Abstract
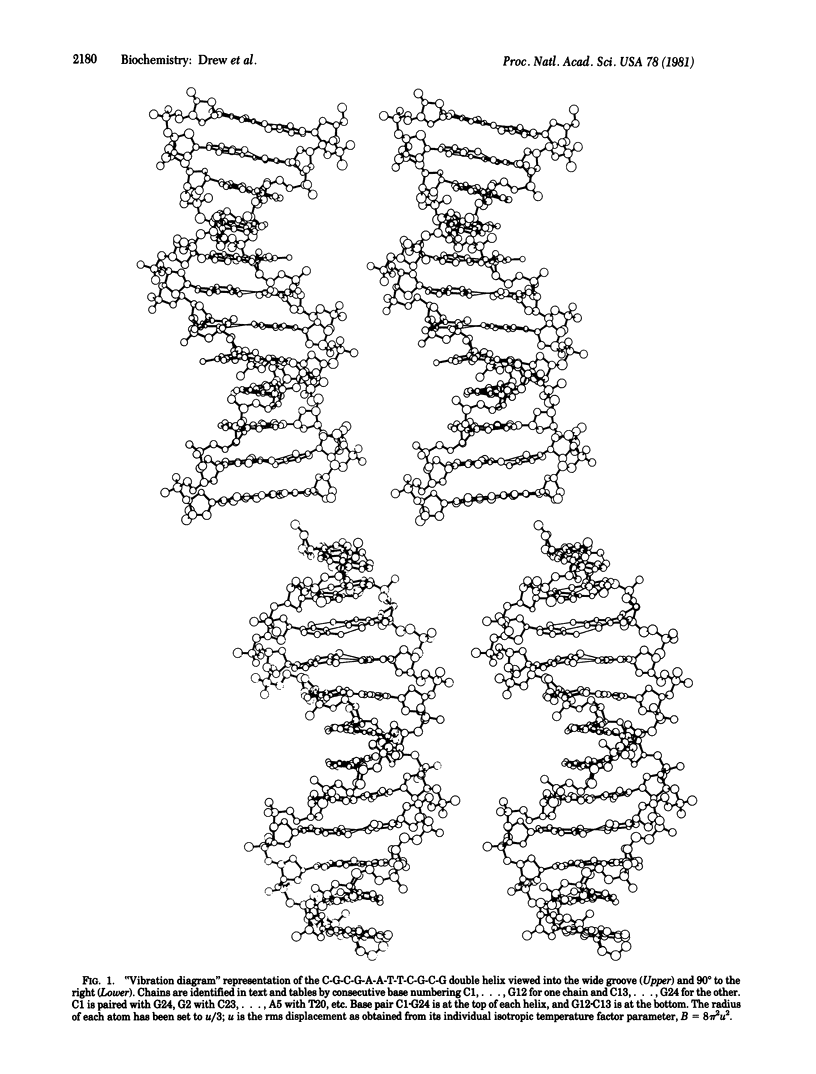
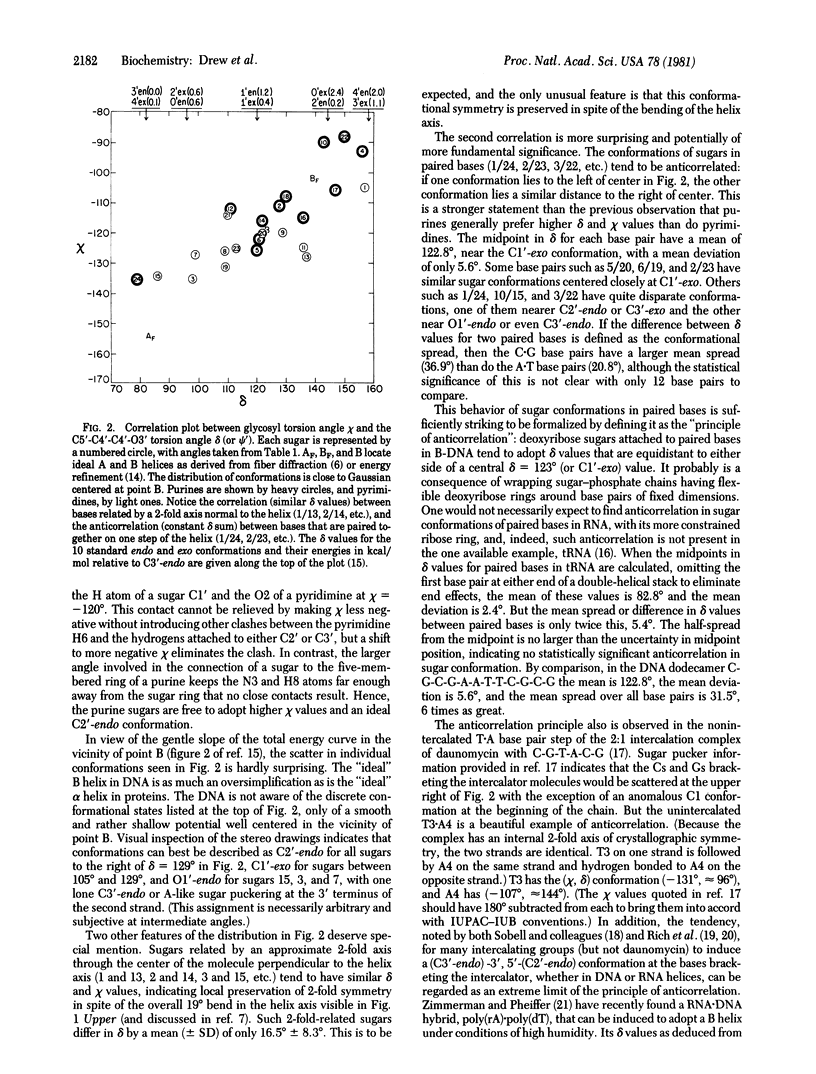
The crystal structure of the synthetic DNA dodecamer d(CpGpCpGpApApTpTpCpGpCpG) has been refined to a residual error of R = 17.8% at 1.9-A resolution (two-sigma data). The molecule forms slightly more than one complete turn of right-handed double-stranded B helix. The two ends of the helix overlap and interlock minor grooves with neighboring molecules up and down a 2(1) screw axis, producing a 19 degrees bend in helix axis over the 11-base-pair steps of the dodecamer. In the center of the molecule, where perturbation is least, the helix has a mean rotation of 36.9 degrees per step, or 9.8 base pairs per turn. The mean propeller twist (total dihedral angle between base planes) between A . T base pairs in the center of the molecule is 17.3 degrees, and that between C . G pairs on the two ends averages 11.5 degrees. Individual deoxyribose ring conformations as measured by the C5'-C4'-C3'-O3' torsion angle delta, exhibit an approximately Gaussian distribution centered around the C1'-exo position with delta avg = 123 degrees and a range of 79 degrees to 157 degrees. Purine sugars cluster at high delta values, and pyrimidine sugars cluster at lower delta. A tendency toward 2-fold symmetry in sugar conformation about the center of the molecule is detectable in spite of the destruction of ideal 2-fold symmetry by the molecular bending. More strikingly, sugar conformations of paired based appear to follow a "principle of anticorrelation," with delta values lying approximately the same distance to either side of the center value, delta = 123 degrees. This same anticorrelation is also observed in other DNA and DNA . RNA structures.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. The conformation of C-DNA. J Mol Biol. 1975 Oct 15;98(1):265–269. doi: 10.1016/s0022-2836(75)80115-x. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- FULLER W., WILKINS M. H., WILSON H. R., HAMILTON L. D. THE MOLECULAR CONFIGURATION OF DEOXYRIBONUCLEIC ACID. IV. X-RAY DIFFRACTION STUDY OF THE A FORM. J Mol Biol. 1965 May;12:60–76. doi: 10.1016/s0022-2836(65)80282-0. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Jardetzky O. Internal motions in deoxyribonucleic acid II. Biochemistry. 1980 Jul 22;19(15):3460–3468. doi: 10.1021/bi00556a009. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Jain S. C., Tsai C. C., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. II. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodocytidylyl (3'-5') guanosine. J Mol Biol. 1977 Aug 15;114(3):317–331. doi: 10.1016/0022-2836(77)90253-4. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Wang A. H., Ughetto G., van der Marel G., van Boom J. H., Rich A. Molecular structure of an anticancer drug-DNA complex: daunomycin plus d(CpGpTpApCpG). Proc Natl Acad Sci U S A. 1980 Dec;77(12):7204–7208. doi: 10.1073/pnas.77.12.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. A RNA.DNA hybrid that can adopt two conformations: an x-ray diffraction study of poly(rA).poly(dT) in concentrated solution or in fibers. Proc Natl Acad Sci U S A. 1981 Jan;78(1):78–82. doi: 10.1073/pnas.78.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]



