Abstract
Human longevity and healthy aging show moderate heritability (20–50%). We conducted a meta-analysis of genome-wide association studies from nine studies from the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium for two outcomes: a) all-cause mortality and b) survival free of major disease or death. No single nucleotide polymorphism (SNP) was a genome-wide significant predictor of either outcome (p < 5 × 10−8). We found fourteen independent SNPs that predicted risk of death, and eight SNPs that predicted event-free survival (p < 10−5). These SNPs are in or near genes that are highly expressed in the brain (HECW2, HIP1, BIN2, GRIA1), genes involved in neural development and function (KCNQ4, LMO4, GRIA1, NETO1) and autophagy (ATG4C), and genes that are associated with risk of various diseases including cancer and Alzheimer’s disease. In addition to considerable overlap between the traits, pathway and network analysis corroborated these findings. These findings indicate that variation in genes involved in neurological processes may be an important factor in regulating aging free of major disease and achieving longevity.
Introduction
The recent, remarkable extension of life expectancy is largely attributed to the postponement of mortality at old age (Vaupel, 1997, Vaupel, 2010). The years of life gained in the older population residing in developed nations are a success story of public health measures and improved health care. In addition to such external factors, longevity and healthy aging consistently show a modest heritability between 20 to 50% and aging associated genetic research may provide further insights into the mechanisms of aging (Herskind, et al., 1996, McGue, et al., 1993, Reed and Dick, 2003). It has been postulated that genes involved in pathways associated with aging identified in animal models, such as IGF-insulin signalling, regulation of lipoprotein metabolism, the mTOR pathway, and the oxidative stress response may also influence survival to old or even exceptionally old age in humans (Christensen, et al., 2006, Kenyon, 2010, Vellai, et al., 2003). However, in humans, common variants within genes involved in these pathways have not been consistently associated with lifespan (Christensen, et al., 2006, Kenyon, 2010, Kuningas, et al., 2008, Vijg and Suh, 2005).
The lack of success in the identification of genes related to aging in humans may be due to the complexity of the phenotype. One approach to investigate aging and longevity is to compare frequencies of genetic variants between nonagenarians or centenarians and the general population. This approach led to the discovery of an association between APOE (Deelen, et al., 2011, Ewbank, 2007, Gerdes, et al., 2000) and more recently FOXO3A (Anselmi, et al., 2009, Flachsbart, et al., 2009, Li, et al., 2009a, Pawlikowska, et al., 2009, Willcox, et al., 2008) and human aging and longevity. However, a recent GWAS of individuals reaching the age of 90 or older failed to identify genome-wide significant variants (Newman, et al., 2010).
Prospective follow-up studies with a continuous outcome such as time to death are more powerful than case-control analyses. A study of time to death simultaneously addresses the effects of genetic variation related to life span, the progression towards death, and disease specific mortality. This design has been successfully applied in animal models (Finch and Ruvkun, 2001, Kenyon, 2010) and also in human genetics research of blood pressure (Levy, et al., 2009, Newton-Cheh, et al., 2009, van Rijn, et al., 2007), a trait with heritability similar to longevity, where examination of a continuous outcome has been more successful in identifying genetic loci than studies that have solely used hypertension as a dichotomous trait. Frailty and survival free of disease have been suggested as more promising phenotypes for studies of aging since mortality is a very heterogeneous outcome caused by multiple chronic conditions (Vijg and Suh, 2005).
This study addresses the genetics of aging in a broad, sequential way using data from cohort studies participating in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. First, we aimed to identify SNPs associated with all cause mortality (time to death) in a hypothesis-free GWAS in ~ 25,000 unselected persons of European ancestry. Second, we performed GWAS of time to event, defined by major incident events (myocardial infarction, heart failure, stroke, dementia, hip fracture or cancer) or death, as an alternative phenotype for healthy aging. Last, we analyzed the SNPs along with their respective most likely associated genes identified in the GWAS meta-analyses to identify pathways and networks associated with aging and longevity.
Methods
Participants
The participants are of recent European ancestry and stem from cohorts of the CHARGE Consortium (Psaty, et al., 2009). All cohorts are follow-up studies periodically assessing the health and vital status of their participants. Although some of the cohorts included multiple ethnic groups, only data from self-reported Caucasians was used. In addition, population structure was assessed using principal components in each CHARGE study and outliers were removed. Any remaining within-study structure was adjusted for using appropriate methods.(Price, et al., 2006) All participants included in this analysis were at least 55 years of age at the time of blood draw for DNA and provided written informed consent. A brief description of each population is given in the Supplementary Information.
Phenotype
We conducted a survival analysis, adjusted for age at baseline and sex, to model continuous time to death or end of follow-up in 25,007 participants (deceased (“cases”) = 8,444, mean follow-up time = 10.6 (SD 5.4) years) that were older than 55 years at baseline. As research demonstrated that the likelihood of incident disease is genetically determined, we defined a second phenotype: survival free of major disease or mortality (Atzmon, et al., 2004, Lunetta, et al., 2007, Vijg and Suh, 2005). The outcome was defined as time to the first of the following adjudicated events: myocardial infarction, heart failure, stroke, dementia, hip fracture, cancer, or death. For this analysis, participants at baseline were older than 55 years of age and free of any of the aforementioned conditions. Inclusion in the study required complete follow-up information on mortality and at least 4 out of 6 of the health conditions. Genome-wide information on polymorphisms was available for 16,995 participants free of disease at the beginning of the study. These participants were followed for 8.8 (SD 5.7) years and we registered 7,314 major events.
Genotyping and Imputation
As different genotyping platforms were used across studies, we imputed to 2.5 million SNPs using the HapMap 22 CEU (build 36) genotyped samples as a reference. For details on the study specific quality control procedures for genotyping and imputation please consult Table S1 in the Supplementary Information (SI).
Statistical Analysis
We used the semi-parametric Cox proportional hazard to model time to event for both phenotypes in each study. Follow-up time since baseline was used as time scale. An additive genetic model was used in this analysis. We subsequently combined the individual study results in a meta-analysis using a fixed effects model that combined the study specific regression parameters and standard errors using inverse variance weighting. We included SNPs that had a minor allele frequency (MAF) of at least 1% and an imputation quality ratio (de Bakker, et al., 2008) (equivalent to the MaCH r2 statistic (Li, et al., 2009b)) of at least 0.3. The study specific inflation factors (λGC) were computed using the set of chi-square statistics used for the meta-analysis for each study. The inflation factor is computed as the median of all chi-square statistics divided by the expected median of the statistics (approximately 0.456) for a chi-square distribution with 1 degree of freedom. SNP associations at p<5 × 10−8 were considered to be genome-wide significant. SNPs with p<5 × 10−5 were considered suggestive associations. The combined meta-analysis hazard ratio (HR) can be interpreted as the increase in the risk of dying or having a major event during follow-up per additional copy of the coded allele. Power analysis revealed 80% statistical power to detect SNPs with a minor allele frequency of 5% and relative risk of 1.10 using a nominal significance level of 0.05 (Supplementary Table 2).
In addition, we incorporated gene annotation information, a technique that has successfully been applied in the field of aging research (de Magalhaes, et al., 2009a, de Magalhaes, et al., 2009b). Protein ANalysis THrough Evolutionary Relationships (PANTHER)(Mi, et al., 2007, Thomas, et al., 2003) and Ingenuity Pathway Analysis (IPA) (www.ingenuity.com) were used for identification and classification of networks, pathways, biological processes and molecular functions of the genes identified in this study. For both phenotypes we generated lists of candidate genes. These genes were the closest reference genes to the SNPs associated with the outcome at p < 1 × 10−3. PANTHER compares these gene lists to the reference list using the binomial test for each molecular function, biological process, or pathway term. IPA builds networks by searching the Ingenuity Pathways Knowledge Base for interactions between the identified genes and all other gene objects stored in the knowledge base.
Results
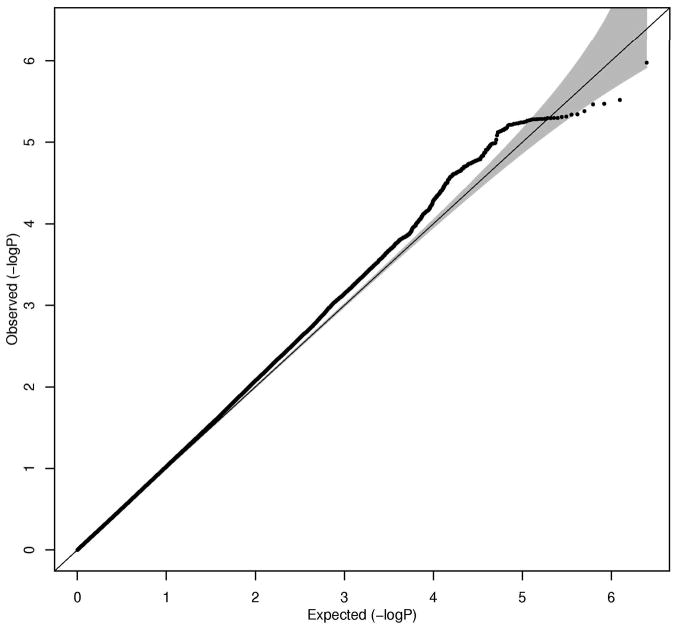
We conducted a meta-analysis of GWAS on time to death adjusted for baseline age and sex in participants of European origin, 55 years of age or older from nine longitudinal cohort studies participating in the CHARGE Consortium (Psaty, et al., 2009). In total, we observed 8,444 deaths (mean age at death: 81.1, Standard Deviation (SD) 8.4)) in 25,007 participants (55% female) after an average follow up of 10.6 (SD 5.4) years. Descriptive characteristics of participants and Manhattan plots showing genome wide p-values for association are displayed in the Supplementary Information, (Figure S1, Tables S3–4). The quantile-quantile plot (Q-Q plot) of observed versus expected p-values showed only a small deviation from the null hypothesis, indicating no significant population stratification (Figure 1a, λGC = 1.066). Although there were no genome-wide significant findings (p < 5 × 10−8), 14 independent SNPs were associated with time to death at a suggestive threshold of p < 1 × 10−5 (Table 1). Among these SNPs, rs4936894 (chromosome 11, near the von Willebrand factor A domain containing 5A gene (VWA5A)) had the strongest association with time to death (p = 3.4 × 10−7). We sought replication for 5 of the 14 top SNPs with the strongest association with time to death in 4 independent samples (n=10,411, deaths= 1,295) of the same ancestry. None of the SNPs were consistently associated with time to death at a nominally significant level of p < 0.05 across all replication samples (Table S5–S8). In the combined meta-analysis of the discovery and replication studies only rs1425609 in the vicinity of otolin-1 (OTOL1) showed a stronger association (1.61 × 10−6).
Figure 1.
Figure 1a Quantile–quantile (Q-Q) plot after meta-analysis for time to death
Figure 1b Quantile–quantile (Q-Q) plot after meta-analysis for time to event
Table 1.
Top 14 SNPs (p-value < 10−5) for time to death ranked by p-value, from meta-analysis of 9 cohorts†
| Nr. | SNP | Chr | Position | Closest Reference Gene | Distance (bp) from closest gene | Coded Allele | Non-coded Allele | Frequency coded allele | HR | P-value | Study Effect Direction | Number of Supporting SNPs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs4936894 | 11 | 123522703 | VWA5A | 123 | A | G | 0.226 | 1.11 | 3.38E-07 | ++++−++−+ | 224 |
| 2 | rs1425609 | 3 | 164164689 | OTOL1 | 1460265 | A | G | 0.381 | 0.92 | 1.46E-06 | 399 | |
| 3 | rs766903 | 12 | 49990101 | BIN2 | 14104 | A | G | 0.834 | 0.90 | 1.61E-06 | + | 7 |
| 4 | rs12042640 | 1 | 63139384 | ATG4C | 36747 | T | C | 0.284 | 1.09 | 1.71E-06 | ++++−+−+− | 19 |
| 5 | rs17149227 | 7 | 75073485 | HIP1 | 72141 | T | G | 0.959 | 0.79 | 3.56E-06 | −??+−? | 0 |
| 6 | rs3128591 | 9 | 136741940 | COL5A1 | 68468 | A | G | 0.754 | 0.92 | 3.64E-06 | 20 | |
| 7 | rs11582903 | 1 | 87618642 | LMO4 | 34804 | A | C | 0.150 | 1.12 | 3.94E-06 | ++−++++++ | 38 |
| 8 | rs4850695 | 2 | 196861504 | HECW2 | 89283 | A | G | 0.766 | 1.09 | 4.62E-06 | +++++++++ | 95 |
| 9 | rs10259086 | 7 | 103680248 | ORC5L | 44549 | T | G | 0.686 | 1.08 | 5.16E-06 | ++++++−++ | 72 |
| 10 | rs2769255 | 1 | 41017941 | KCNQ4 | 4329 | T | C | 0.374 | 1.08 | 5.17E-06 | ++++++−++ | 95 |
| 11 | rs17291546 | 6 | 2660681 | LOC340156 | 35472 | A | G | 0.957 | 0.82 | 7.65E-06 | −? | 8 |
| 12 | rs12606100 | 18 | 69102967 | NETO1 | 417177 | T | C | 0.202 | 1.11 | 8.72E-06 | +??−++++− | 4 |
| 13 | rs1274214 | 11 | 122979741 | GRAMD1B | 18987 | T | C | 0.500 | 0.93 | 8.87E-06 | 42 | |
| 14 | rs10811679 | 9 | 2224701 | SMARCA2 | 41080 | T | C | 0.330 | 1.08 | 9.53E-06 | +++++++++ | 37 |
N = 25,007 participants with 8,444 deaths, only SNPs with MAF > 3% presented
p-values are for the inverse variance-weighted meta-analysis.
Distances to genes are given in base pairs. Position is for NCBI Build 36.
Chr = chromosome, Hazard Ratios (HR) are for each additional coded allele
Number of supporting SNPs: the number of SNPs within 500 kb of the top SNP that are in LD with the top SNP in the HapMap CEU release 22 (r2>=0.10) and have association p-value<0.05.
Study Effect Direction: study-specific information is presented in the order: RS, CHS, FHS, ARIC, AGES, HABC, BLSA, InCHIANTI, SHIP
Direction: “+” = coded allele increases risk of mortality, “−” = coded allele decreases risk of mortality, ?= not tested
For information on all SNP associations with p-value<10−4 see Table S2
Likewise, no genome-wide significant findings were identified in the time to event analysis following 16,995 participants free of disease at baseline and registering 7,314 events over an average of 8.8 (SD 5.7) years of follow-up (Table 2). Events included incident myocardial infarction, heart failure, stroke, dementia, hip fracture, and cancer or death. The Q-Q plot (Figure 1b, λGC = 1.019) showed no evidence of inflation of type I error. In total, there were 8 independent SNPs associated with event-free survival at p < 10−5. The SNP with the strongest association was rs10412199 (chromosome 19, p = 3.02 × 10−6), which is in close proximity to ataxia, cerebellar, Cayman type (ATCAY). Additional descriptive information including definitions of each event and association results with p < 10−4 are provided in the Figure S2, Tables S9–S12.
Table 2.
Top 8 SNP (p-value < 10−5) associations from meta-analysis of 8 cohorts for time to event, ranked by p-value (n = 16,995 with 7,314 events)
| Nr. | SNP | Chr | Position | Closests Reference Gene | Distance (bp) from closest gene | Coded Allele | Non-coded Allele | Frequency coded allele | HR | P-value | Study Effect Direction | Number of Supporting SNPs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs10412199 | 19 | 3878771 | ATCAY | 307 | A | G | 0.33 | 0.91 | 3.02E-06 | −−?−++−− | 6 |
| 2 | rs16852912 | 3 | 170169370 | MECOM | 114610 | T | C | 0.08 | 1.18 | 3.37E-06 | ++++−+−+ | 72 |
| 3 | rs8001976 | 13 | 47285723 | SUCLA2 | 129069 | T | C | 0.44 | 1.09 | 3.43E-06 | +++−++−− | 173 |
| 4 | rs11162963 | 1 | 80507169 | ELTD1 | 1262086 | T | C | 0.63 | 1.09 | 4.15E-06 | ++++++++ | 40 |
| 5 | rs4764043 | 12 | 14006749 | GRIN2B | 17570 | T | C | 0.08 | 1.17 | 6.10E-06 | ++++++++ | 2 |
| 6 | rs3112530 | 5 | 152619870 | GRIA1 | 230628 | A | G | 0.08 | 0.85 | 6.79E-06 | + | 130 |
| 7 | rs10202497 | 2 | 237935633 | COL6A3 | 38233 | A | C | 0.14 | 0.89 | 8.22E-06 | 36 | |
| 8 | rs2367725 | 1 | 43988415 | ST3GAL3 | 42611 | T | C | 0.42 | 1.08 | 9.31E-06 | ++++++−+ | 119 |
p-values are for the inverse variance-weighted meta-analysis. Distances to genes are given in base pairs. Position is for NCBI Build 36.
Chr=chromosome, Hazard Ratios (HR) are for each additional coded allele
Number of supporting SNPs: the number of SNPs within 500 kb of the top SNP that are in LD with the top SNP in the HapMap CEU release 22 (r2>=0.10) and have association p-value<0.05.
Study Effect Direction: study-specific information is presented in the order: RS, CHS, FHS, ARIC, AGES, HABC, BLSA, InCHIANTI
Direction: + = coded allele increases risk of event, - = coded allele decreases risk of event, ?= not tested
For information on all SNP associations with p-value<10−4 see Supplementary Information, Table S11
As both phenotypes may provide different but complimentary information about the aging process, we evaluated the overlap between their association results (Table 3). Interpretation of the overlap between the phenotypes requires caution as both phenotypes are correlated, nevertheless it helps to focus on specific loci and put them into the context of aging. From the 14 loci passing the pre-specified, suggestive threshold of p < 1 × 10−5 in the time to death analysis, 5 had corresponding SNPs within 500 kb distance, in linkage disequilibrium (LD, r2 > 0.1) associated with p < 1 × 10−4 and the same overall direction of the effect in the time to event analysis. These 5 regions were in the vicinity of the following genes: OTOL1 (3q26.1), bridging integrator 2 (BIN2, 12q13), ATG4 autophagy related 4 homolog C (ATG4C, 1p31.3), origin recognition complex, subunit 5-like (ORC5L, 7q22.1), and potassium voltage-gated channel, KQT-like subfamily, member 4 (KCNQ4, 1p34). Similarly, in the time to event analysis three of the eight top SNPs showed considerable overlap and the same direction of effect in the time to death analysis. These SNPs were close to the following genes: MDS1 and EVI1 complex locus (MECOM, 3q24–q28), succinate-CoA ligase, ADP-forming, beta subunit (SUCLA2, 13q12.2–q13.3), and ST3 beta-galactoside alpha-2,3-sialyltransferase 3 (ST3GAL3, 1p34.1).
Table 3.
Overlap between the associations of time to death and time to event*
| Top Hit | SNP | Chr | Closest Reference Gene | time to death | time to event | Top SNPs from time to death (time to event) analysis associated with different p-values in time to event (time to death) analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | Effect | P | Effect | TOTAL | P >=0.05 | P<0.05 | P<0.01 | P<0.001 | P<0.0001 | |||||
| Time to death | ||||||||||||||
|
|
||||||||||||||
| 1 | rs1425609 | 3 | OTOL1 | 1.46E-06 | − | 0.005704 | − | 1119 | 693 | 235 | 132 | 37 | 22 | |
| 2 | rs766903 | 12 | BIN2 | 1.61E-06 | − | 0.01315 | − | 37 | 27 | 4 | 5 | 0 | 1 | |
| 3 | rs12042640 | 1 | ATG4C | 1.71E-06 | + | 0.03701 | + | 93 | 60 | 19 | 4 | 0 | 10 | |
| 4 | rs11582903 | 1 | LMO4 | 3.94E-06 | + | 0.7336 | − | 133 | 91 | 8 | 12 | 21 | 1 | |
| 5 | rs10259086 | 7 | ORC5L | 5.16E-06 | + | 0.03266 | + | 239 | 154 | 56 | 21 | 4 | 4 | |
| 6 | rs2769255 | 1 | KCNQ4 | 5.17E-06 | + | 0.01322 | + | 287 | 151 | 68 | 56 | 7 | 5 | |
| 7 | rs17291546 | 6 | LOC340156 | 7.65E-06 | − | 0.01624 | − | 29 | 19 | 9 | 1 | 0 | 0 | |
| 8 | rs12606100 | 18 | NETO1 | 8.72E-06 | + | 0.02853 | + | 23 | 16 | 5 | 2 | 0 | 0 | |
| 9 | rs1274214 | 11 | GRAMD1B | 8.87E-06 | − | 0.0567 | − | 101 | 39 | 28 | 17 | 17 | 0 | |
| Time to event | ||||||||||||||
| 1 | rs16852912 | 3 | MECOM | 0.00589 | + | 3.37E-06 | + | 169 | 67 | 49 | 49 | 2 | 2 | |
| 2 | rs8001976 | 13 | SUCLA2 | 0.01473 | + | 3.43E-06 | + | 433 | 198 | 91 | 46 | 59 | 39 | |
| 3 | rs4764043 | 12 | GRIN2B | 0.0017 | + | 6.10E-06 | + | 45 | 42 | 2 | 1 | 0 | 0 | |
| 4 | rs10202497 | 2 | COL6A3 | 0.00035 | − | 8.22E-06 | + | 135 | 83 | 27 | 12 | 9 | 4 | |
| 5 | rs2367725 | 1 | ST3GAL3 | 0.0274 | + | 9.31E-06 | + | 459 | 317 | 56 | 37 | 31 | 18 | |
P: p-values are for the inverse variance-weighted meta-analysis.
Chr: chromosome, Effect = meta-analysis direction of effect
Total: the number of SNPs in time to death (time to event) analysis within 500 kb of SNPs from the time to event (time to death) analysis that are in LD with the top SNPs from the time to death (time to event) analysis in the HapMap CEU release 22 (r2>=0.10) and have association p-value<0.05.
only SNPs that were nominally significant (p<0.05) for both traits are shown.
Finally, we evaluated candidate genes for aging by identification and classification of networks, pathways, biological processes and molecular functions. The candidate genes were derived from the meta-analyses of GWAS and included the reference genes closest to the SNPs associated with p < 1 × 10−3 (time to death: 862 genes, time to event: 704 genes). We used PANTHER (Mi, et al., 2007, Thomas, et al., 2003, Thomas, et al., 2006) and Ingenuity Pathway Analysis (IPA) software (www.ingenuity.com) for these analyses. PANTHER compares these gene lists to the reference list using the binomial test for each molecular function, biological process, or pathway term. IPA builds networks by searching the Ingenuity Pathways Knowledge Base for interactions between the identified genes and all other gene objects stored in the knowledge base.
For the analysis of time to death, the relevant biological processes overrepresented in the PANTHER analysis were developmental processes, neuronal activities, signal transduction, neurogenesis, ectoderm development, and cell adhesion. For the analysis of time to incident event, developmental processes and neuronal activities were overrepresented among other biological process (Table 4). The analyses also highlighted the Wnt signalling pathway. The Wnt signalling pathway is ubiquitous and know to be involved in cancer but also plays an important role in the early stages of the development of the central nervous system, in synaptic formation by axon guidance, and in modulating fibrosis during muscle repair scored high in both traits under study (Brack, et al., 2007, Inestrosa and Arenas, 2010, Keeble, et al., 2006, Ulloa and Marti, 2010). For extended tables see Supplementary Information Table S13 and Table S14. In addition, Ingenuity identified one network with p = 10−31 containing 26 genes involved in processes related to nervous system development and function for the analysis of time to death (Figure 2) and one network with p = 10−40 containing 28 genes involved in cellular function and development for time to event (Supplementary Information, Figure S3).
Table 4.
Results from the gene annotation analysis using PANTHER
| Biological Process | H. sapiens (Reference) | Nr genes observed | Nr genes expected | −/+ | p-value unadjusted | p-value adjusted* |
|---|---|---|---|---|---|---|
| Time to Death: | ||||||
| Biological process unclassified | 11321 | 238 | 367,71 | − | 1,29E-20 | 4,00E-19 |
| Developmental processes | 2152 | 152 | 69,9 | + | 1,39E-19 | 4,32E-18 |
| Neuronal activities | 569 | 65 | 18,48 | + | 8,94E-18 | 2,77E-16 |
| Signal transduction | 3406 | 199 | 110,63 | + | 9,09E-17 | 2,82E-15 |
| Neurogenesis | 587 | 64 | 19,07 | + | 1,43E-16 | 2,84E-14 |
| Ectoderm development | 692 | 68 | 22,48 | + | 2,33E-15 | 3,38E-13 |
| Cell adhesion | 622 | 57 | 20,2 | + | 7,00E-12 | 2,17E-10 |
| Time to Event: | ||||||
| Developmental processes | 2152 | 115 | 57,46 | + | 1,02E-12 | 3,16E-11 |
| Biological process unclassified | 11321 | 214 | 302,27 | − | 2,93E-12 | 9,08E-11 |
| Neuronal activities | 569 | 47 | 15,19 | + | 2,28E-11 | 7,08E-10 |
Legend: Candidate genes (genes observed) were in the neighbourhood of SNPs associated with p-value < 1 × 10−3. For time to death 862 candidate genes were identified; 826 could be matched to the PANTHER gene list. For time to event 704 candidate genes were identified; 679 could be matched to the PANTHER gene list. Extended lists of PANTHER pathways, biological processes, and molecular functions are listed in the Supplementary Information (S12, S13).
Bonferroni correction multiplying the single-test P-value by the number of independent tests to obtain an expected error rate
Figure 2. Network describing neuronal activities related to time to death.

Pathway analysis of genes (SNPs) associated with time to death. Genes are represented as nodes; edges indicate known interactions (solid lines depict direct and hatched lines depict indirect interaction). Human gene functions are color-coded as follows: Red= Unknown, Yellow= Transmembrane Receptor and G-Protein Coupled Receptor, Magenta (Pink-Purple)= Group/Complex/Other, Bright Green= Ion Channel, Hunter Green (Dark Green) = Peptidase, Navy Blue = transcription regulator, Light Blue=Transporter, Beige= Enzyme, Orange= Kinase, Light green= Cytokine, Light Purple= Phosphate, Gray= Translation Regulator, Olive Green=Ligand-dependent nuclear receptor.
IPA analysis highlighted the following genes associated with the time to death trait: NTRK2 (neurotrophic tyrosine kinase, receptor, type 2), - a member of the neurotrophic tyrosine receptor kinase family. This kinase is a membrane-bound receptor that, upon neurotrophin binding, phosphorylates itself and members of the MAPK pathway. Signaling through this kinase leads to cell differentiation. Second in line were NCAM1 (neural cell adhesion molecule 1), - a cytoskeletal binding protein, GRID2 (glutamate receptor, ionotropic, delta 2), - a relatively new member of the family of ionotropic glutamate receptors which are the predominant excitatory neurotransmitter receptors in the mammalian brain, and have a role in neuronal apoptotic death, and RIMS1 (regulating synaptic membrane exocytosis 1), which regulates synaptic vesicle exocytosis and may be part of the protein scaffold of the cell.
Among the genes that were highlighted through the IPA analysis in the analysis of time to event was MYC (v-myc myelocytomatosis viral oncogene homolog), - a multifunctional, nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis and cellular transformation. MYC functions as a transcription factor that regulates transcription of specific target genes. Second in line were E2F1 (E2F transcription factor 1), EGFR (epidermal growth factor receptor), and CEBPA (CCAAT/enhancer binding protein (C/EBP), alpha). EF21, a transcription factor, plays a crucial role in the control of cell cycle and action of tumor suppressor proteins, can mediate both cell proliferation and p53-dependent/independent apoptosis. EGFR is a transmembrane glycoprotein that serves as a receptor for members of the epidermal growth factor family and supports cell proliferation. CEBP-Alpha, a bZIP transcription factor, can bind as a homodimer to certain promoters and enhancers. CEBPA also forms heterodimers with the related proteins CEBP-beta and CEBP-gamma and modulates the expression of leptin, interacts with CDK2 and CDK4, and thereby inhibits these kinases and causes growth arrest in cultured cells.
Discussion
In our analyses of over 25,000 individuals of 55 years and older followed for an average of 11 years, we did not identify genome-wide significant associations for all-cause mortality and survival free of major diseases. However, both traits highlighted loci with suggestive significance that were in the neighbourhood of genes related to neural regulation. In addition, our pathway and network analyses identified an enrichment of genes associated with cellular and neural development and function, and cell communication that may contribute to variation in human aging. Brain development might be responsible for the creation of redundancy in brain circuitry, which is associated with functional reserve and resiliency. Brain function regulates most of the compensatory strategy supporting maintenance of homeostatic equilibrium. Both of these processes are essential to healthy aging and longevity.
Several explanations are possible for the lack of genome-wide significant findings. First, mortality is arguably one of the most complex phenotypes, and several trajectories towards extreme old age have been identified (Evert, et al., 2003). Multiple genes could mediate the aging process but would have their effects through numerous different pathophysiological processes and diseases that act as intermediate factors on the pathway to death (de Magalhaes, et al., 2009b). Therefore, any common variation in genes associated with aging probably has a small effect.
Second, the largely negative findings of this and other studies contrast with the intriguing animal studies of longevity. Very large effects of single genes on lifespan have indeed been observed in laboratory animals, but humans often have several homologues of these genes which might significantly differ in function or compensate for mutated genes through redundant mechanisms (Kuningas, et al., 2008). This could explain why our top findings did not include genes in these pathways found in animal models. Animal models also represent genetically homogenous populations and are exposed to controlled environmental influences. The lack of replication of animal model findings in humans suggests that the use of knock out animals may not provide the optimal approach to understanding the variation in survival in humans as interactions with environmental factors may obscure the associations and prevent the identification of loci in humans.
Third, our study is based on common genetic variants and therefore we cannot exclude effects due to low frequency and rare variants (< 5%) or due to the presence of structural variation, such as copy number polymorphisms. Our discovery set may lack the power to identify all the relevant loci, even though we had sufficient power to detect common SNPs (MAF = 5% or more) with a relative risk of 1.10 (SI, Table S2).
Last, we cannot exclude that phenotypic heterogeneity influenced our findings. While all cohorts had prospectively-collected information on major health events and diagnoses, heterogeneity in the methods of assessment and classification might have limited the ability to identify true effects.
Complex diseases may result from the effects of a large number of low frequency variants, with substantial allelic heterogeneity at disease-causing loci (Pritchard, 2001, Pritchard and Cox, 2002, Swarbrick and Vaisse, 2003). Theoretical modelling that incorporates mutation, random genetic drift, and purifying selection suggests that many of the variants that affect complex traits may be in the 1–5% frequency range (Pritchard, 2001). Indeed, sequencing of candidate genes in an attempt to capture such low frequency variants, has led to the identification of rare variants with modest effects on body mass index (Ahituv, et al., 2007, Cone, 2000, Challis, et al., 2002), triglyceride levels (Romeo, et al., 2007), HDL- (Cohen, et al., 2004, Romeo, et al., 2007) and LDL-cholesterol levels (Cohen, et al., 2005, Cohen, et al., 2006, Kotowski, et al., 2006).
It is impossible to determine the functional variant of a gene by GWAS. Moreover, we cannot conclude from the location of a SNP that this variation is involved in the expression of the closest gene. However, our top results suggested a possible role of genes involved in neurological processes in human longevity and aging. Ten of the 22 suggestive associations identified in our analyses are in or near genes that are highly expressed in the brain (HECW2(Rotin and Kumar, 2009), HIP1(Blanpied, et al., 2003), BIN2, GRIA1), were previously related to the regulation of neuronal excitability and plasticity (KCNQ4(Van Eyken, et al., 2006), LMO4(Joshi, et al., 2009, Leuba, et al., 2004), GRIA1), and the maintenance of neural circuitry and synaptic plasticity(NETO1), or are associated with neurological diseases such as Alzheimer’s disease (LMO4(Leuba, et al., 2004), BIN2, GRIA1, GRIN2B) and amyotrophic lateral sclerosis (GRIN2B). In addition, 6 of the 22 SNPs were in close proximity to genes associated with other phenotypes of aging such as autophagy (ATG4C(Kenyon, 2010)), cancer (ATG4C(Maiuri, et al., 2009), HIP1(Bradley, et al., 2007), HECW2(Rotin and Kumar, 2009), VWA5A(Zhou, et al., 2009), MECOM), and mitochondrial depletion syndrome (SUCLA2). Notably, BIN2, ATG4C, KCNQ4, MECOM and SUCLA2 showed associations with both traits in our study.
Using the expression quantitative trait loci (eQTL) browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/) we detected eQTL associated with HIP1, COL5A1, LOC340156, and SMARCA2 in time to death only.
Interestingly, SNPs known to be associated with longevity and disease in the neighbourhood of APOE(Deelen, et al., 2011) or FOXO3A(Flachsbart, et al., 2009, Willcox, et al., 2008) only reached nominal significance (results not shown). These genes were originally identified in studies of centenarians; it is possible that our study of cohorts comprised of individuals from the general populations did not have sufficient statistical power to identify these genes with certainty.(Tan, et al., 2008)
While meta-analysis of GWAS has the power to detect small changes of allele frequencies between groups with the analyzed trait, true association signals may not be revealed based on a stringent genome-wide significance threshold. This situation, although limiting false positive findings, performs poorly in identifying false negatives as they may fall below the threshold. Network analyses using a less stringent significance threshold do not amend the overall negative finding of this study. However, it is well-recognized that within the many associations that failed to attain this level of significance lie true positive associations. Network analyses can provide useful information exploring multiple gene effects and their interactions.
In fact the interpretation of most GWAS results is difficult because individual results may involve many seemingly unrelated genes. Since PANTHER and IPA are built on different conceptual approaches, database sources and different pathway classifications, they can be seen as complementary approaches. Our pathway and network analyses highlighted neuronal activities and organism developmental processes as major biological processes involved aging. In addition, it highlighted Wnt signalling and showed that those genes that were involved in most pathways indeed had substantial effects within the analyzed trait. NTRK2(Rico, et al., 2002), NCAM1(Rutishauser, et al., 1988), GRID2(Hirai, et al., 2003), and RIMS1(Johnson, et al., 2003, Schoch, et al., 2002) are associated with neuronal development and disease pathways that were highlighted in the analysis of time to death. MYC(Cole, 1986, Goga, et al., 2007), E2F1(Nevins, 2001), EGFR(Wang, et al., 2004), and CEBPA(Menard, et al., 2002, Wang, et al., 2001) are associated with “cancer”, “cell function” and “development” pathways.
Few if any of the top hits from the GWAS were involved in common pathways of aging, typically addressed in candidate gene studies. For example, there was no specific evidence for genes involved in IGF-insulin signalling. However, this negative finding cannot be interpreted as evidence against the importance of IGF-insulin signalling, as well as other processes such as inflammation, oxidative stress, cellular damage and repair, growth hormone, and cell proliferation in aging. Moreover, it is possible that polymorphisms in related genes have an effect in the oldest old, who were represented by fewer numbers in our study population such that our study design would be underpowered to detect it. It is also conceivable that the neurological pathways identified by our analysis interact with the known candidate genes involved in aging (Bishop, et al., 2010, Finch and Ruvkun, 2001). It is feasible that the traditional aging pathways are hierarchically controlled by neurons and that the brain might be the location coordinating physiological changes (Bishop, et al., 2010, Finch and Ruvkun, 2001). Because neurons are particularly susceptible to damage caused by reactive oxygen species, limitations in cellular maintenance and repair might reinforce these pathways and accelerate aging (Finch and Ruvkun, 2001). An increased ability of neuronal cells to prevent or repair oxidative damage might result in beneficial hormonal signalling, otherwise deregulated with age, thus delaying the onset of age-related disease and directly regulating cognitive aging and life span (Bishop, et al., 2010, Cutler and Mattson, 2006, de Magalhaes and Sandberg, 2005).
In conclusion, our analysis did provide suggestive evidence that aging is under neuronal control. Unfortunately, we have no relevant tissue or expression experiment available to further underscore or validate our findings. Future investigations of changes of gene expression with age at cellular and population levels are warranted.
Supplementary Material
Acknowledgments
Funding Statement:
Rotterdam Study:
The Rotterdam Study is supported by Netherlands Genomics Initiative/Netherlands Consortium for Healthy Aging (050-060-810); Netherlands Organisation for Scientific Research (NWO) (904-61-090, 904-61-193, 480-04-004, 400-05-717, SPI 56-464-1419, 175.010.2005.011, 911-03-012 and 017.106.370); Netspar – Living longer for a good health; the Erasmus Medical Center and Erasmus University in Rotterdam; the Netherlands Organization for Health Research and Development; the Netherlands Research Institute for Diseases in the Elderly; the Dutch Ministry of Education, Culture and Science, and the Ministry for Health, Welfare and Sports; the European Commission; and the Municipality of Rotterdam, the Netherlands.
Cardiovascular Health Study:
The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, the National Institute of Aging R01 AG031890 with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01-RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Framingham Heart Study:
The Framingham Study phenotype-genotype analyses were supported by the National Institute of Aging grant number R01AG029451 (PI: JMM). “The Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine were supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study Contract No. N01-HC-25195 and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Dr. Kiel’s effort as well as all hip fracture data from the Framingham Osteoporosis Study was supported by a grant from the National Institute of Arthritis, Musculoskeletal and Skin Diseases and the National Institute on Aging; R01 AR/AG 41398. This research was additionally supported by the following grants: AG033193, AG081220, NS17950, P30AG013846, 1R01AG028321.
Atherosclerosis Risk in Communities Study:
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022. The authors thank the staff and participants of the ARIC study for their important contributions.
Age, Gene/Environment Susceptibility -Reykjavik Study:
The Age, Gene/Environment Susceptibility-Reykjavik Study is funded by NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). Genotyping was conducted at the NIA IRP Laboratory of Neurogenetics.
Invecchiare nel Chianti:
The InCHIANTI study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336).
Baltimore Longitudinal Study of Aging:
The BLSA was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. A portion of that support was through a R&D contract with MedStar Research Institute.
Health, Aging and Body Composition:
This research is supported in part by the Intramural Research Program of the NIH, National Institute on Aging. This research was supported by NIA contracts N01AG62101, N01AG62103, N01AG62106 and NIA grant 1R03AG032498-01. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C.
Study of Health in Pomerania:
SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg- West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG.
Replication Samples:
Whitehall II:
Whitehall II has been supported by grants from the Medical Research Council; Economic and Social Research Council; British Heart Foundation; Health and Safety Executive; Department of Health; National Heart Lung and Blood Institute (HL36310), US, NIH: National Institute on Aging (AG13196), US, NIH; Agency for Health Care Policy Research (HS06516); and the John D and Catherine T MacArthur Foundation Research Networks on Successful Midlife Development and Socio-economic Status and Health.
English Longitudinal Study of Aging:
Samples from the English Longitudinal Study of Ageing (ELSA) DNA Repository (EDNAR), received support under a grant (AG1764406S1) awarded by the National Institute on Ageing (NIA). ELSA was developed by a team of researchers based at the National Centre for Social Research, University College London and the Institute of Fiscal Studies. The data were collected by the National Centre for Social Research.
Religious Order Study:
Grants P30AG10161, R01AG15819, and R01AG30146 from the National Instiute on Aging, and the Translation Genomics Research Institute.
Memory and Aging Project:
Grants R01AG17917 and R01AG15819 from the National Instiute on Aging, and the Translation Genomics Research Institute.
The Longevity Consortium, funded by the National Institute of Aging, grant number U19 AG023122, provided administrative resources to CHARGE investigators for this phenotype as well as scientific opportunity funds for de novo genotyping of the five selected SNPs in the ELSA and Whitehall II replication studies.
Footnotes
Disclosure Statement
The authors declare that no competing interests exist. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Ahituv N, Kavaslar N, Schackwitz W, Ustaszewska A, Martin J, Hebert S, Doelle H, Ersoy B, Kryukov G, Schmidt S, Yosef N, Ruppin E, Sharan R, Vaisse C, Sunyaev S, Dent R, Cohen J, McPherson R, Pennacchio LA. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80(4):779–91. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12(2):95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- Atzmon G, Schechter C, Greiner W, Davidson D, Rennert G, Barzilai N. Clinical phenotype of families with longevity. J Am Geriatr Soc. 2004;52(2):274–7. doi: 10.1111/j.1532-5415.2004.52068.x. 52068 [pii] [DOI] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464(7288):529–35. doi: 10.1038/nature08983. nature08983 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Scott DB, Ehlers MD. Age-related regulation of dendritic endocytosis associated with altered clathrin dynamics. Neurobiol Aging. 2003;24(8):1095–104. doi: 10.1016/j.neurobiolaging.2003.04.004. S0197458003001751 [pii] [DOI] [PubMed]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317(5839):807–10. doi: 10.1126/science.1144090. 317/5839/807 [pii] [DOI] [PubMed] [Google Scholar]
- Bradley SV, Holland EC, Liu GY, Thomas D, Hyun TS, Ross TS. Huntingtin interacting protein 1 is a novel brain tumor marker that associates with epidermal growth factor receptor. Cancer Res. 2007;67(8):3609–15. doi: 10.1158/0008-5472.CAN-06-4803. 67/8/3609 [pii] [DOI] [PubMed] [Google Scholar]
- Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–5. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103(6):1810–5. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MD. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–84. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Cone RD. Haploinsufficiency of the melanocortin-4 receptor: part of a thrifty genotype? J Clin Invest. 2000;106(2):185–7. doi: 10.1172/JCI10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler RG, Mattson MP. The adversities of aging. Ageing Res Rev. 2006;5(3):221–38. doi: 10.1016/j.arr.2006.05.002. S1568-1637(06)00047-X [pii] [DOI] [PubMed] [Google Scholar]
- Challis BG, Pritchard LE, Creemers JW, Delplanque J, Keogh JM, Luan J, Wareham NJ, Yeo GS, Bhattacharyya S, Froguel P, White A, Farooqi IS, O’Rahilly S. A missense mutation disrupting a dibasic prohormone processing site in pro-opiomelanocortin (POMC) increases susceptibility to early-onset obesity through a novel molecular mechanism. Hum Mol Genet. 2002;11(17):1997–2004. doi: 10.1093/hmg/11.17.1997. [DOI] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7(6):436–48. doi: 10.1038/nrg1871. nrg1871 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17(R2):R122–8. doi: 10.1093/hmg/ddn288. ddn288 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Budovsky A, Lehmann G, Costa J, Li Y, Fraifeld V, Church GM. The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell. 2009a;8(1):65–72. doi: 10.1111/j.1474-9726.2008.00442.x. ACE442 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Finch CE, Janssens G. Next-generation sequencing in aging research: Emerging applications, problems, pitfalls and possible solutions. Ageing Res Rev. 2009b doi: 10.1016/j.arr.2009.10.006. S1568-1637(09)00074-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Sandberg A. Cognitive aging as an extension of brain development: a model linking learning, brain plasticity, and neurodegeneration. Mech Ageing Dev. 2005;126(10):1026–33. doi: 10.1016/j.mad.2005.04.004. S0047-6374(05)00110-7 [pii] [DOI] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van de Breggen R, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert J, Lawler E, Bogan H, Perls T. Morbidity profiles of centenarians: survivors, delayers, and escapers. J Gerontol A Biol Sci Med Sci. 2003;58(3):232–7. doi: 10.1093/gerona/58.3.m232. [DOI] [PubMed] [Google Scholar]
- Ewbank DC. Differences in the association between apolipoprotein E genotype and mortality across populations. J Gerontol A Biol Sci Med Sci. 2007;62(8):899–907. doi: 10.1093/gerona/62.8.899. 62/8/899 [pii] [DOI] [PubMed] [Google Scholar]
- Finch CE, Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet. 2001;2:435–62. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106(8):2700–5. doi: 10.1073/pnas.0809594106. 0809594106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes LU, Jeune B, Ranberg KA, Nybo H, Vaupel JW. Estimation of apolipoprotein E genotype-specific relative mortality risks from the distribution of genotypes in centenarians and middle-aged men: apolipoprotein E gene is a “frailty gene,” not a “longevity gene”. Genet Epidemiol. 2000;19(3):202–10. doi: 10.1002/1098-2272(200010)19:3<202::AID-GEPI2>3.0.CO;2-Q. [pii] 10.1002/1098-2272(200010)19:3<202::AID-GEPI2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13(7):820–7. doi: 10.1038/nm1606. nm1606 [pii] [DOI] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97(3):319–23. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- Hirai H, Launey T, Mikawa S, Torashima T, Yanagihara D, Kasaura T, Miyamoto A, Yuzaki M. New role of delta2-glutamate receptors in AMPA receptor trafficking and cerebellar function. Nat Neurosci. 2003;6(8):869–76. doi: 10.1038/nn1086. nn1086 [pii] [DOI] [PubMed] [Google Scholar]
- Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci. 2010;11(2):77–86. doi: 10.1038/nrn2755. nrn2755 [pii] [DOI] [PubMed] [Google Scholar]
- Johnson S, Halford S, Morris AG, Patel RJ, Wilkie SE, Hardcastle AJ, Moore AT, Zhang K, Hunt DM. Genomic organisation and alternative splicing of human RIM1, a gene implicated in autosomal dominant cone-rod dystrophy (CORD7) Genomics. 2003;81(3):304–14. doi: 10.1016/s0888-7543(03)00010-7. S0888754303000107 [pii] [DOI] [PubMed] [Google Scholar]
- Joshi K, Lee S, Lee B, Lee JW, Lee SK. LMO4 controls the balance between excitatory and inhibitory spinal V2 interneurons. Neuron. 2009;61(6):839–51. doi: 10.1016/j.neuron.2009.02.011. S0896-6273(09)00123-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble TR, Halford MM, Seaman C, Kee N, Macheda M, Anderson RB, Stacker SA, Cooper HM. The Wnt receptor Ryk is required for Wnt5a-mediated axon guidance on the contralateral side of the corpus callosum. J Neurosci. 2006;26(21):5840–8. doi: 10.1523/JNEUROSCI.1175-06.2006. 26/21/5840 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–12. doi: 10.1038/nature08980. nature08980 [pii] [DOI] [PubMed] [Google Scholar]
- Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78(3):410–22. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuningas M, Mooijaart SP, van Heemst D, Zwaan BJ, Slagboom PE, Westendorp RG. Genes encoding longevity: from model organisms to humans. Aging Cell. 2008;7(2):270–80. doi: 10.1111/j.1474-9726.2008.00366.x. ACE366 [pii] [DOI] [PubMed] [Google Scholar]
- Leuba G, Vernay A, Vu D, Walzer C, Belloir B, Kraftsik R, Bouras C, Savioz A. Differential expression of LMO4 protein in Alzheimer’s disease. Neuropathol Appl Neurobiol. 2004;30(1):57–69. doi: 10.1046/j.0305-1846.2003.00511.x. 511 [pii] [DOI] [PubMed] [Google Scholar]
- Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009 doi: 10.1038/ng.384. ng.384 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, Li W, Zheng GY, Cui H, Chen X, Zhu Z, He H, Dong B, Mo X, Zeng Y, Tian XL. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009a;18(24):4897–904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009b;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetta KL, D’Agostino RB, Sr, Karasik D, Benjamin EJ, Guo CY, Govindaraju R, Kiel DP, Kelly-Hayes M, Massaro JM, Pencina MJ, Seshadri S, Murabito JM. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1):S13. doi: 10.1186/1471-2350-8-S1-S13. 1471-2350-8-S1-S13 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16(1):87–93. doi: 10.1038/cdd.2008.131. cdd2008131 [pii] [DOI] [PubMed] [Google Scholar]
- McGue M, Vaupel JW, Holm N, Harvald B. Longevity is moderately heritable in a sample of Danish twins born 1870–1880. J Gerontol. 1993;48(6):B237–44. doi: 10.1093/geronj/48.6.b237. [DOI] [PubMed] [Google Scholar]
- Menard C, Hein P, Paquin A, Savelson A, Yang XM, Lederfein D, Barnabe-Heider F, Mir AA, Sterneck E, Peterson AC, Johnson PF, Vinson C, Miller FD. An essential role for a MEK-C/EBP pathway during growth factor-regulated cortical neurogenesis. Neuron. 2002;36(4):597–610. doi: 10.1016/s0896-6273(02)01026-7. S0896627302010267 [pii] [DOI] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35(Database issue):D247–52. doi: 10.1093/nar/gkl869. gkl869 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10(7):699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Newman AB, Walter S, Lunetta KL, Garcia ME, Slagboom PE, Christensen K, Arnold AM, Aspelund T, Aulchenko YS, Benjamin EJ, Christiansen L, D’Agostino RB, Sr, Fitzpatrick AL, Franceschini N, Glazer NL, Gudnason V, Hofman A, Kaplan R, Karasik D, Kelly-Hayes M, Kiel DP, Launer LJ, Marciante KD, Massaro JM, Miljkovic I, Nalls MA, Hernandez D, Psaty BM, Rivadeneira F, Rotter J, Seshadri S, Smith AV, Taylor KD, Tiemeier H, Uh HW, Uitterlinden AG, Vaupel JW, Walston J, Westendorp RG, Harris TB, Lumley T, van Duijn CM, Murabito JM. A Meta-analysis of Four Genome-Wide Association Studies of Survival to Age 90 Years or Older: The Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J Gerontol A Biol Sci Med Sci. 2010 doi: 10.1093/gerona/glq028. glq028 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009 doi: 10.1038/ng.361. ng.361 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8(4):460–72. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. ng1847 [pii] [DOI] [PubMed] [Google Scholar]
- Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69(1):124–37. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11(20):2417–23. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. 2/1/73 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed T, Dick DM. Heritability and validity of healthy physical aging (wellness) in elderly male twins. Twin Res. 2003;6(3):227–34. doi: 10.1375/136905203765693889. [DOI] [PubMed] [Google Scholar]
- Rico B, Xu B, Reichardt LF. TrkB receptor signaling is required for establishment of GABAergic synapses in the cerebellum. Nat Neurosci. 2002;5(3):225–33. doi: 10.1038/nn808. nn808 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39(4):513–6. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10(6):398–409. doi: 10.1038/nrm2690. nrm2690 [pii] [DOI] [PubMed] [Google Scholar]
- Rutishauser U, Acheson A, Hall AK, Mann DM, Sunshine J. The neural cell adhesion molecule (NCAM) as a regulator of cell-cell interactions. Science. 1988;240(4848):53–7. doi: 10.1126/science.3281256. [DOI] [PubMed] [Google Scholar]
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Sudhof TC. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415(6869):321–6. doi: 10.1038/415321a. 415321a [pii] [DOI] [PubMed] [Google Scholar]
- Swarbrick MM, Vaisse C. Emerging trends in the search for genetic variants predisposing to human obesity. Curr Opin Clin Nutr Metab Care. 2003;6(4):369–75. doi: 10.1097/01.mco.0000078997.96795.03. [DOI] [PubMed] [Google Scholar]
- Tan Q, Zhao JH, Zhang D, Kruse TA, Christensen K. Power for genetic association study of human longevity using the case-control design. Am J Epidemiol. 2008;168(8):890–6. doi: 10.1093/aje/kwn205. kwn205 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–41. doi: 10.1101/gr.772403. 13/9/2129 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34(Web Server issue):W645–50. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa F, Marti E. Wnt won the war: antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Dev Dyn. 2010;239(1):69–76. doi: 10.1002/dvdy.22058. [DOI] [PubMed] [Google Scholar]
- Van Eyken E, Van Laer L, Fransen E, Topsakal V, Lemkens N, Laureys W, Nelissen N, Vandevelde A, Wienker T, Van De Heyning P, Van Camp G. KCNQ4: a gene for age-related hearing impairment? Hum Mutat. 2006;27(10):1007–16. doi: 10.1002/humu.20375. [DOI] [PubMed] [Google Scholar]
- van Rijn MJ, Schut AF, Aulchenko YS, Deinum J, Sayed-Tabatabaei FA, Yazdanpanah M, Isaacs A, Axenovich TI, Zorkoltseva IV, Zillikens MC, Pols HA, Witteman JC, Oostra BA, van Duijn CM. Heritability of blood pressure traits and the genetic contribution to blood pressure variance explained by four blood-pressure-related genes. J Hypertens. 2007;25(3):565–70. doi: 10.1097/HJH.0b013e32801449fb. 00004872-200703000-00013 [pii] [DOI] [PubMed] [Google Scholar]
- Vaupel JW. The remarkable improvements in survival at older ages. Philos Trans R Soc Lond B Biol Sci. 1997;352(1363):1799–804. doi: 10.1098/rstb.1997.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel JW. Biodemography of human ageing. Nature. 2010;464(7288):536–42. doi: 10.1038/nature08984. nature08984 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426(6967):620. doi: 10.1038/426620a. 426620a [pii] [DOI] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Genetics of longevity and aging. Annu Rev Med. 2005;56:193–212. doi: 10.1146/annurev.med.56.082103.104617. [DOI] [PubMed] [Google Scholar]
- Wang H, Iakova P, Wilde M, Welm A, Goode T, Roesler WJ, Timchenko NA. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8(4):817–28. doi: 10.1016/s1097-2765(01)00366-5. S1097-2765(01)00366-5 [pii] [DOI] [PubMed] [Google Scholar]
- Wang K, Yamamoto H, Chin JR, Werb Z, Vu TH. Epidermal growth factor receptor-deficient mice have delayed primary endochondral ossification because of defective osteoclast recruitment. J Biol Chem. 2004;279(51):53848–56. doi: 10.1074/jbc.M403114200. M403114200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;1050801030105 [pii](37):13987–92. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YQ, Chen SL, Ju JY, Shen L, Liu Y, Zhen S, Lv N, He ZG, Zhu LP. Tumor suppressor function of BCSC-1 in nasopharyngeal carcinoma. Cancer Sci. 2009;100(10):1817–22. doi: 10.1111/j.1349-7006.2009.01261.x. CAS1261 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.