Abstract
Background
Numerous features are derived from the asymmetry, border irregularity, color variegation, and diameter of the skin lesion of dermatology for diagnosing malignant melanoma. Feature selection for the development of automated skin lesion discrimination systems is an important consideration.
Methods
In this research, a systematic heuristic approach is investigated for feature selection and lesion classification. The approach integrates statistical-, correlation-, histogram-, and expert system-based components. Using statistical and correlation measures, interrelationships among features are determined. Expert system analysis is performed to identify redundant features. The feature selection process is applied to 19 shape and color features for a clinical image data set containing 355 malignant melanomas, 125 basal cell carcinomas, 177 dysplastic nevi, 199 nevocellular nevi, 139 seborrheic keratoses, and 45 vascular lesions.
Results
Experimental results show reduced lesion classification error rates based on condensing the shape and color feature set from 19 features to 13 features using the feature selection process. Specifically, average test lesion classification error rates for discriminating malignant melanoma from non-melanoma lesions were reduced from 26.6% for 19 features to 23.2% for 13 features over five randomly generated training and test sets.
Conclusions
The experimental results show that the systematic heuristic approach for feature reduction can be successfully applied to achieve improved lesion discrimination. The feature reduction technique facilitates the elimination of redundant information that may inhibit lesion classification performance. The clinical application of this result is that automated skin lesion classification algorithm development can be fostered with systematic feature selection techniques.
Keywords: color analysis, image analysis, malignant melanoma, skin lesion
Physicians’ low accuracy in diagnosing pigmented lesions has been well documented (1–4). In three previous studies of diagnostic accuracy, sensitivity for malignant melanoma detection by dermatologists without aids such as dermoscopy was 89%, 77%, and 81% (2–4). Many physicians rely on guidelines such as asymmetry, border irregularity, color variegation, and diameter of the skin lesion, called the ABCD criteria (5). Conventional educational programs can improve diagnostic accuracy, but some discriminations such as melanoma vs. dysplastic nevi remain difficult (6).
Many factors affect the success of automated melanoma recognition systems. The quality of the data is one such factor. If information is irrelevant or redundant, or the data are noisy and unreliable, then knowledge discovery during training is more difficult. The performance of the classification process depends on the quality of the features used in the process. So feature selection is often an essential data-processing step prior to applying a classification algorithm. Feature subset selection (feature reduction) is the process of identifying and removing as much of the irrelevant and redundant information as possible. The purpose of this feature subset selection is to select the smallest subset of features without degrading lesion classification performance. By carrying out feature selection before feeding the whole feature set into a classification system, the performance of the classification system can often be improved. In this research, shape and color descriptors for clinical skin lesion images are evaluated to determine which features contribute most successfully to discriminating melanoma from benign lesions. Statistical-, histogram-, and rule-based approaches are utilized for feature evaluation and feature set reduction to improve lesion discrimination.
Numerous computer-based techniques have been applied to pigmented lesion images, both clinical and dermoscopy, for investigating features to detect malignant melanoma (7–25). In the research presented here, skin lesion shape and color feature analysis is examined for clinical images. Skin lesion border irregularity has been analyzed by Golston et al. (13) using circular indices. Skin lesion asymmetry has been analyzed by Stoecker et al. (12). Irregularity and asymmetry measures have been reported to correlate with dermatologists’ measures of these features in 92% and 93% of instances, respectively (12, 13). Clinical examination of color provides discriminating information in the diagnosis of malignant melanoma (5). There are colors generally associated with melanocytic lesions, including shades of tan, brown, or black and occasional patches of red, white, or blue. Colors characteristic of melanoma can be represented using various color spaces (11, 17). Several color descriptors have been applied to melanoma detection or discrimination, including variation of hues (8), analytical color techniques for detecting color variegation (17), and RGB color channel statistical parameters (23–25). Ercal et al. (11) utilized relative chromaticity, spherical color coordinates, and (L, a*, b*) color coordinate features as part of an overall neural network approach for melanoma detection. Ganster et al. (14) also used a neural network-based melanoma detection scheme that utilized absolute, unnormalized color-based percent melanoma color features for melanoma color discrimination in dermoscopy images. The color features included the percentages of absolute color shades of reddish, bluish, grayish, and blackish areas found within the skin lesion as well as the number of these colors present within the skin lesion. Color quantization was performed using the median-cut color quantization algorithm (26).
The remainder of the paper is organized as follows: (1) description of skin lesion features, (2) feature evaluation experiments performed, (3) experimental results, (4) discussion, and (5) conclusions.
Skin Lesion Features
Based on the clinical ABCDs for melanoma, 19 shape and color features have been examined and evaluated for discriminating melanoma from benign lesions. Table 1 provides a listing of shape and color features examined in this research. For feature calculations, skin lesion borders were determined using manually chosen points along the border that were joined with a least mean squares distance second-order spline curve. A dermatologist working with a group of students found all borders, making every attempt to find accurate borders, in spite of the absence of a known gold standard for border accuracy. Using the manually determined borders, shape, and color features were computed as follows.
TABLE 1.
Shape and color features examine for lesion discrimination
| Shape feature | Label | Color feature | Label |
|---|---|---|---|
| Border irregularity index | F1 | Percent melanoma color | F4 |
| Asymmetry index | F2 | Percent non-melanoma color | F5 |
| Solid pigment asymmetry index | F3 | Color clustering ratio | F6 |
| Variance red, green, blue | F7–F9 | ||
| Relative chromaticity of red, green, blue | F10–F12 | ||
| Ratio in red, green, blue | F13–F15 | ||
| Difference in lightness, chroma, color, hue | F16–F19 |
Shape Features
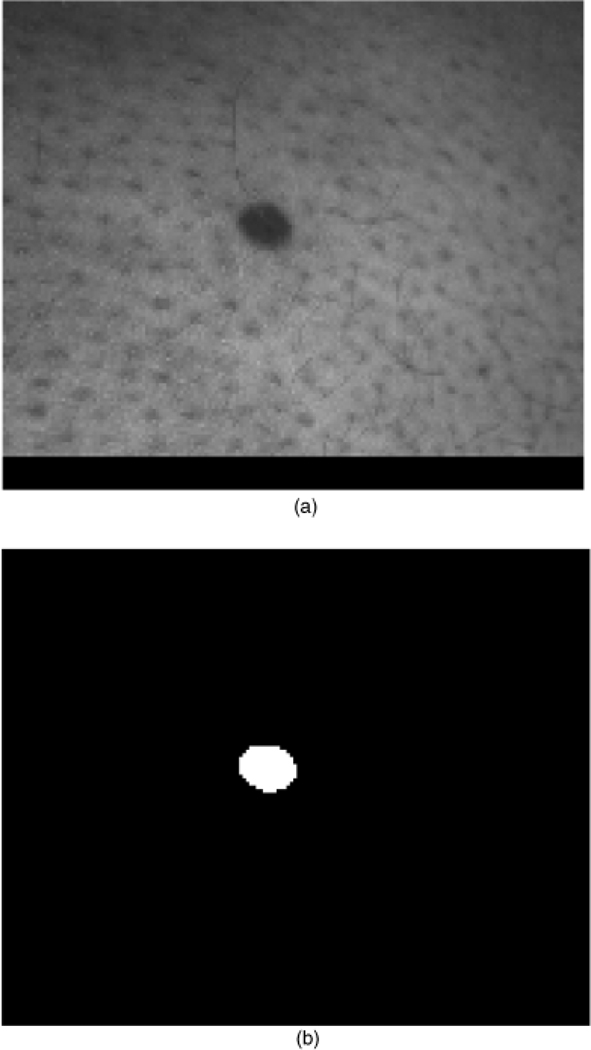
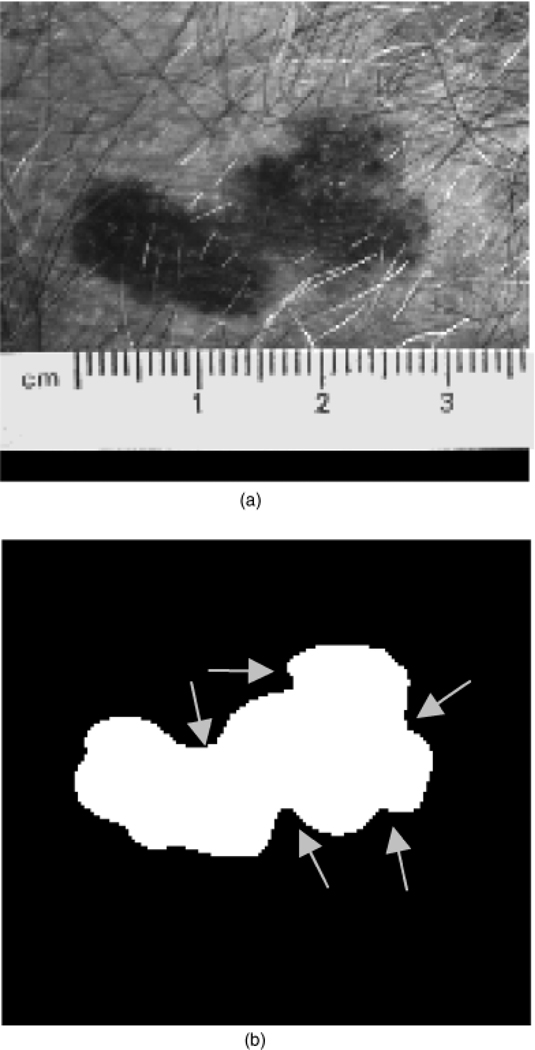
The shape features examined in this research are based on quantifying border irregularity and lesion asymmetry. The lesion in Fig. 1, a nevocellular nevus, is an example of a symmetric, regular lesion. Notice that it is more or less elliptical, without many protrusions. The lesion in Fig. 2 is an example of an irregular lesion, an invasive melanoma, with the border containing indentations and notches, often indicative of malignant melanoma.
Fig. 1.
A regular skin lesion (nevocellular nevus) (a) and corresponding manually segmented lesion (b).
Fig. 2.
An irregular skin lesion (invasive melanoma) (a) and corresponding manually segmented lesion (b). The arrows points to notches and indentions in the lesion border.
Border irregularity index
In order to quantify border irregularity, an irregularity index is computed for each lesion. The irregularity index is given by
| (1) |
where a and b are the lengths of the major and minor axes of the best-fit ellipse, respectively, P is the perimeter of the lesion border, and A is the area of the lesion (26). The correction factor provides the same index for similarly shaped lesions at different sizes. In preliminary studies using this index, dermatologists’ assessment in 60 clinical image lesions yielded two errors.
Lesion asymmetry measure
The second shape feature investigated is a lesion asymmetry measure. From Figs 1 and 2, there is an axis about which a symmetrical lesion can be folded so the two halves match. The symmetrical lesion in Fig. 1 is relatively elliptical and can be folded along its principal axis of inertia with little difference between the two halves of the lesion. Folding the melanoma lesion in Fig. 2 along its principal axis of inertia provides a significant disparity between the two halves of the lesion, compared with the symmetrical lesion in Fig. 1. Accordingly, an asymmetry measure is defined based on computing the principal axis of inertia and reflecting half of the lesion across its principal axis. Let AX denote the exclusive OR area between the two halves of the lesion resulting from the folding process. Then, the asymmetry measure is given by
| (2) |
where At is the lesion area.
Solid pigment asymmetry index
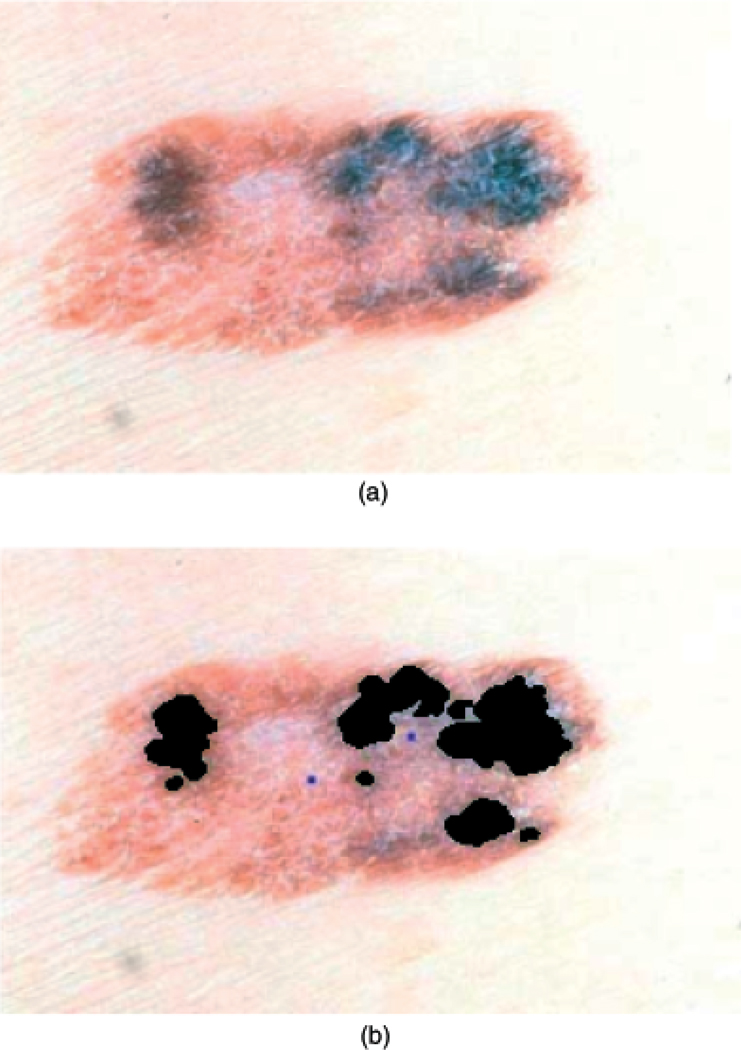
The third shape feature examined addresses symmetry of solid pigment within the lesion, which is quantified in the pigment asymmetry index (27). In some regions of the lesion, the pigment network under the epidermis is dominant, imparting a dark color to the lesion. The incidence of asymmetric patches of dark pigment is indicative of the irregular growth of melanin cells that is characteristic of melanoma. Using histogram-based thresholding of the luminance image, solid pigment areas are identified within the lesion. An example of solid pigmented region segmentation is shown in Fig. 3. Figure 3 presents the original lesion in (a) and the segmented solid pigment regions in (b) shown in black. The pigment asymmetry index is used to quantify the symmetry of the location of the solid pigmented regions within the lesion.
Fig. 3.
Solid pigment asymmetry lesion example (invasive melanoma). (a) Original lesion. (b) Marked solid pigment regions (shown in black).
The following procedure is used to compute the pigment asymmetry index. Rather than simply compute the distance of pigment centroids from lesion centroids, pigment asymmetry is computed by quadrants. This gives extra weighting to two-axis asymmetry, which is more indicative of melanoma than single-axis asymmetry. First, compute the first-order, second-order, and product inertia moments for the segmented lesion. Let mx and mxx represent the first- and second-order moments with respect to the horizontal axis, respectively, and my and myy denote the first- and second-order moments with respect to the vertical axis, respectively. Let mxy represent the product inertia for the lesion based on the manually segmented border. Second, computing the axes of inertia and the angle of elevation of the axis of symmetry for the lesion. Third, partition the lesion into four quadrants based on the principal axes of inertia. Fourth, compute the area and centroid location of the lesion within each quadrant. For quadrant i (i = 1, …,4), let denote the area and centroid of the lesion region contained in quadrant i, respectively. Fifth, compute the area and centroid location of the segmented dark pigment region within each quadrant. For quadrant i, let denote the area and centroid of the segmented dark pigment region contained in quadrant i, respectively. Sixth, compute the distance from the lesion and dark pigment centroid locations in each quadrant to the centroid of the entire lesion. Let represent the Euclidean distances from the centroids of the lesion and dark pigment in quadrant i, respectively, to the centroid of the entire lesion (mx, my). Seventh, compute the pigment asymmetry index for each quadrant. Let λi represent the asymmetry index for quadrant i, which is given by
| (3) |
Finally, determine the pigment asymmetry index (α) for the lesion as the variance of the pigment asymmetry indices given by
| (4) |
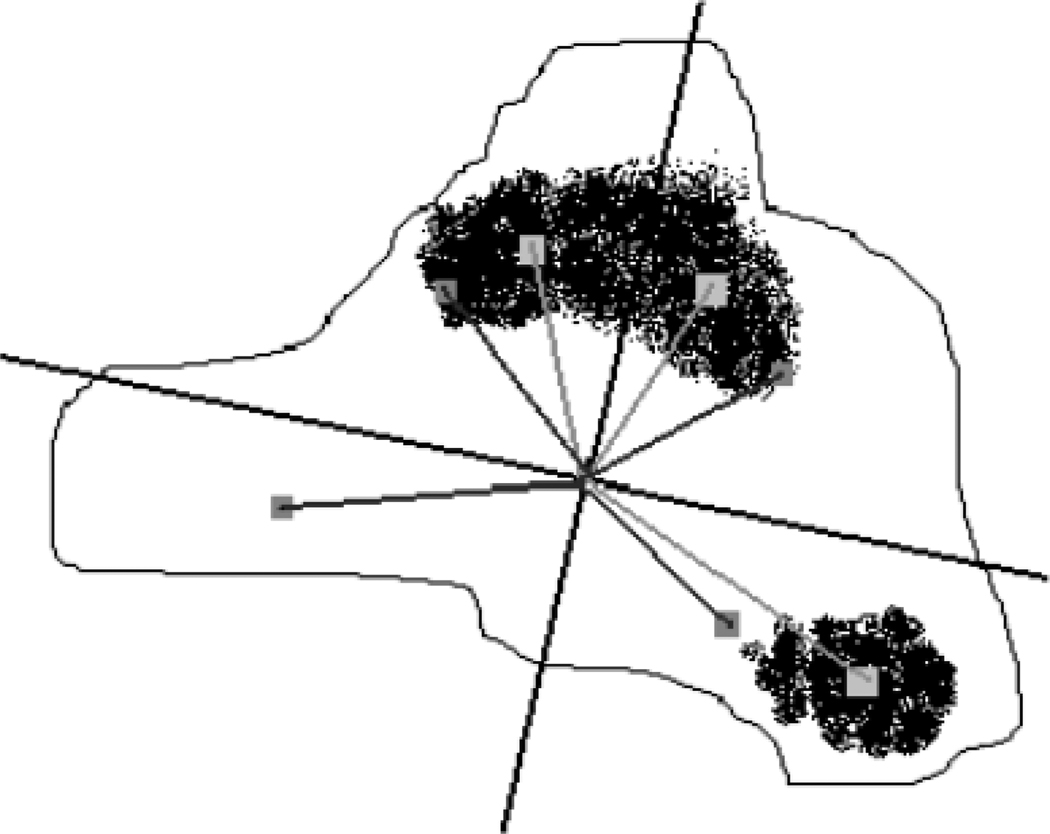
where µi is the mean pigment asymmetry index over the four (N = 4) quadrants. Figure 4 gives a visual representation of the pigment asymmetry feature based on identifying the lesion and dark pigment regions in each quadrant.
Fig. 4.
Pigment asymmetry calculation example. The principal axes of inertia are shown along with the quadrant lesion centroids (boxes in dark gray) and quadrant pigment dark pigment centroids (boxes in light gray).
Color Features
From the ABCDs of dermatology, color is an important discriminator of malignant melanoma from benign lesions. Table 1 lists the color features evaluated in this research, which are presented in the following sections.
Relative color and surrounding skin color
The relative color of a skin lesion pixel is the difference between the actual pixel value and the average color value of representative surrounding skin. For a Y row by X column RGB image, I, the actual value of each pixel is given by I(x, y) = (r(x, y), g(x, y), b(x, y)), where 1 ≤ x ≤ X and 1 ≤ y ≤ Y. The algorithm to determine the surrounding skin color has been applied in other research (28, 29). Surrounding skin color is represented as the average RGB value of pixels outside the lesion but within a circular region with center point at the lesion centroid. The circular region size is dependent on the skin lesion size (28, 29). Within the surrounding skin region, non-skin colored pixels such as from clothing, hair, teeth, direct camera flash reflections, shadows, etc., must be removed in order to achieve a reliable sample of the surrounding skin color. Based on prior research for skin lesion border segmentation, normal skin color appears to fall within a small region of the RGB color space with R>80, R>G, and R>B (17). Let O denote the skin lesion region within the color clinical image, defined as
| (5) |
Then, the relative color for all skin lesion pixels is given as
| (6) |
where −255 ≤ rrel(x, y), grel(x, y), brel(x, y) ≤ 255. rskin, gskin, and bskin are the average RGB values computed from the surrounding skin. Thus, the relative color space is of size 511 × 511 × 511, where each (rrel, grel, brel) refers to a bin in a relative color histogram. Requantizing the relative RGB histogram bins is performed in order to represent discrete ranges of colors that are characteristic of melanoma and benign lesions. The resolution of the relative color histogram is reduced by combining 4 × 4 × 4 blocks of bins into a single bin, making the resulting histogram of size 128 × 128 × 128. The range of requantized relative color bins for each color is −63 to 64. Each relative color bin contains 64 relative colors (4 × 4 × 4) except for the bin (−63, −63, −63), which contains 27 relative colors (3 × 3 × 3). This irregularity in bin size causes no problem because those relative colors are infrequently observed in skin lesions.
Percent melanoma color, percent non-melanoma color, color clustering ratio features
The percent melanoma color, percent non-melanoma color, and color clustering ratio features require the mapping of colors within the relative color histogram as melanoma and benign colors (28, 29). For the algorithm, training images are used to populate the relative color histogram one image at a time. For each training image, a minimum percentage of the skin lesion area (0.125%) must be contained within a histogram bin for that bin to be considered populated by that image (28).
After bin population for each melanoma and benign training image is completed, melanoma and benign relative color bin probability densities are determined based on the number of melanoma and benign images that populate each histogram bin. Each bin is labeled as melanoma, benign, uncertain, or unpopulated based on comparing the corresponding melanoma and benign probabilities at each bin. The rules for bin labeling are given in detail in (28). The percent melanoma color, percent non-melanoma color, and color clustering ratio features are computed after the relative color histogram bins have been labeled based on a training set of images and the lesion region for feature analysis of the test image has been identified. For the percent melanoma color feature, a count denoted as U is maintained for the number of pixels within the lesion region of interest with relative color values that map as melanoma colors. Let At denote the area in pixels of the lesion region of interest. The percent melanoma color within a lesion region of interest of area At is given as
| (7) |
The percentage of non-melanoma colored pixels is given by
| (8) |
where Q is the number of benign-colored pixels within the skin lesion region of interest.
The color clustering ratio feature gives an indication of the grouping of melanoma-colored pixels within the lesion region. The color clustering ratio feature is computed as follows. Let M denote the set of relative color values that map into relative color histogram bins labeled as melanoma colors from the training set of images. Let L denote the set of pixel locations within the skin lesion region of interest with relative color O that map into melanoma colors, formally
| (9) |
Let N(x, y) denote the number of eight-connected neighbors and NM(x, y) denote the number of melanoma-colored eight-connected neighbors that are contained in the lesion region of interest for pixel (x, y)∈L. The eight-connected neighbors for (x, y)∈L that lie outside of the lesion region of interest are excluded from calculating N(x, y) and NM(x, y). Then,
| (10) |
represents the total number of melanoma color neighbors for all pixels within the skin lesion with relative color values that map as melanoma colors. The cumulative total number of eight-connected neighbors for all (x, y)∈L is denoted as
| (11) |
T includes all neighbors of melanoma color pixels within the skin lesion regardless of whether the neighbor is mapped to a melanoma color. The color clustering ratio for a skin lesion is given as
| (12) |
Red, green, and blue variance
Variegated coloring plays an important role as one of the most predictive features in identification of malignant melanoma. The variance of red within the lesion is defined as
| (13) |
Variances of green and blue are defined similarly.
Relative chromaticity of red, green, and blue
The next set of color features are relative chromaticity measures. The relative chromaticity of red is defined as
| (14) |
where rLesionr, gLesion, bLesion are the average red, green, and blue of the lesion area, respectively.
Ratio in red, green, and blue
The ratio of colors is the average color of the lesion divided by the average color of the background skin. The ratio of red is defined as
| (15) |
The ratios of green and blue are defined similarly.
Difference in lightness, chroma, color, and hue
The final set of color indices are the differences in lightness, chroma, color, and hue between the average lesion values and the corresponding surrounding skin values. Lightness, chroma, and hue are denoted as L*, C*, and H, respectively, and are defined (30). The differences in lightness, chroma, color, and hue are given as
| (16) |
| (17) |
| (18) |
| (19) |
where
| (20) |
and
| (21) |
Experiments Performed
Overview of feature evaluation
In this research, statistical-, histogram-, and expert system-based approaches are used to evaluate the 19 shape and color features for feature reduction and rule-based discrimination between malignant melanoma and benign lesions. A correlation-based expert system filter is used as the feature selection method. It uses a correlation-based heuristic to evaluate the worth of features. This heuristic takes into account the usefulness of individual features for predicting the class label along with the level of correlation among them. The hypothesis on which the heuristic is based is that good feature subsets contain features highly correlated with the class, yet uncorrelated with each other.
Data set description
The relative color technique attempts to address some of the variations in the acquired photographs (because of lighting, film types, and digitizing techniques, etc.) and also helps in correcting for different skin coloration (pigmentation).
Three image data sets were used in the experiments here. Data set 1 was photographed and digitized at the University of Sydney. Data set 2 was photographed at New York University and Dr Stoecker’s clinic in Rolla, Missouri and digitized with a Nikon Coolscan®. Data set 3 was acquired at a private practice in New York and New York University and similarly digitized. All sets used 35mm slides, with the Rolla Clinic using Kodachrome and the other sites using Ektachrome and Kodachrome. Accordingly, there was no way to directly control color variations from the different light sources, film types, and film processing used for the acquired 35mm slides. The following lesion types were included in the data sets: invasive malignant melanoma (Mel), basal cell carcinoma (bcc), dysplastic nevus (dys), nevus (n), and seborrheic keratosis (sk), and vascular lesions consisting of hemangiomas and pyogenic granulomas (Vasc). Table 2 presents the three data sets examined, including the number of each lesion type. A total of 1040 lesions were examined. For expert system evaluation, the total data set was randomly separated into five disjoint subsets, each with the same percentage from each lesion category. Four subsets were combined to form the training set (80% of the total data set) while the remaining subset was used as the test set. This process was repeated a total of five times, with each subset used exactly one time as a test set.
TABLE 2.
Data set summary
| Data set 1 | Data set 2 | Data set 3 | Total | |
|---|---|---|---|---|
| Mel | 50 | 136 | 169 | 355 |
| bcc | 3 | 43 | 79 | 125 |
| dys | 24 | 49 | 104 | 177 |
| n | 12 | 107 | 80 | 199 |
| sk | 6 | 57 | 76 | 139 |
| Vasc | 1 | 0 | 44 | 45 |
| Total | 96 | 392 | 552 | 1040 |
Feature evaluation experiments
Preliminary histogram, statistical, and correlation analysis was performed on the 19 shape and color features to assess the interrelationships among features and the discrimination capability of those features to distinguish melanomas from benign lesions. In order to evaluate the separability of individual features for melanoma and benign lesion discrimination, histogram and statistical analysis was carried out on the entire data set. The statistics examined for each feature included the minimum, maximum, mean, and standard deviation values. Pearson’s correlation was used to assess the interrelationships among features based on the evaluation of the entire data set.
Finally, expert system analysis was performed to assess feature interrelationships and discrimination capability. C4.5 is an algorithm introduced by Quinlan for extracting decision trees from data (31). C4.5 works with a set of records, each of which has the same structure, consisting of a number of attribute/value pairs. One of these attributes represents the goal of the record, i.e. the attribute whose values are most significant. The problem is to determine a decision tree that, on the basis of answers to questions about the non-goal attributes, predicts correctly the value of the goal attribute.
The feature selection process is carried out in a backward elimination fashion. It is a multiple step procedure. At each step, correlation-based expert system evaluation of a subset of features is performed. The data are randomly selected from the whole data set. And, the selected data set is divided into a randomly picked training set and a testing set. Using this procedure, error rates were determined using the C4.5 algorithm from training and test sets for each of the data sets for different feature combinations.
Experimental Results
Evaluation of 19 shape and color features was performed over a data set of 1040 clinical images containing 355 melanomas and 685 non-melanoma lesions. Statistical-, histogram-, correlation-, and expert system-based approaches were investigated to determine which features contributed most successfully to lesion discrimination.
Results from statistical analysis of shape and color features
In order to assess the distribution of the shape and color features, statistical analysis was performed over the combined 1040 image data set. Table 3 presents the minimum, maximum, mean, and standard deviation values for each of the 19 features. Because of the wide clinical variation of the lesions, even within a diagnostic class, the different sources of the data, and the noise introduced during the extraction process, several features presented a wide range and large standard deviation. In particular, the color variance features (variance in red, green, blue) yielded the widest range and standard deviation of the features inspected. The wide range of feature values makes feature normalization for classifier development more difficult. The solid pigment asymmetry index and the relative chromaticity of red, green, and blue features have relatively narrow ranges, compared with the other features, which makes separation of benign lesions and melanomas based on these features more difficult. Although the classifier can accommodate to various feature ranges and distributions, normalization might yield some improvement in the classifier.
TABLE 3.
Summary statistics of features
| Minimum | Maximum | Mean | SD | |
|---|---|---|---|---|
| Irregularity index (F1) | 0.940 | 5.877 | 1.291 | 0.353 |
| Asymmetry index (F2) | 0.000 | 32.652 | 6.911 | 4.361 |
| Solid pigment asymmetry index (F3) | 0.000 | 0.462 | 0.063 | 0.078 |
| Percentage of melanoma (F4) | 0.060 | 100.000 | 25.383 | 26.676 |
| Percentage of non-melanoma (F5) | 0.000 | 99.920 | 71.823 | 26.968 |
| Clustering ratio (F6) | 0.000 | 99.580 | 44.609 | 26.747 |
| Variance red (F7) | 23.496 | 4653.328 | 1157.177 | 838.205 |
| Variance green (F8) | 2.493 | 3600.404 | 839.636 | 618.305 |
| Variance blue (F9) | 0.063 | 4233.125 | 863.551 | 727.933 |
| Relative chromaticity of red (F10) | −0.150 | 0.286 | 0.009 | 0.066 |
| Relative chromaticity of green (F11) | −0.194 | 0.091 | −0.013 | 0.032 |
| Relative chromaticity of blue (F12) | −0.118 | 0.133 | 0.003 | 0.037 |
| Ratio red (F13) | 0.231 | 2.032 | 0.800 | 0.226 |
| Ratio green (F14) | 0.014 | 2.494 | 0.780 | 0.314 |
| Ratio blue (F15) | 0.001 | 2.979 | 0.820 | 0.346 |
| Difference in lightness (F16) | −37.815 | 25.115 | −7.847 | 8.979 |
| Difference in chroma (F17) | −26.552 | 29.257 | −0.468 | 8.462 |
| Difference in color (F18) | 0.004 | 25.925 | 4.719 | 4.489 |
| Difference in hue (F19) | 0.924 | 41.150 | 14.141 | 7.508 |
Results from Pearson’s correlation analysis
The second set of experiments focused on correlation analysis of the 19 features over the image data set. There are three purposes for these experiments: (1) determine interrelationships among variables, (2) evaluate which individual variables are most strongly correlated with melanoma, and (3) evaluate which individual variables are most strongly correlated with the six different non-melanoma lesion categories. Pearson’s correlation is performed between each feature and the other features, each feature and the melanoma class, and each feature and the non-melanoma lesion categories. The resulting Pearson’s correlations are presented in Table 4 (a) and (b). ‘Class 1’ corresponds to the exact diagnosis, i.e. the diagnosis related to the six lesion categories shown in Table 2. ‘Class 2’ refers to categorizing lesions as melanomas/non-melanomas, i.e. the bcc, dys, n, sk, and Vasc lesion types are combined into the non-melanoma category.
TABLE 4.
Pearson’s correlation results between individual features and the other features and melanoma and benign lesion categories. The table is presented in two parts (a) and (b)
| Class 1 | Class 2 | F1 | F3 | F2 | F7 | F8 | F9 | F10 | F11 | |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) | ||||||||||
| Class 1 | 1 | |||||||||
| Class 2 | −0.8 | 1 | ||||||||
| F1 | −0 | 0.09 | 1 | |||||||
| F3 | −0.2 | 0.17 | 0.06 | 1 | ||||||
| F2 | 0.03 | 0.03 | 0.56 | −0.1 | 1 | |||||
| F7 | 0.01 | 0.05 | 0.11 | 0.02 | 0.13 | 1 | ||||
| F8 | 0.19 | −0.1 | 0.13 | −0.1 | 0.09 | 0.66 | 1 | |||
| F9 | 0.24 | −0.1 | 0.11 | −0.1 | 0.07 | 0.54 | 0.94 | 1 | ||
| F10 | −0.1 | 0.07 | −0.1 | −0 | −0 | −0.3 | −0.5 | −0.6 | 1 | |
| F11 | 0.09 | −0 | 0.15 | 0.07 | 0.02 | 0.27 | 0.43 | 0.49 | −0.9 | 1 |
| F12 | 0.17 | −0.1 | 0.14 | 0.01 | 0.05 | 0.33 | 0.51 | 0.59 | −1 | 0.8 |
| F13 | −0 | 0.03 | −0 | −0 | −0.2 | −0.4 | −0 | 0.05 | −0.1 | 0.2 |
| F14 | 0.01 | 0.04 | 0.08 | 0.05 | −0.2 | −0.1 | 0.23 | 0.33 | −0.6 | 0.67 |
| F15 | 0.05 | 0.01 | 0.09 | 0.03 | −0.1 | −0 | 0.28 | 0.4 | −0.7 | 0.67 |
| F16 | 0.07 | −0.1 | 0.06 | 0 | −0.1 | −0.2 | 0.16 | 0.28 | −0.5 | 0.6 |
| F17 | −0.1 | −0 | −0.2 | −0.1 | −0.1 | −0.4 | −0.4 | −0.5 | 0.89 | −0.8 |
| F18 | 0.09 | 0.04 | −0.1 | −0.1 | 0.01 | 0.07 | 0.1 | 0.04 | 0.09 | −0.3 |
| F19 | −0.2 | 0.24 | −0.1 | 0.09 | −0.1 | 0.06 | −0.1 | −0.2 | 0.4 | −0.5 |
| F4 | −0.2 | 0.34 | −0.1 | 0 | 0.01 | −0.1 | −0.3 | −0.4 | 0.54 | −0.6 |
| F5 | 0.24 | −0.3 | 0.05 | −0 | −0 | 0.13 | 0.29 | 0.37 | −0.5 | 0.55 |
| F6 | −0.2 | 0.33 | −0.1 | −0 | 0.01 | −0.1 | −0.3 | −0.4 | 0.51 | −0.5 |
| F12 | F13 | F14 | F15 | F16 | F17 | F18 | F19 | F4 | F5 | F6 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (b) | |||||||||||
| F12 | 1 | ||||||||||
| F13 | 0.05 | 1 | |||||||||
| F14 | 0.53 | 0.82 | 1 | ||||||||
| F15 | 0.65 | 0.75 | 0.97 | 1 | |||||||
| F16 | 0.44 | 0.88 | 0.95 | 0.92 | 1 | ||||||
| F17 | −0.8 | 0.11 | −0.4 | 0.5 | −0.3 | 1 | |||||
| F18 | 0.07 | −0.2 | −0.2 | 0.2 | −0.3 | 0.11 | 1 | ||||
| F19 | −0.3 | −0.4 | −0.4 | 0.4 | −0.6 | 0.22 | 0.56 | 1 | |||
| F4 | −0.5 | −0.3 | −0.5 | 0.5 | −0.5 | 0.32 | 0.31 | 0.58 | 1 | ||
| F5 | 0.42 | 0.31 | 0.45 | 0.45 | 0.52 | −0.3 | −0.3 | −0.6 | −1 | 1 | |
| F6 | −0.4 | −0.3 | −0.4 | −0.4 | −0.5 | 0.33 | 0.28 | 0.51 | 0.9 | −0.9 | 1 |
From Table 4, there are several observations. First, some features have higher correlation with the melanoma/benign lesion classes compared with other features, including the percentage of melanoma color (F4) and color clustering ratio (F6) with correlation values of 0.34 and 0.33, respectively, with class 2. The percentage of melanoma color and color clustering ratio features have been shown to achieve high melanoma discrimination in clinical images as individual features (28, 29). Second, there are many features that are strongly interrelated. The features that are strongly interrelated (or strongly inversely related) include: (1) relative chromaticity of red (F10) and green (F11) with difference of chroma (F17), (2) relative chromaticity of red (F10), green (F11), blue (F12), (3) variance in green (F8) and blue (F9), (4) percentage of melanoma color (F4) and color clustering ratio (F6), (5) ratio of red (F13), green (F14), and blue (F15), (6) ratio of red (F13), green (F14), and blue (F15) with the difference in lightness (F16), and (7) relative chromaticity of blue (F12) with the difference in chroma (F17). The strongly correlated (or inversely correlated) features provide sources of redundant information that can be eliminated for determining a final feature set used for classifier development.
Feature histogram analysis
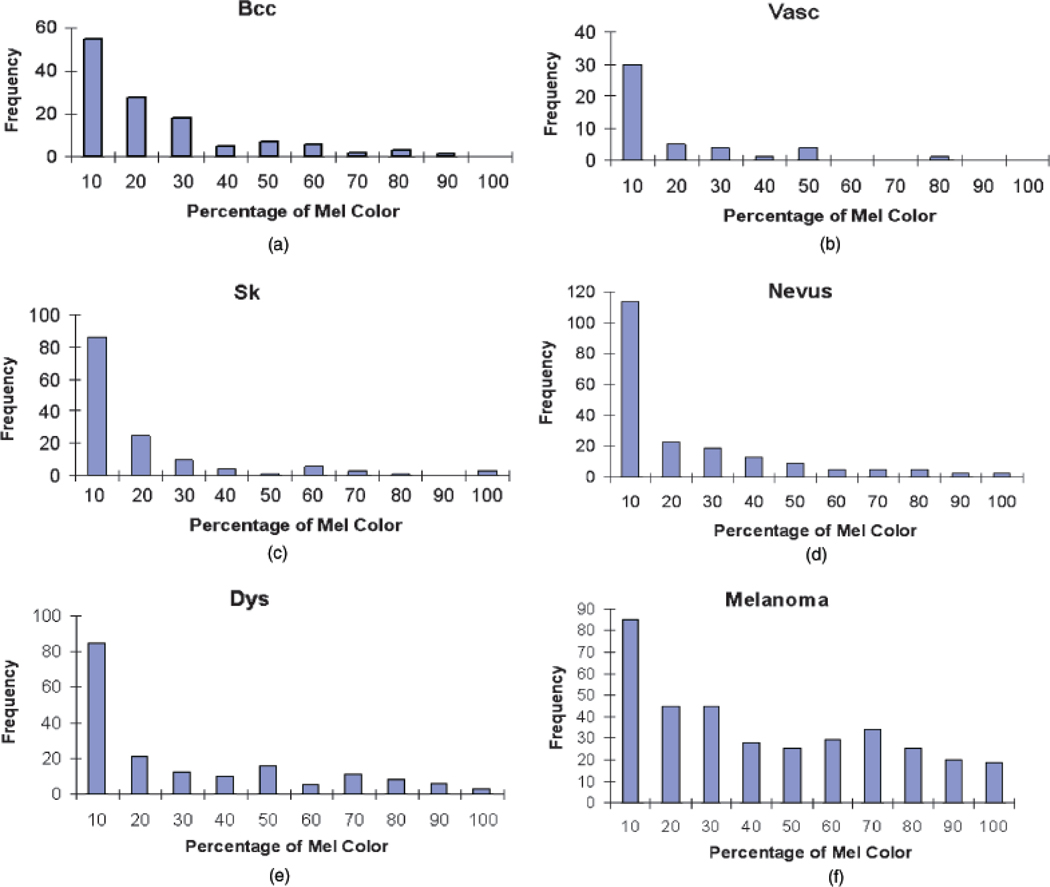
Based on the correlation analysis, interrelationships among features and the relationships between the individual features and the melanoma and benign lesion classes were examined. The next goal with feature analysis was to determine the discrimination capability of features found to be most strongly correlated with melanoma/benign lesion classification. The percentage of melanoma color (F4) feature was found to have the strongest correlation with melanoma/benign lesion discrimination. Figure 5 presents the histograms for the percentage of melanoma color feature for the different benign lesion categories and melanoma. The histograms present the number of images for the lesion type in the data set with the percentage of melanoma color feature that falls within the specified range on the horizontal axis. The histograms for the bcc lesions are given in (a), Vasc in (b), sk in (c), nevus in (d), dys in (e), and melanomas in (f).
Fig. 5.
Histograms for percentage of melanoma color for different lesion categories. (a) Basal cell carcinomas (bcc). (b) Vascular lesions (Vasc). (c) Seborrheic keratoses (sk). (d) Nevocellular nevi (Nevus). (e) Dysplastic nevi (dys). (f) Melanoma. Frequency represents the number of lesions satisfying the feature constraint.
From Fig. 5, there is a different distribution of the percentage of melanoma color feature among the different categories. Melanoma lesions have higher frequencies for greater than 20% for the percentage of melanoma color than the benign lesion categories. For nevus, a large number of lesions of this type have the percentage of melanoma color falling into the less than 10% category. Other features have similar histograms. But certainly one feature does not contain enough information for the classification task. There are a certain number of melanoma cases with percentage of melanoma color falling into the less than 20% bins. These are mostly amelanotic/hypomelanotic melanomas and are difficult cases to diagnose.
Expert system example
In the previous sections, statistical-, correlation-, and histogram-based approaches were examined for evaluating the 19 shape and color features. The final set of experiments focused on expert system-based feature evaluation and lesion discrimination using C4.5. Figure 6 presents an example of the output from C4.5 for developing a tree structure for feature selection and lesion discrimination using the entire data set.
Fig. 6.
Example of decision tree from C4.5 algorithm.
In this example, a decision tree is built based on the data. At each level, one attribute is selected to divide subgroups. For example, at the first level, Var Red is selected and then the data are divided into two groups according to the threshold 2139.310. The breakdown of the decision tree shows the features and thresholds used to achieve the best classification at each level of the tree.
Expert system feature evaluation and discrimination results
The first step in the feature evaluation process using C4.5 was to examine the error rates for melanoma discrimination using all 19 features. First, an expert system evaluation of the original feature space is carried out. Table 5 shows the error rate results for all 19 features for five randomly generated training/test sets over the entire data set.
TABLE 5.
Training and test error rate results for 19 features using C4.5
| Error rate | ||
|---|---|---|
| 19 features | Train set (%) | Test set (%) |
| Set 1 | 3.3 | 26.7 |
| Set 2 | 5.2 | 25.5 |
| Set 3 | 2.6 | 26.7 |
| Set 4 | 2.6 | 28.6 |
| Set 5 | 2.2 | 25.5 |
Based on Pearson’s correlation results shown in Table 4 and the error results given in Table 5, several highly correlated features were found. The absolute values of correlation among percentage of melanoma color (F4), percentage of non-melanoma color (F5), and clustering ratio (F6) are greater than or equal to 0.90. From the hypothesis for a good feature subset, only a subset of highly correlated features should be used as a predictor. Comparison on performance is carried out first among these three features using C4.5. Table 6 shows the error results for 17 features based on leaving out the percentage of non-melanoma color and color clustering ratio features. Table 7 gives the error results for 17 features based on leaving out the percentage of melanoma color and color clustering ratios. Table 8 presents the error results for 17 features based on leaving out the percentage of melanoma color and percentage of non-melanoma color features. The error results in Tables 6–8 are shown for five randomly generated training and test sets and are given as the combined melanoma and non-melanoma training and test error percentages.
TABLE 6.
Error rate results using C4.5 using 17 features including the percentage of melanoma color feature and leaving out the percentage of non-melanoma and color clustering ratio features
| Error rate | ||
|---|---|---|
| 17 features | Train set (%) | Test set (%) |
| Set 1 | 2.1 | 21.4 |
| Set 2 | 6.2 | 23.2 |
| Set 3 | 2.4 | 25.5 |
| Set 4 | 4.3 | 30.3 |
| Set 5 | 2.4 | 25.5 |
TABLE 7.
Error rate results using C4.5 using 17 features including the percentage of non-melanoma included and leaving out the percentage of melanoma color and clustering ratio features
| Error rate | ||
|---|---|---|
| 17 features | Train set (%) | Test set (%) |
| Set 1 | 3.6 | 25.5 |
| Set 2 | 5.2 | 25.5 |
| Set 3 | 5.0 | 23.2 |
| Set 4 | 2.6 | 32.1 |
| Set 5 | 2.6 | 23.2 |
TABLE 8.
Error rate results using C4.5 including the clustering ratio feature and leaving out the percentage of melanoma color and percentage of non-melanoma color features
| Error rate | ||
|---|---|---|
| 17 features | Train set (%) | Test set (%) |
| Set 1 | 3.0 | 25.5 |
| Set 2 | 5.2 | 26.7 |
| Set 3 | 3.2 | 17.9 |
| Set 4 | 3.9 | 23.2 |
| Set 5 | 3.7 | 26.7 |
From Tables 6–8, the percentage melanoma color feature yielded overall error rates lower than the other two features. For determining the feature set used for classifier development, the percentage of melanoma color feature is retained over the percentage of non-melanoma color and clustering ratio features. A similar process was carried out for all the features, so after the backward elimination process, a sub-optimum subset of features was chosen to achieve better performance in the sense of correct classification rate. Accordingly, the final feature set contained 13 features, including irregularity index (F1), asymmetry index (F2), solid pigment asymmetry index (F3), percentage of melanoma color (F4), variance of red (F7), variance of blue (F9), relative chromaticity in red (F10), relative chromaticity in blue (F12), ratio in red (F13), ratio in blue (F15), difference in lightness (F16), difference in chroma (F17), and difference in hue (F19). Note that the selection process is not unique. There is a compromise on the performance and time needed for the feature selection stage. Table 9 presents the 13-feature error rate results using C4.5 over the five randomly generated training and test sets. From Table 9, the error rate results using 13 features provide slight improvement over the 19- and 17-feature results given in Tables 6–8. These results show the impact of feature selection on discrimination capability. The feature selection process fosters the development of more sophisticated classifier development to improve lesion discrimination.
TABLE 9.
C4.5 error rate results using 13 features
| Error rate | ||
|---|---|---|
| 13 features | Train (%) | Test (%) |
| Set 1 | 4.5 | 21.4 |
| Set 2 | 2.3 | 17.9 |
| Set 3 | 2.7 | 26.7 |
| Set 4 | 5.5 | 26.7 |
| Set 5 | 3.4 | 23.2 |
Discussion
This research was conducted using data sets acquired from different sources, and diagnostic results from individual sets were higher than that of the combined set. Although this lowered diagnostic accuracy somewhat, non-homogenous sets may better represent the real-world variability in lesion acquisition, and better represent the problem of diagnosis at a distance, increasingly encountered with internet transmission of images.
In recent years, computerized automatic diagnosis of pigmented lesions has been mostly applied to digital dermoscopy images (24, 32) and hyperspectral images, including visible light and infrared images (33). However, most American dermatologists still do not routinely use dermoscopy in clinical practice, and those who do, tend to favor non-contact dermoscopy because of the ease of image acquisition. And for now, hyperspectral imaging is largely confined to pigmented lesion centers. Thus, the images that are exchanged in formats such as the dermatology internet discussion group are almost all either clinical images, such as this report describes, or non-contact dermoscopy images, which share some features with the images discussed here. Therefore, the techniques discussed here may be directly applied to these images. It has been shown that the combination of clinical examination and dermoscopy allows a higher diagnostic accuracy in screening, than either method alone, as the two techniques play a complementary role (34). Thus, techniques presented here for clinical image analysis could potentially be combined with dermoscopy image analysis in screening.
There are several limitations with the expert system approach. First, the systematic expert system approach may be feasible when d, the number of features, is small. However, the feature selection results depend on the value of d. There are many ways to select m features out of d, namely, d!/(m!(d − m)!). Second, the results depend on the test sets selected. If one test data set is repeatedly used, features obtained might well be suited for that particular test set, but might not be the best in general. Third, the feature selection results also depend on the value of m. The process with various values of m may also have to be repeated to make a choice. However, there are some heuristic approaches that are often useful. A common practice for avoiding the consideration of all subsets is to use stepwise selection using forward selection or backward elimination. Finally, the expert system-based method for feature selection does not guarantee the optimum feature subset. The measure may be good in some classes but bad in other classes. Other methods or measures could be used to select a good feature set. However, it is unclear how to perform this optimization systematically. A more systematic method for feature subset selection may yield further improvements.
Conclusions
In this research, a statistical-, correlation-, histogram-, and expert system-based approach was used for feature selection for malignant melanoma discrimination. Statistical and correlation measures provide an initial basis to find interrelated features. The expert system was used for decision tree-based lesion discrimination for feature evaluation in a backward elimination manner. From the initial 19 shape and color features examined, the systematic approach yielded 13 features that provided lower error classification rates than the original feature set. The systematic approach is heuristic and suboptimal. However, the approach provides the basis for feature selection that can be applied to more sophisticated lesion discrimination techniques.
Acknowledgements
This work was supported in part by NIH SBIR grant 1R43 CA-101639–01. The authors wish to thank Scott Menzies, M.D. and Ashfaq Marghoob, M.D. for supplying clinical images. Jason Hagerty helped as system administrator.
References
- 1.Cassileth BR, Clark WH, Jr, Lusk EJ, Frederick BE, Thompson CJ, Walsh P. How well do physicians recognize melanoma and other problem lesions? J Am Acad Dermatol. 1986;14:555–560. doi: 10.1016/s0190-9622(86)70068-6. [DOI] [PubMed] [Google Scholar]
- 2.Kopf A, Mintzis M, Bar R. Diagnostic accuracy in malignant melanoma. Arch Dermatol. 1975;111:1291–1292. [PubMed] [Google Scholar]
- 3.Grin C, Kopf AW, Welkovich B, Bar R, Levenstein M. Accuracy in the clinical diagnosis of melanoma. Arch Dermatol. 1990;126:763–766. [PubMed] [Google Scholar]
- 4.Lindelof B, Hedblad MA. Accuracy in the clinical diagnosis and pattern of malignant melanoma at a dermatological clinic. J Dermatol. 1994;21:461–464. doi: 10.1111/j.1346-8138.1994.tb01775.x. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RJ, Rigel DS, Kopf AW. Early detection of malignant melanoma: the role of physician examination and self-examination of the skin. Ca-A Cancer J Clin. 1985;35:130–151. doi: 10.3322/canjclin.35.3.130. [DOI] [PubMed] [Google Scholar]
- 6.Marks R, Jolley D, McCormack C, Dorevitch AP. Who removes pigmented skin lesions? J Am Acad Dermatol. 1997;36(5 Part 1):721–726. doi: 10.1016/s0190-9622(97)80324-6. [DOI] [PubMed] [Google Scholar]
- 7.Dhawan AP. An expert system for the early detection of melanoma using knowledge-based image analysis. Anal Quant Cytol Histol. 1989;10:405–416. [PubMed] [Google Scholar]
- 8.Landau M, Matz H, Tur E, Dvir M, Brenner S. Computerized system to enhance the clinical diagnosis of pigmented cutaneous malignancies. Int J Dermatol. 1999;38:443–446. doi: 10.1046/j.1365-4362.1999.00629.x. [DOI] [PubMed] [Google Scholar]
- 9.Schindewolf T, Stolz W, Albert R, Abmayr R, Abmayr W, Harms H. Classification of melanocytic lesions with color and texture analysis using digital image processing. Anal Quant Cytol Histol. 1993;15:101–111. [PubMed] [Google Scholar]
- 10.Andreassi L, Perotti R, Burroni M, Dell’Eva G, Biagioli M. Computerized image analysis of pigmented lesions. Chronica Dermatol. 1995;1:11–24. [Google Scholar]
- 11.Ercal F, Chawla A, Stoecker WV, Lee HC, Moss RH. Neural network diagnosis of malignant melanoma from color images. IEEE Trans Biomed Eng. 1994;41:837–845. doi: 10.1109/10.312091. [DOI] [PubMed] [Google Scholar]
- 12.Stoecker WV, Li WW, Moss RH. Automatic detection of asymmetry in skin tumors. Comput Med Image Graphics. 1992;16:191–197. doi: 10.1016/0895-6111(92)90073-i. [DOI] [PubMed] [Google Scholar]
- 13.Golston JE, Stoecker WV, Moss RH, Dhillon IPS. Automatic detection of irregular borders in melanoma and other skin tumors. Comput Med Image Graphics. 1992;16:199–203. doi: 10.1016/0895-6111(92)90074-j. [DOI] [PubMed] [Google Scholar]
- 14.Ganster H, Pinz A, Rohrer R, Wilding E, Binder M, Kittler H. Automated melanoma recognition. IEEE Trans Med Imag. 2001;20:233–238. doi: 10.1109/42.918473. [DOI] [PubMed] [Google Scholar]
- 15.Xu L, Jackowski M, Goshtasby A, Roseman D, Bines S, Yu C, Dhawan A, Huntley A. Segmentation of skin cancer images. Image Vis Comput. 1999;17:65–74. [Google Scholar]
- 16.Sober AJ, Burstein JM. Computerized digital image analysis: an aid for melanoma diagnosis-preliminary investigations and brief review. J Dermatol. 1994;21:885–890. doi: 10.1111/j.1346-8138.1994.tb03307.x. [DOI] [PubMed] [Google Scholar]
- 17.Umbaugh SE, Moss RH, Stoecker WV. Automatic color segmentation of images with application to detection of variegated coloring in skin tumors. IEEE Eng Med Biol. 1989;8:43–50. doi: 10.1109/51.45955. [DOI] [PubMed] [Google Scholar]
- 18.Claridge E, Hall PN, Keefe M, et al. Shape analysis for classification of malignant melanoma. J Biomed Eng. 1992;14:229–234. doi: 10.1016/0141-5425(92)90057-r. [DOI] [PubMed] [Google Scholar]
- 19.Moss RH, Stoecker WV, Lin S-J, et al. Skin cancer recognition by computer vision. Comput Med Image Graphics. 1989;13:31–36. doi: 10.1016/0895-6111(89)90076-1. [DOI] [PubMed] [Google Scholar]
- 20.Donohoe GW, Nemeth S, Soliz P. ART-based image analysis for pigmented lesions of the skin; IEEE Symposium Computer-Based Medical Systems; Los Alamitos, CA: IEEE Comp Soc; 1998. pp. 293–298. [Google Scholar]
- 21.Lee T, Ng V, McLean D, Coldman A, Gallagher R, Sale J. A multi-stage segmentation method for images of skin lesions; IEEE Pacific Rim Conference on Communications, Computers, and Signal Processing Proceedings; Piscataway, NJ: IEEE; 1995. pp. 602–605. [Google Scholar]
- 22.Hance GA, Umbaugh SE, Moss RH, Stoecker WV. Unsupervised color image segmentation with application to skin tumor borders. IEEE Eng Med Biol. 1996;15:104–111. [Google Scholar]
- 23.Green A, Martin N, Pfitzner J, O’Rourke M, Knight N. Computer image analysis in the diagnosis of melanoma. J Am Acad Dermatol. 1994;31:958–964. doi: 10.1016/s0190-9622(94)70264-0. [DOI] [PubMed] [Google Scholar]
- 24.Seidenari S, Burroni M, Dell’Eva G, Pepe P, Belletti B. Computerized evaluation of pigmented skin lesion images recorded by a videomicroscope: comparison between polarizing mode observation and oil/slide mode observation. Skin Res Technol. 1995;1:187–191. doi: 10.1111/j.1600-0846.1995.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 25.Aitken JF, Pfitzner J, Battistutta D, O’Rourke PK, Green AC, Martin NG. Reliability of computer image analysis of pigmented skin lesions of Australian adolescents. Cancer. 1996;78:252–257. doi: 10.1002/(SICI)1097-0142(19960715)78:2<252::AID-CNCR10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 26.Subbiah P. MS Thesis. Department of Electrical and Computer Engineering, University of Missouri-Rolla; 2001. Automatic detection of border irregularity in skin lesions. [Google Scholar]
- 27.Murali M, Stoecker WV, Moss RH. Detection of solid pigment in dermatoscopy images using texture analysis. Skin Res Technol. 2000;6:193–198. doi: 10.1034/j.1600-0846.2000.006004193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faziloglu Y, Stanley RJ, Moss RH, Stoecker WV, McLean RP. Color histogram analysis for melanoma discrimination in clinical images. Skin Res Technol. 2003;9:147–155. doi: 10.1034/j.1600-0846.2003.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Stanley R, Moss RH, Stoecker WV. Color analysis of skin lesion regions for melanoma discrimination in clinical images. Skin Res Technol. 2003;9:94–104. doi: 10.1034/j.1600-0846.2003.00024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Celenk M. A color clustering technique for image segmentation. Comput Vision, Graphics Image Process. 1991;52:145–170. [Google Scholar]
- 31.Quinlan JR. C4.5: programs for machine learning. San Mateo, CA: Morgan Kauffman; 1993. [Google Scholar]
- 32.Rubegni P, Burroni M, Cevenini G, et al. Digital dermoscopy analysis and artificial neural network for the differentiation of clinically atypical pigmented skin lesions: a retrospective study. J Invest Dermatol. 2002;19:471–474. doi: 10.1046/j.1523-1747.2002.01835.x. [DOI] [PubMed] [Google Scholar]
- 33.Elbaum M, Kopf AW, Rabinovitz HS, Langley RG, Kamino H, Mihm MC, Jr, et al. Automatic differentiation of melanoma from melanocytic nevi with multispectral digital dermoscopy: a feasibility study. J Am Acad Dermatol. 2001;44:207–218. doi: 10.1067/mjd.2001.110395. [DOI] [PubMed] [Google Scholar]
- 34.Bono A, Bartoli C, Baldi M, Tomatis S, Bifulco C, Santinami M. Clinical and dermatoscopic diagnosis of small pigmented skin lesions. Eur J Dermatol. 2002;12:573–576. [PubMed] [Google Scholar]