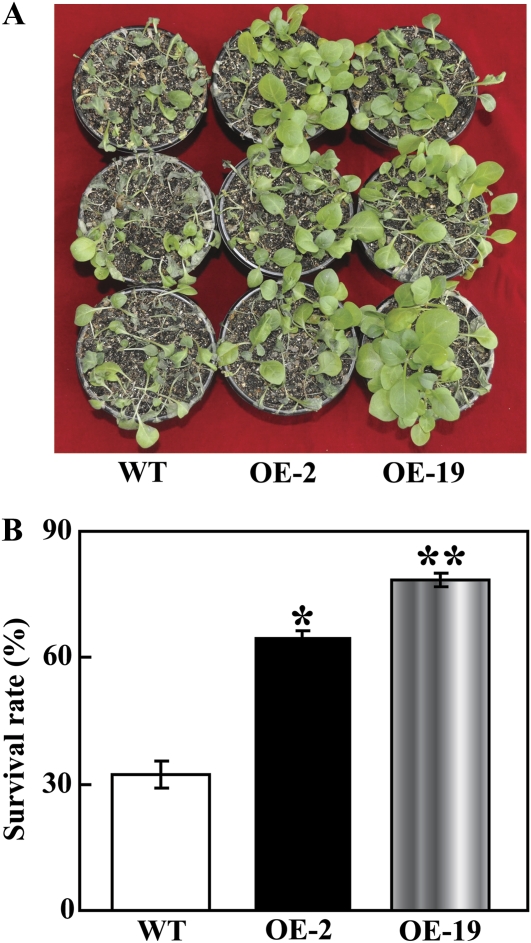
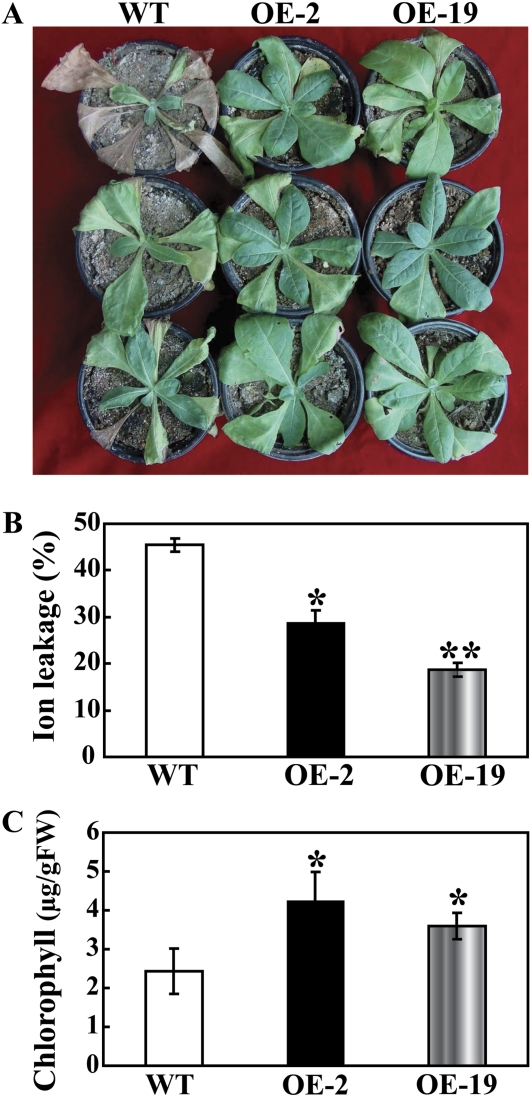
To assess the effect of PtrMAPK overexpression on drought tolerance, the WT and transgenic plants were deprived of water for 20 d, followed by re-watering for 3 d. After 20 d without water, leaf wilting was more evident in the WT relative to the two transgenic lines (data not shown). After watering was resumed, the transgenic plants recovered their growth more rapidly than did the WT plants (Fig. 7A). Three days after the resumption of watering and growth under normal conditions, the survival rate in OE-2 (64.5%) and OE-19 (78.4%) was significantly higher than that of the WT (32%, Fig. 7B). In another experiment, 2-month-old plants were exposed to 21 d drought stress. Similarly, the morphological appearances of OE-2 and OE-19 were better than that of WT, as more leaves remained green in the transgenic lines (Fig. 8A). In order to compare the physiological differences, IL and total chlorophyll content were measured in the samples collected after 21 d of drought. IL of OE-2 (28.93%) and OE-19 (18.68%) was significantly lower than the 45.56% found in the WT (Fig. 8B), while total chlorophyll contents in the two transgenic lines (3.59 μg g−1 FW and 4.25 μg g−1 FW for OE-2 and OE-19, respectively) were 1.3–1.7 times higher than that of the WT (2.43 μg g−1 FW, Fig. 8C). These results suggest that overexpression of PtrMAPK had a significant effect on the improvement of drought resistance in the transgenic tobacco plants.
Fig. 7.
The transgenic lines exhibited a higher survival rate under drought and recovery when compared with the WT. (A) The photographs are of representative plants after drought for 20 d and a subsequent recovery for 3 d. (B) The survival rate in OE-2, OE-19, and the WT after the 3 d re-watering following the drought. Data are means ±SD calculated from three replicates. * (P <0.05) and ** (P <0.01) indicate that the value in the transgenic lines is significantly different from that of the WT. (This figure is available in colour at JXB online.)
Fig. 8.
Overexpression of PtrMAPK enhances drought tolerance in transgenic tobacco. (A) The photographs are of representative WT, OE-2, and OE-19 plants after drought for 21 d. (B and C) Ion leakage (B) and total chlorophyll content (C) of the WT, OE-2, and OE-19 after drought stress for 21 d. A significant difference from the WT is indicated by asterisks (*P <0.05; **P <0.01). (This figure is available in colour at JXB online.)
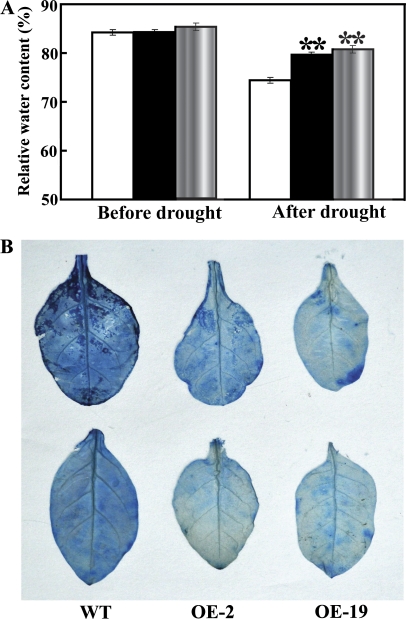
RWC and cell death in the WT and transgenic lines
RWC in the WT was close to that of the two transgenic lines prior to drought stress. Water deprivation for 7 d led to a reduction of RWC in all three lines, but OE-2 and OE-19 had significantly higher RWC relative to the WT (Fig. 9A). Trypan blue staining showed that the WT leaves were stained to a greater extent relative to the transgenic lines, suggesting that drought resulted in more serious cell death in the WT (Fig. 9B).
Fig. 9.
Relative water content (RWC) and cell death staining in the WT and transgenic lines (OE-2 and OE-19) under drought. (A) The RWC in the leaves of the three lines before and after 7 d drought treatment. (B) Trypan blue staining of the leaves from the three lines after 7 d drought treatment.
Analysis of ROS accumulation by histochemical staining
A lower IL and MDA level and less extensive cell death in the two transgenic lines imply that they might be subjected to less serious oxidative stress than the WT. Thus, it was of interest to determine the ROS accumulation in the tested lines after water stress. Histochemical staining by DAB and NBT was used to reveal in situ accumulation of H2O2 and O2−, respectively. Before dehydration, no drastic differences were observed between the WT and transgenic lines, whereas the leaves of transgenic plants were stained less intensely after 90 min dehydration (Fig. 10A, B). Following this, the ROS level in the leaves sampled from the potted plants exposed to drought for 21 d was also checked. Likewise, OE-2 and OE-19 exhibited clearly less intense DAB and NBT staining in comparison with the WT (Fig. 10C). Taken together, these data suggest that the transgenic lines accumulated lower levels of ROS (H2O2 and O2−) under water stress.
Fig. 10.
Accumulation of O2− and H2O2 in the WT and transgenic lines (OE-2 and OE-19) under dehydration or drought, as measured by histochemical staining with NBT and DAB, respectively. (A and B) Representative photographs showing staining of O2− (A) and H2O2 (B) in the leaves before (0, upper panel) and after dehydration (90, lower panel). (C) Representative photographs showing accumulation of O2− (upper panel) and H2O2 (lower panel) in leaves that have been subjected to drought for 21 d. (This figure is available in colour at JXB online.)
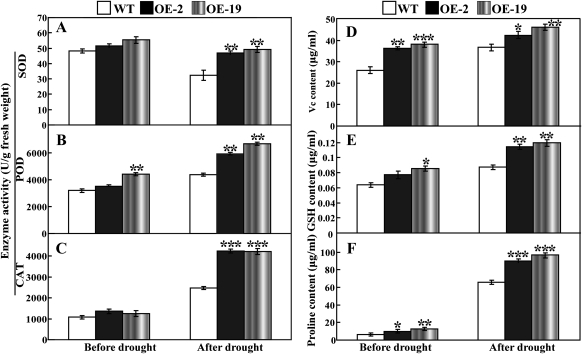
Analysis of antioxidant enzyme activity and metabolite levels
Enzymatic and non-enzymatic antioxidants play significant roles in ROS scavenging and thus influence the cellular ROS level. Since the two transgenic lines contained fewer ROS relative to the WT, the activity of three significant antioxidant enzymes (SOD, CAT, and POD) and the level of several important metabolites were assessed in the leaves sampled from the potted plants before and after drought treatment. As can be seen in Fig. 11, under normal growth conditions, the activities of the three enzymes in the two transgenic lines were slightly higher than in the WT, except the significant difference in POD activity between OE-19 and the WT. After 7 d of withholding water, it is noticeable that the two transgenic lines contained significantly higher enzyme activity than did the WT (Fig. 11A–C). In the absence or presence of drought, the transgenic plants had higher levels of ascorbic acid, GSH, and proline in comparison with the WT (Fig. 11D–F). These data indicate that PtrMAPK overexpression had a remarkable impact on the antioxidant enzymes and metabolites in the transgenic plants.
Fig. 11.
Analysis of enzyme activity and metabolite levels in the WT and the transgenic lines (OE-2 and OE-19) before and after drought treatment for 7 d. (A–C) Activity of SOD (A), POD (B), and CAT (C) in the three lines. (D–F) Content of ascorbic acid (D), GSH (E), and proline (F) in the three lines. Asterisks show that the values are significantly different between the transgenic lines and the WT at the same time point (*P <0.05; **P <0.01; ***P <0.001).
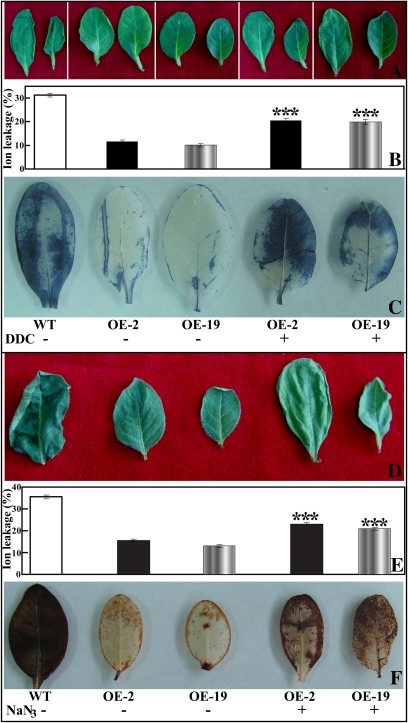
SOD and POD inhibitors attenuated stress tolerance of the transgenic lines
To determine whether activation of the antioxidant enzymes was involved in regulating stress tolerance of the transgenic lines, the transgenic plants were pre-treated with either DDC (a SOD inhibitor) or NaN3 (a POD inhibitor). The leaves were then subjected to dehydration for 50 min together with the untreated WT and transgenic lines, followed by IL measurement and ROS staining. After 50 min of dehydration, more severe leaf withering was observed in the DDC- or NaN3-treated OE-2 and OE-19 plants when compared with the untreated plants (Fig. 12A, D). The IL of OE-2 and OE-19 was enhanced by 43.6% and 49.5% with DDC treatment (Fig. 12B), and by 32.2% and 37.7% with NaN3 treatment (Fig. 12E), relative to the untreated OE-2 and OE-19, respectively. Histochemical staining showed that more intense NBT (Fig. 12C) and DAB (Fig. 12F) staining was detected in the DDC- and NaN3-treated leaves, respectively, suggesting that generation of O2− and H2O2 was greatly promoted after application of the antioxidant enzyme inhibitors. These results demonstrate that pre-treatment of the transgenic plants with the antioxidant enzyme inhibitors enhanced ROS generation and concurrent attenuation of stress tolerance.
Fig. 12.
Effect of a SOD inhibitor (DDC) and a POD inhibitor (NaN3) on the dehydration response of the transgenic lines. (A and D) Representative photographs showing the morphology of leaves from WT, OE-2, OE-19, and DDC- (A) or NaN3- (D) treated OE-2 and OE-19 plants (from the left to the right). (B and E) Ion leakage of the samples indicated in A and D, respectively. Asterisks show that the value is significantly different between the transgenic line treated or not with the enzyme inhibitor. (C and F) NBT (C) and DAB (F) staining of the leaves from WT, OE-2, OE-19, and DDC- or NaN3-treated OE-2 and OE-19 plants. (This figure is available in colour at JXB online.)
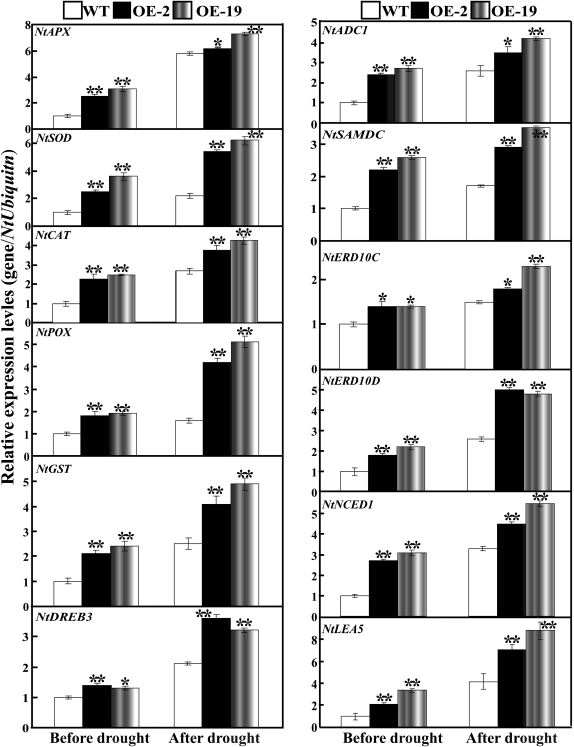
Expression analysis of ROS-related or stress-responsive genes in the WT and transgenic lines before and after drought
To gain further insight into the molecular mechanism underlying the enhanced drought resistance in the transgenic lines, the transcript abundance of 12 ROS-related or stress-responsive genes was examined in the WT and transgenic lines before and after 7 d drought stress (Fig. 13). These genes encode enzymes for direct ROS detoxification (NtSOD, NtAPX, NtCAT, NtPOX, and NtGST), enzymes involved in biosynthesis of polyamine (NtADC1 and NtSAMDC) or abscisic acid (ABA; NtNCED1), stress defensive proteins (NtERD10C, NtERD10D, and NtLEA5), or significant regulatory protein (NtDREB3). Under normal conditions, mRNA levels of all 12 genes in OE-2 and OE-19 were higher than those in the WT. Exposure to drought caused up-regulation of the transcript levels of the analysed genes in all lines, but OE-2 and OE-19 still had a higher expression level in comparison with the WT. These results demonstrate that overexpression of PtrMAPK in tobacco enhances the transcript levels of the ROS-related and stress-responsive genes with or without water stress.
Fig. 13.
Quantitative real-time PCR analysis of expression levels of ROS-related and stress-responsive genes in the WT and the transgenic lines (OE-2 and OE-19) under normal and drought conditions. Data represent the means ±SE of four replicates. Asterisks show that the values are significantly different between the transgenic lines and the WT under the same growth conditions (*P <0.05; **P <0.01).
Discussion
Abiotic stress responses require the concerted and coordinated action of a myriad of important signalling members, including the MAPK cascade. As the terminal module of the hierarchical organization of the MAPK cascade, MAPK plays a paramount role in amplifying, integrating, and channelling information between the extracellular stimuli and intracellular response (Nadarajah and Sidek, 2010). The importance of this signalling component in the regulation of cellular processes prompted the cloning and characterization of the function of a MAPK gene from P. trifoliata, in which the MAPK cascade has never been explored.
PtrMAPK shares a high degree of sequence similarity and identity with other plant MAPKs. It contains the 11 evolutionarily conserved kinase domains that may assume biological functions related to substrate specificity or protein interaction (Nadarajah and Sidek, 2010). In addition, similar to the aligned MAPKs, PtrMAPK possesses the consensus sequence of TEY, a MAPK signature motif in the kinase activation T-loop, located close to kinase domain VIII. As T (threonine) and Y (tyrosine) are two important phosphorylation residues, TEY provides a protein-binding platform for the activation of MAPKs (Rohila and Yang, 2007). At the C-terminus of PtrMAPK there is a CD domain ([LH][LHY]DXX[DE]EPXC), which is thought to function as the binding site for MAPKK (Wang et al., 2010).
Phylogenetic analysis showed that the MAPKs were categorized into four distinct groups, in line with earlier reports (Nadarajah and Sidek, 2010; Kumar and Kirti, 2010). PtrMAPK belongs to the first group (group I), which consists of several well-characterized MAPK genes, including AtMPK3, AtMPK6, and OsMPK5. It has been documented that MAPK members in groups I and II are predominantly implicated in signalling biotic and abiotic stress responses (Nadarajah and Sidek, 2010). For example, AtMPK3, OsMPK5, and NtWIPK were activated by various pathogens and environmental stresses (Zhang and Klessig, 2001; Hamel et al., 2006; Rohila and Yang, 2007). OsMPK5 has been demonstrated to be a key element for orchestrating the resistance to blast disease (Song and Goodman, 2002). AtMPK6, an important MAPK gene of Arabidopsis, has been suggested to be a universal regulator in plant stress responses since it can be activated by various abiotic and biotic stresses (Ichimura et al., 2000; Yuasa et al., 2001; Feilner et al., 2005). Transcript levels of PtrMAPK were increased by both dehydration and cold, in particular the former, indicating that PtrMAPK may be involved in the signal transduction of these two types of stresses. Clustering of these genes in the same group suggests that they may have a similar or the same biological function in abiotic stress response.
The stronger induction of the PtrMAPK transcript level by dehydration led to the elucidation of its function in dehydration/drought tolerance by the gene overexpression approach in tobacco. The stress tolerance assay demonstrates that the two transgenic lines exhibited improved tolerance to either short- or long-term water stress (dehydration and drought) as compared with the WT. The line OE-19 showed better performance than OE-2, which may be due to the higher expression of the transgene (data not shown). Based on the physiological and biochemical analysis, it can be determined that the enhanced tolerance in the transgenic lines is correlated with the maintenance of better water status and less accumulation of ROS.
It is well known that plants have to maintain their ROS pools at low levels in order to minimize cellular damage caused by oxidative stress (Harb et al., 2010; Foyer and Shigeoka, 2011). ROS accumulation depends greatly on the balance between production and concurrent scavenging (Buchanan and Balmer, 2005; Pitzschke et al., 2009). Plant cells possess a complex antioxidant defence system for ROS detoxification, which consists of non-enzymatic antioxidants and ROS-scavenging enzymes (Miller et al., 2010). It was found that OE-2 and OE-19 contained higher levels of antioxidants such as ascorbic acid, GSH, and proline, and higher activities of SOD, POD, and CAT in comparison with the WT before drought stress. This indicates that overexpression of the PtrMAPK gene has facilitated the activation of the antioxidant defence system even in the absence of stresses. These results agree with earlier reports working on MAPK genes in other plants. In maize, A Zhang et al. (2006) reported that the total activity of antioxidant enzymes was enhanced by ABA- or H2O2-induced MAPK. In a recent study, overexpression of the ZmMAPK gene in tobacco has also been shown to cause an elevation of POD activity (Zong et al., 2009). Under drought conditions the antioxidant defence systems were activated in both the WT and transgenic lines, which may be due to a dramatically elevated rate of ROS generation. However, it is clear that the levels are still higher in the two transgenic lines, implying that they displayed more robust ROS scavenging capacity, which perfectly matched with the lower ROS level. Interestingly, pre-treatment of OE-2 and OE-19 with a SOD or POD inhibitor increased the accumulation of O2− or H2O2, respectively, accompanied by compromised dehydration tolerance as compared with the untreated transgenic lines. This provides convincing evidence to show that the PtrMAPK functions in dehydration/drought tolerance by, at least partially, the activation of the antioxidant system. The present data corroborate earlier work suggesting that the MAPK cascade is implicated in ROS signalling or homeostasis regulation (Pitzschke et al., 2009; Zong et al., 2009; Ning et al., 2010).
To gain a deeper understanding of the function of PtrMAPK in stress tolerance, steady-state mRNA levels of the stress-associated genes were analysed. It was found that transcript levels of the genes encoding ROS-scavenging enzymes were up-regulated in the PtrMAPK-overexpressing lines under normal or drought conditions, consistent with the greater activity of these antioxidant enzymes. This may presumably explain the activation of the antioxidant enzymes and the lower ROS levels in the transgenic lines. Although the definitive mechanism underlying the induction of these antioxidant genes could not be deciphered, several earlier studies have demonstrated that the MAPK cascade regulates the expression of a set of ROS-related genes in maize (A Zhang et al., 2006) and tobacco (Samuel and Ellis, 2002). Most recently, Pitzschke et al. (2009) reported that mutation of the Arabidopsis MPK4 gene led to transcriptional repression of POD and CAT genes. On the other hand, two genes (NtADC1 and NtSAMDC) involved in polyamine synthesis were also induced to a higher level in the transgenic lines relative to the WT. Polyamines are important stress molecules that play critical roles in abiotic stress tolerance due to chemical and physical interactions with macromolecules including nucleic acids, phospholipids, and proteins (Liu et al., 2007; Wang et al., 2011). Stronger induction of these genes suggests that the transgenic lines may produce higher levels of polyamines, which constitutes another strategy for the enhanced tolerance in the transgenic lines. Despite the paucity of knowledge connecting the MAPK cascade and polyamine biosynthesis, recent work by Jang et al. (2009) showed that the MAPK cascade was involved in the signalling for polyamine biosynthesis of tobacco.
NtERD10 (C/D) and NtLEA5 encode group 2 and group 5 late embryogenesis abundant (LEA) proteins, respectively. Circumstantial evidence has demonstrated that LEA proteins function in dehydration tolerance by binding water, stabilizing labile enzymes, and protecting cellular and macromolecular structures (Hundertmark and Hincha, 2008; X Liu et al., 2009). Greater induction of these genes suggests that more LEA proteins might be synthesized in the transgenic lines, which is partially supported by the greater water retention and less extensive membrane damage. The phytohormone ABA plays an essential role in adaptive responses to environmental stress including drought. Biochemical and genetic evidence reveals that ABA biosynthesis under stress is largely regulated by the rate-limiting enzyme, 9-cis-epoxycarotenoid dioxygenase (NCED) (Qin and Zeevaart, 1999). Interestingly, the transcript level of NtNCED1 was enhanced in OE-2 and OE-19 as compared with the WT before and after drought, suggesting that de novo ABA synthesis may be promoted due to PtrMAPK overexpression. This assumption sounds reasonable since an increase in the ABA level by overexpression of NCED genes has been shown to improve drought tolerance in various plants (Qin and Zeevaart, 2002; Aswath et al., 2005).
Accumulating experimental evidence demonstrates that dehydration-responsive element-binding proteins (DREBs) play important roles in regulating stress responses. Overexpression of DREB genes has been shown to confer drought tolerance in a variety of plants (Umezawa et al., 2006). Herein, the expression level of a DREB family member, NtDREB3, was higher in the transgenic lines than in the WT, implying that PtrMAPK overexpression had a stimulatory impact on the transcriptional level of the DREB genes. Although the present study did not clarify if NtDREB3 is a direct target of PtrMAPK, it is known that a mode of action of MAPKs is related to phosphorylation of transcription factors in the nucleus, by which they regulate gene expression (Rohila and Yang, 2007). The subcellular localization of PtrMAPK in the nucleus makes it reasonable to speculate that there might be a transcription factor functioning as a bridge between PtrMAPK and the downstream genes, including those illustrated above.
In conclusion, a trifoliate orange group-I MAPK gene, PtrMAPK, was cloned whose mRNA abundance was dramatically induced by dehydration and slightly by cold, but not by salt. Overexpression of PtrMAPK conveyed tolerance to both dehydration and drought in tobacco transgenic plants. The enhanced stress tolerance is partially correlated with activation of ROS-related antioxidant genes/enzymes, leading to more efficient scavenging of ROS under stress. Meanwhile, stress-responsive genes were found to be up-regulated by PtrMAPK overexpression. In the future, it will be of paramount significance to identify interacting genes located upstream and downstream of PtrMAPK, which will shed light on elucidation of the molecular mechanism of action underlying the PtrMAPK-mediated stress tolerance.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Information on the proteins used for construction of the phylogenetic tree.
Acknowledgments
This work was supported by National Natural Science Foundation of China (30871685, 30921002, 31071778), the Research Fund for the Doctoral Program of Higher Education (20090146110010), the Fok Ying Tong Education Foundation (114034), the National High Technology Research and Development Program (863 Program) of China (2011AA100205), the Wuhan Municipal Project for Academic Leaders (201150530148) and the Hubei Provincial Natural Science Foundation (2009CDA080). We are grateful to Xiao-Yi Zhu for his assistance with protein kinase activity analysis.
References
- Aswath CR, Kim SH, Mo SY, Kim DH. Transgenic plants of creeping bent grass harboring the stress inducible gene, 9-cis-epoxycarotenoid dioxygenase, are highly tolerant to drought and NaCl stress. Plant Growth Regulation. 2005;47:129–139. [Google Scholar]
- Buchanan BB, Balmer Y. Redox regulation: a broadening horizon. Annual Review of Plant Biology. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- Eichberg J, Iyer S. Phosphorylation of myelin protein: recent advances. Neurochemical Research. 1996;21:527–535. doi: 10.1007/BF02527718. [DOI] [PubMed] [Google Scholar]
- Feilner T, Hultschig C, Lee J, et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Molecular and Cellular Proteomics. 2005;4:1558–1568. doi: 10.1074/mcp.M500007-MCP200. [DOI] [PubMed] [Google Scholar]
- Fiil BK, Petersen K, Petersen M, Mundy J. Gene regulation by MAP kinase cascades. Current Opinion in Plant Biology. 2009;12:615–621. doi: 10.1016/j.pbi.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Shigeoka S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiology. 2011;155:93–100. doi: 10.1104/pp.110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends in Plant Science. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Harb A, Krishnan A, Ambavaram MMR, Pereira A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiology. 2010;154:1254–1271. doi: 10.1104/pp.110.161752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsh RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SC, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Huang XS, Chen XJ, Liu JH. Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biology. 2010;10:230. doi: 10.1186/1471-2229-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. The Plant Journal. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- Jang EK, Min KH, Kim SH, Nam SH, Zhang S, Kim YC, Cho BH, Yang KY. Mitogen-activated protein kinase cascade in the signaling for polyamine biosynthesis in tobacco. Plant and Cell Physiology. 2009;50:658–664. doi: 10.1093/pcp/pcp009. [DOI] [PubMed] [Google Scholar]
- Kosetsu K, Matsunaga S, Nakagami H, Colcombet J, Sasabe M, Soyano T, Takahashi Y, Hirt H, Machida Y. The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. The Plant Cell. 2010;22:3778–3790. doi: 10.1105/tpc.110.077164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KR, Kirti PB. A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiology and Biochemistry. 2010;48:481–486. doi: 10.1016/j.plaphy.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Liu HY, Feng DR, Liu B, He YM, Wang HB, Wang JF. Studies on subcellular localization of MpASR in onion epidermal cells mediated by Agrobacterium (in Chinese) Journal of Tropical and Subtropical Botany. 2009;17:218–222. [Google Scholar]
- Liu JH, Kitashiba H, Wang J, Ban Y, Moriguchi T. Polyamines and their ability to provide environmental stress tolerance to plants. Plant Biotechnology. 2007;24:117–126. [Google Scholar]
- Liu X, Wang Z, Wang L, Wu R, Phillips J, Deng X. LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobacco. Plant Science. 2009;176:90–98. [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant, Cell and Environment. 2010;33:453–457. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- Morris PC. MAP kinase signal transduction pathways in plants. New Phytologist. 2001;151:67–89. doi: 10.1046/j.1469-8137.2001.00167.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nadarajah K, Sidek HM. The green MAPKs. Asian Journal of Plant Sciences. 2010;9:1–10. [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signaling. Trends in Plant Science. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Ning J, Li X, Hicks LM, Xiong L. A raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiology. 2010;152:876–890. doi: 10.1104/pp.109.149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Masia D, Perez-Amador MA, Carbonell P, Aniento F, Carbonell J, Marcote MJ. Characterization of PsMPK2, the first C1 subgroup MAP kinase from pea (Pisum sativum L.) Planta. 2008;227:1333–342. doi: 10.1007/s00425-008-0705-5. [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB. Role of mitogen-activated protein kinases in plant immunity. Current Opinion in Plant Biology. 2005;8:541–547. doi: 10.1016/j.pbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Bitton F, Hirt H. A major role of the MEKK1–MKK1/2–MPK4 pathway in ROS signalling. Molecular Plant. 2009;2:120–137. doi: 10.1093/mp/ssn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogány M, Koehl J, Heiser I, Elstner EF, Barna B. Juvenility of tobacco induced by cytokinin gene introduction decreases susceptibility to Tobacco necrosis virus and confers tolerance to oxidative stress. Physiological and Molecular Plant Pathology. 2004;65:39–47. [Google Scholar]
- Qin X, Zeevaart JAD. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proceedings of the National Academy of Sciences, USA. 1999;96:15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiology. 2002;128:544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila JS, Yang YN. Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. Journal of Integrative Plant Biology. 2007;49:751–759. [Google Scholar]
- Samuel MA, Ellis BE. Double jeopardy: both overexpression and suppression of a redox-activated plant mitogen-activated protein kinase render tobacco plants ozone sensitive. The Plant Cell. 2002;14:2059–2069. doi: 10.1105/tpc.002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Fu XZ, Peng T, Huang XS, Fan QJ, Liu JH. Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiology. 2010;30:914–922. doi: 10.1093/treephys/tpq030. [DOI] [PubMed] [Google Scholar]
- Song F, Goodman RM. OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta. 2002;215:997–1005. doi: 10.1007/s00425-002-0794-5. [DOI] [PubMed] [Google Scholar]
- Stulemeijer IJE, Stratmann JW, Joosten MHAJ. Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiology. 2007;144:1481–1494. doi: 10.1104/pp.107.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annual Review of Plant Biology. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu WL, Sheen J. Plant mitogen-activated protein kinase signaling cascades. Current Opinion in Plant Biology. 2001;4:392–400. doi: 10.1016/s1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Ding H, Zhang A, Ma F, Cao J, Jiang M. A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. Journal of Integrative Plant Biology. 2010;52:442–452. doi: 10.1111/j.1744-7909.2010.00906.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun PP, Chen CL, Wang Y, Fu XZ, Liu JH. An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. Journal of Experimental Botany. 2011;62:2899–2914. doi: 10.1093/jxb/erq463. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes unlock the future. Current Opinion in Biotechnology. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Xiong LZ, Yang YN. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. The Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa T, Ichimura K, Mizoguchi T, Shinozaki K. Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant and Cell Physiology. 2001;42:1012–1016. doi: 10.1093/pcp/pce123. [DOI] [PubMed] [Google Scholar]
- Zaïdi I, Ebel C, Touzri M, Herzog E, Evrard JL, Schmit AC, Masmoudi K, Hanin M. TMKP1 is a novel wheat stress responsive MAP kinase phosphatase localized in the nucleus. Plant Molecular Biology. 2010;73:325–338. doi: 10.1007/s11103-010-9617-4. [DOI] [PubMed] [Google Scholar]
- Zhang A, Jiang M, Zhang J, Tan M, Hu X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiology. 2006;141:475–487. doi: 10.1104/pp.105.075416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Klessig DF. MAPK cascades in plant defense signaling. Trends in Plant Science. 2001;11:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- Zhang T, Yang T, Zhang L, Xu S, Xue L, An L. Diverse signals converge at MAPK cascades in plant. Plant Physiology and Biochemistry. 2006;44:274–283. doi: 10.1016/j.plaphy.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Zhao L, Liu F, Xu W, Di C, Zhou S, Xue Y, Yu J, Su Z. Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnology Journal. 2009;7:550–561. doi: 10.1111/j.1467-7652.2009.00423.x. [DOI] [PubMed] [Google Scholar]
- Zong XJ, Li DP, Gu LK, Li DQ, Liu LX, Hu XL. Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta. 2009;229:485–495. doi: 10.1007/s00425-008-0848-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.