Abstract
ADAM23, a member of a disintegrin and metalloprotease (ADAM) family, has been reported to be expressed in several types of tumours. The exact role of ADAM23 and the possible mechanisms in which it is involved in non-small-cell lung carcinoma (NSCLC) remains unclear. Therefore, this study was designed to explore the expression of ADAM23 and its correlation with promoter methylation in NSCLC. Immunohistochemistry and RT-PCR together with Western blotting methods were used to analyse the expression of ADAM23 in 52 cancer tissue samples and eight benign pulmonary lesions as well as four cell lines. The methylated status of ADAM23 gene was determined with methylation-specific PCR (MSP). The results of immunohistochemistry showed that the expression of ADAM23 protein was lower in NSCLC than that in corresponding normal tissues and benign pulmonary lesions (38.5%vs. 86.5% and 87.5%, P < 0.05), and decreased as NSCLC progressed. Meanwhile, methylation of ADAM23 gene was observed in 21 of 52 NSCLC tissues (40.4%), much higher than that of adjacent normal tissues (7.6%) and benign pulmonary lesions (0/8). In the cancer tissues of ADAM23-negative samples, the rate of ADAM23 gene methylation was 50.3% (17/32). ADAM23 expression and its promoter methylation were negatively associated (r = −0.328, P = 0.017). Moreover, weak expression of ADAM23 in methylated cancer cells increased after treatment with 5-aza-2′-deoxycytidine (5-Aza-2′-dC), confirming that methylation was responsible for the gene downregulation. Our results demonstrate that the expression level of ADAM23 is likely to be involved in the progression of NSCLC and its downregulation is probably correlated with promoter methylation. These findings may provide potential diagnostic and prognostic information about NSCLC.
Keywords: 5-Aza-2′-dC, ADAM23 gene, cancer cells lines, methylation, methylation-specific PCR, non-small-cell lung carcinoma
Lung cancer is the most common cause of cancer-related death worldwide (Jemal et al. 2009). It accounts for over a million deaths annually and still has a poor prognosis. Non-small-cell lung cancer (NSCLC) is the main form of lung cancer, which has three major histological types: squamous cell carcinoma (SCC), adenocarcinoma (ADC) and large cell carcinoma (LCC) (Travis et al. 2004). Despite significant advances in the treatment of NSCLC over the past years, the prognosis of the patients with advanced or metastatic disease is poor with a median survival for the newly diagnosed stage IV patients ranging from 8 to 11 months, and 5 to 7 months for those with relapsed disease (Longo-Sorbello et al. 2007). It is well known that lung carcinogenesis, like many other cancers, is a multistep process characterized by the accumulation of genetic alterations and by epigenetic alterations, which are manners to modify gene expression without altering the DNA sequence (Kim et al. 2009). Recently, evidence has emerged that epigenetic changes, especially the DNA methylation of 5′-CpG islands, can lead to the inactivation of tumour suppressor genes (TSGs) in human malignant tumours playing significant roles in tumour initiation and progression including NSCLC (Yanagawa et al. 2003; Minna et al. 2007). Many studies have shown that TSGs were frequently silenced by promoter hypermethylation in NSCLC and premalignant lung lesions, indicating that aberrant methylation correlation with TSGs expression could be used for diagnosis, prognosis and therapeutic responsiveness and warrants further investigation in NSCLC (Belinsky 2005; Suzuki & Yoshino 2010). Therefore, it is necessary to identify new TSGs that are silenced by tumour-specific methylation in NSCLC, which could serve as valuable biomarkers and also provide new clues to the molecular pathogenesis of the tumour (Qureshi et al. 2010).
The ADAM23 gene, a member of the disintegrin and metalloprotease (ADAM) protein family, is located on chromosome 2q33, encodes ADAM domain 23 and was first cloned by Sagane et al. (1998). Although it exhibits the typical structure of the ADAM family members, the metalloproteinase domain of ADAM23 is inactive, suggesting that its disintegrin plays the more significant role in biological behaviour (Sagane et al. 1998), especially in cell adhesion and cell-matrix interactions. Furthermore, an increasing body of evidence indicates that alteration in cell adhesion is causally involved in tumour progression and metastases formation rather than merely being a consequence of it (Neal et al. 2009). Indeed, the loss of expression of ADAM23 gene and its correlation with promoter methylation have been frequently reported in pancreatic cancer, breast cancer, brain cancer and gastric cancer (Costa et al. 2004, 2005; Hagihara et al. 2004; Takada et al. 2005), and recently the role of this gene was also examined in colorectal cancer (Choi et al. 2009). However, the expression level and promoter methylation status of ADAM23 in NSCLC remain unclear.
In this study, we evaluated the expression level of ADAM23 both in tissues and cells lines. Then, the methylated status of ADAM23 gene was determined with MSP in the lung cancer samples and cell lines. In addition, 5-aza-2′-dC was used to study whether demethylation could lead to re-expression ADAM23 gene and to analyse further the relationship between the expression of ADAM23 and methylation. Our findings suggest ADAM23 promoter methylation correlation with its expression may be used as a potential biomarker in NSCLC.
Materials and methods
Tissue samples and cell lines
We analysed 52 pairs of NSCLC specimens and corresponding adjacent lung tissues (5 cm away considered as normal tissues) together with eight benign pulmonary lesions obtained at the time of surgery in the First Affiliated Hospital of Xiangya from June 2009 to May 2010. One section of the samples was frozen immediately in liquid nitrogen and stored at −80 °C until required while the other was fixed with formalin and used for further histological examination to confirm the diagnosis postoperatively. The age of the patients ranged from 39 to 74, with a median 54, and the numbers of them in stage I, II, III and IV were 12, 15, 12 and 13 respectively. None of the patients had received preoperative radiation, chemotherapy or immunotherapy. All diagnoses were based on pathological and/or cytological evidence. Histological classification was conducted according to the 1999 ‘Histological typing of lung and pleural tumours: third edition’ of the WHO, and tumour stage was determined according to the 2004 TNM staging guideline suggested by the American Joint Committee on Cancer (AJCC) and the Union Internationale Contre le Cancer (UICC). Ethical approval was obtained from the hospital and fully informed consent from all patients prior to sample collection. Histological grade, tumour size and lymph node metastasis in these cases are summarized in Table 2.
Table 2.
Relationship between ADAM23 protein expression and clinicopathological characteristics of non-small-cell lung carcinoma
| ADAM23 | ||||
|---|---|---|---|---|
| Category | n | + | − | P value |
| Age (years) | ||||
| ≥50 | 31 | 11 | 20 | 0.592 |
| <50 | 21 | 9 | 12 | |
| Sex | ||||
| Male | 32 | 12 | 20 | 0.857 |
| Female | 20 | 8 | 12 | |
| Diagnosis | ||||
| Squamous cell carcinoma | 30 | 11 | 19 | 0.756 |
| Adenocarcinoma | 22 | 9 | 13 | |
| Histological grading | ||||
| Well/moderately differentiated | 29 | 15 | 14 | 0.027 |
| Poorly differentiated | 23 | 5 | 18 | |
| Tumour size (cm) | ||||
| ≥5 cm | 18 | 7 | 11 | 0.963 |
| <5 cm | 34 | 13 | 21 | |
| Lymph node metastasis | ||||
| Yes | 28 | 4 | 24 | 0.000 |
| No | 24 | 16 | 8 | |
| UICC stage | ||||
| I | 12 | 8 | 4 | 0.000* |
| II | 15 | 9 | 6 | |
| III | 12 | 2 | 10 | |
| IV | 13 | 1 | 12 | |
Linear-by-linear association (χ2 = 12.892, P = 0.000).
All cell lines (A549, SPC-A1, LTEP-a-2, SK-MES-1, MKN-45, MKN-1, SW480) were obtained from Shanghai Cell Line Bank. MKN-45, SW480 and MKN-1 were used as positive and negative controls respectively (Takada et al. 2005; Choi et al. 2009). Most of the cells were maintained in RPMI1640 (Invitrogen, Carlsbad, CA, USA), except for SK-MES-1 that was maintained in DMEM (Gibco, Carlsbad, CA, USA) at 37 °C, 5% CO2 and 95% humidified air. All media were supplemented with 10% foetal bovine serum (Invitrogen).
Immunohistochemistry
Immunohistochemistry was performed to detect the expression of ADAM23 in NSCLC specimens and adjacent normal tissues as well as benign control samples as follows. Briefiy, sections (5 μm) were processed for immunohistochemistry according to the manufacturer's recommendations with the ADAM23 antibody (polyclonal H-65; Santa Cruz, CA, USA). This ADAM23 antibody was diluted 1:200 with antibody diluent. The immune reactions were carried out with microwave heat-induced epitope retrieval system in citrate buffer (pH 8.0) for 30 min, using the Envision kit (Dako Antigen retrieval, Dako, Glostrup, Denmark). Subsequently, incubations were performed with biotinylated secondary antibodies (15 min at room temperature; labelled horseradish peroxidase; DakoCytomation). In all cases, negative controls were included in which specific antibody was substituted by the primary mouse negative control (DakoCytomation). The staining intensity was classified by the number of stained cells as follows: negative (−), faint (+; 0–4% positive cells), positive (++; 5–49% positive cells), or high positive (+++; >50% positive cells). +, ++ and +++ were merged into positive in a statistical table. An overall immunoreactivity score was evaluated according to the methods previously described (Takanami et al. 2008; Wang et al. 2008; Karim et al. 2009).
DNA/RNA isolation
Tissue samples, which were frozen in liquid nitrogen, and cell lines were dissolved in lysis buffer for subsequent DNA isolation using the DNA Extraction kit (Takara, Otsu, Japan). Genomic DNA was then subjected to sodium bisulphate treatment using the EZ DNA Methylation kit (Zymo Research, Orange, CA, USA), converting unmethylated cytosine to thymidine and leaving methylated cytosine unchanged. For total RNA isolation, the TRIzol Reagent (Invitrogen) was used according to the protocol supplied by the manufacturer.
RT-PCR analysis
After RNA was quantified, cDNA was synthesized from 5 μg of total RNA according to the manufacturer's guidelines in a total volume of 20 μl (Fermentas, Hanover, MD, USA). Primers used for RT-PCR are as follows: ADAM23-f, 5′-TATGAGCAGCTGTCCACTCG-3′, and ADAM23-r, 5′-CCCCAGCCTGTGCCCCCAAG-3′; β-actin-f, 5′-CACGCACGATTTCCCGCTCGG-3′, and β-actin-r, 5′-CAGGCTGTGCTATCCTGTAC-3′. The PCR was performed at 94 °C for 5 min followed by 35 cycles at 94 °C for 30 s, 56 °C for 30 s and 72 °C for 40 s with a PCR Thermal Cycler Dice (Takara); β-actin was used as an internal control.
Western blotting
Western blotting to detect ADAM23 protein was performed on one SCC (SK-MES-1) and three lung ADCs (A549, SPC-A1 and LTEP-a-2). Protein lysates were obtained from approximately 1.2 × 107 cells in lysis buffer (50 mM Tris–HCl, pH 7.4, 0.5% Triton X-100 and 0.2% sodium deoxycholate). Equal amounts of protein were subjected to 8% SDS–polyacrylamide gel electrophoresis and transferred to PVDF membranes by electroblotting. The membranes were blocked with 5% non-fat milk and incubated with primary antibody. After three washes for 15 min in Tris-buffered saline (TBS) supplemented with 0.1% Tween-20 (TBST), the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody, followed by enhanced chemiluminescence. Primary antibodies against ADAM23 and β-actin were applied at the optimized concentrations (Santa Cruz Biotechnology).
Methylation-specific polymerase chain analysis
After the conversion of the DNA, methylation-specific PCR (MSP) amplification was performed to analyse the methylation pattern of the ADAM23 gene s. The primers used in the PCR were specific for methylated and unmethylated DNA (Choi et al. 2009). The sequences were as follows: forward, 5′-AGTAATAGTATTGTATTCGAAGTTTCGT-3′, and reverse, 5′-ACTTAAAACCTCCCCAACGAC-3′, for the amplification of methylated DNA (MSP, located −462 to −249 from the transcription start site); forward, 5′-AGTAATAGTATTAGTATTTGAAGTTTTGT-3′, and reverse, 5′-CAACTTAAAACCTCCCCAACA-3′, for the amplification of unmethylated DNA (USP, located −462 to −247 from the transcription start site). The PCR conditions consisted of 12 min at 94 °C for initial denaturation, followed by 37 cycles of 94 °C (30 s), 60 °C (30 s) and 72 °C (1 min) and a final elongation of 7 min at 72 °C. Amplified PCR products were fractionated in 2.5% agarose gel with ethidium bromide.
Demethylating drug treatment
For 5-aza-2′-dC treatment, cells were seeded in two culture flasks at a density of 2 × 105 cells/75 cm2 on day 0. The cells were treated with and without 1–4 μM of 5-aza-2′-dC (Sigma Chemical, Sigma, USA) for 24 h on days 2 and 5, and the medium was changed 24 h after adding 5-aza-2′-dC. Cells were harvested on day 6 for the analysis of ADAM23 expression. DMSO alone was used as a control to exclude non-specific solvent effects on cells.
Statistical analysis
Statistical analysis was performed using the chi-square or Fisher's exact tests and Spearman correlation. P-values of <0.05 were considered significant. Statistical analysis was performed using spss version 10.0 for Windows (LEAD Technologies Inc. NC, USA).
Results
ADAM23 protein and mRNA expression of ADAM23 in lung tissues
Normal bronchial tissues, pneumocytes, SCC and ADC were labelled with antibody against ADAM23 protein. ADAM23 is weakly expressed or absent in ADCs and SCCs, whereas it is strongly expressed in the basal cells of normal bronchial epithelium (Figure 1). ADAM23 expression was also detected in lung parenchyma, vascular endothelium and inflammatory cells. The status of ADAM23 expression in tumour cells was classified as negative or positive. The positive rate of ADAM23 protein was significantly lower in NSCLC than that in paired normal tissues and benign lung tissues (38.5%vs. 86.5% and 87.5%, P<0.05) (Table 1). The expression level of ADAM23 correlated with the clinicopathological characteristics of the NSCLC, as shown in Table 2. To confirm and extend the observations on the ADAM23 expression in lung SCCs and ADCs, transcript level of ADAM23 was measured by RT-PCR (data not shown). The results showed that the expression level of ADAM23 mRNA in non-cancer tissues were higher than that of cancer tissues, which are in accordance with the results of the immunohistochemistry.
Figure 1.
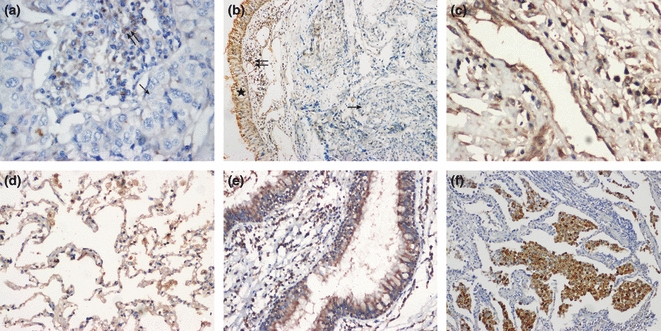
ADAM23 protein was downregulated or silenced in tumour tissues (a, b), while expressed in adjacent normal lung tissues or non-cancer tissues (c, blood vessels; d, normal parenchyma; e, normal human bronchial epithelium; f, inflammatory cells). Single arrows indicate negative cancer cells; double arrows and star indicate positive inflammatory cells and normal bronchial epithelium respectively (a, c, d, e, f ×200 magnification; b ×100 magnification).
Table 1.
The expression of ADAM23 between cancer tissues and non-cancer tissues
| ADAM23* | |||||
|---|---|---|---|---|---|
| Tissue typing | n | + | ++ | − | P† |
| Lung cancer tissues | 52 | 5 | 15 | 32 | P1 = 0.000 |
| Adjacent normal tissues | 52 | 19 | 26 | 7 | P2 = 0.009 |
| Benign pulmonary lesions | 8 | 2 | 5 | 1 | P3 = 0.941 |
ADAM23 expression was significantly lower in lung cancer compared to non-cancer tissues.
P1, P2, P3 represent the correlation between lung cancer tissues and its adjacent normal tissues (P = 0.000), lung cancer tissues and benign pulmonary lesions (0.009), and adjacent normal tissues and benign pulmonary lesions (P = 0.941) respectively.
ADAM23 protein and mRNA expression in lung cancer cell lines
We also evaluated ADAM23 expression at mRNA and protein levels in four cell lines. ADAM23 mRNA expression was lower or not detected in cell lines of SPC-A1 and LTEP-a-2 but much higher in A549 and SK-MES-1 (Figure 2a). Western blotting analysis detected ADAM23 expression at the molecular weights of 92 kDa by using a specific anti-ADAM23 antibody (Figure 2b). The results are consistent with those of RT-PCR.
Figure 2.
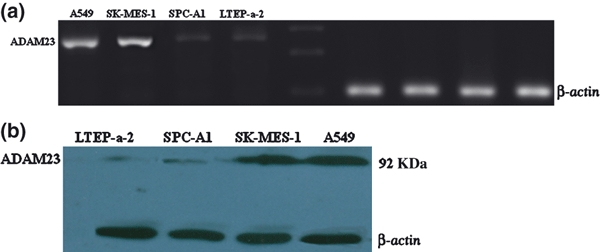
ADAM23 mRNA and protein expression. (a) ADAM23 mRNA in lung cancer cell lines. The expected products corresponding to ADAM23 (249 bp) and β-actin (101 bp) are showed. (b) Consistent with mRNA levels, ADAM23 protein was downregulated in SPC-A1 and LTEP-A-2 while overexpressed in SK-MES-1 and A549. The expected products corresponding to ADAM23 (92 kDa) and β-actin (43 kDa) were used as an endogenous control.
Mechanism of ADAM23 silencing by DNA methylation
In MSP, we tested cancer tissues and adjacent normal tissues as well as the lung cancer cells (A549, SPC-A1, LTEP-a-2, SK-MES-1). The representative results of MSP are shown in Figure 3a and b respectively. Notably, ADAM23 promoter methylation was observed in 21 of 52 (40.4%) cancer specimens and four of its adjacent normal tissues (7.6%; 4/52) but in none of 8 benign pulmonary lesions (0/8; P<0.000), together with two cell lines (SPC-A1, LTEP-a-2). Furthermore, there were 32 cancer samples of ADAM23 which showed negative staining by immunohistochemistry, in which the rate of the ADAM23 gene methylation was 50.3% (17/32). The correlation between ADAM23 protein expression and its methylation in NSCLC is shown in Table 3. Taken together, ADAM23 methylation was observed frequently in cancer tissues and unmethylated ADAM23 was found in most of the tissue samples, especially in adjacent normal tissues. We could not find any significant relationship between ADAM23 methylation status and clinicopathological features.
Figure 3.
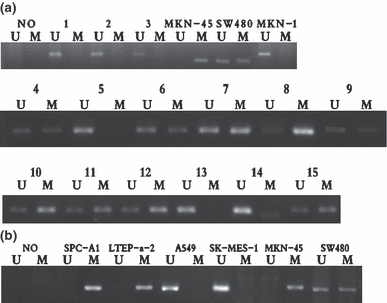
ADAM23 methylation-specific PCR (MSP) in cell lines and lung tissues. (a) MSP of ADAM23 in primary lung cancer tissues. The results from one to twelve are the representative patients of lung cancer tissues; from 13 to 15 indicate the representative patients of adjacent normal tissues. The lanes marked U indicate the presence of the unmethylated ADAM23 gene; the lanes marked M indicate the presence of methylated ADAM23 gene. MKN-45, MKN-1 and SW480 were used as positive and negative control respectively. In the blank control (NO), water was used instead of template. (b) MSP of ADAM23 in lung cancer cell lines.
Table 3.
Correlation between ADAM23 protein expression and methylation of non-small-cell lung carcinoma
| ADAM23 expression | |||
|---|---|---|---|
| ADAM23 methylation | + | − | Total |
| + | 4 | 17 | 21 |
| − | 16 | 15 | 31 |
| Total | 20 | 32 | 52 |
The positive relationship between ADAM23 protein expression and its methylation were negatively associated (r = −0.328, P = 0.017).
ADAM23 mRNA re-expression with 5-aza-2′-dC
Aberrant methylation of DNA in 5′ regulatory regions harbouring a higher than expected number of CpG dinucleotides is a key mechanism by which genes relevant to cancer initiation and progression can be silenced. To investigate whether the mechanism of ADAM23 transcriptional silencing correlates with promoter hypermethylation, we treated these heavily methylated and silenced cells (SPC-A1, LTEP-a-2) (106) incubated with 1–4 μM of 5-aza-2′-dC, a methyltransferase inhibitor, for 4 days. ADAM23 expression was restored and its methylation was reversed in some degree after treatment with 5-aza-2′-dC as detected by RT-PCR and MSP respectively (Figure 4a,b). The expression level of ADAM23 in methylated tumour cell lines increased markedly after treatment with 5-aza-2′-deoxycytidine (5-Aza-2′-dC).
Figure 4.
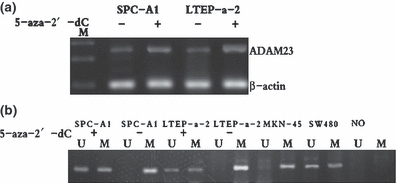
Re-expression and demethylation of ADAM23 in NSCLC cell lines by 5-aza-2′-dC. (a) The results of RT-PCR analysis revealing ADAM23 expression in two cell lines (SPC-A1, LTEP-a-2) with or without 5-aza-2′-dC treatment. Re-expression of ADAM23 was observed in two cell lines; M, molecular marker 100 bp. (b) The results of MSP analysis revealing ADAM23 gene demethylation in two cell lines after treatment with 5-aza-2′-dC. NSCLC, non-small-cell lung cancer.
Discussion
ADAM proteins are a large family of type I transmembrane glycoproteins containing more than 40 members. These proteins exert a wide variety of biological functions modulating cell–cell and cell-extracellular matrix interactions and they have been implicated in cell migration, muscle development, fertilization, and the cell cycle (Takada et al. 2005). All these functions reflect the unique topographic organization of disintegrin and metalloprotease domains (Seals & Courtneidge 2003). At present, many members of this family have been reported as being involved in the tumourigenesis, and most of these molecules are thought to be capable of accelerating tumourigenesis and progression through overexpression, while a few of them, such as ADAM11 and ADAM23, might be associated with tumourigenesis through their inactivation (Emi et al. 1993). In particular, ADAM23 is a new tumour suppressor gene that has an inactive metalloprotease domain, suggesting that it may be exclusively involved in cell adhesion through disintegrin (Takada et al. 2005). Recently, this gene has been studied in several cancers, including colorectal cancer, and showed DNA promoter hypermethylation, which is an epigenetic modification that plays an important role in the control of gene expression and chromosome structure in mammalian cells (Frigola et al. 2005). In this study, we evaluated the expression of ADAM23 and promoter methylation in NSCLC and analysed the correlation. In addition, we examined a gastric cancer cell and a colorectal cell to compare the findings with previously published studies. The results of immunohistochemisty showed that the expression level of ADAM23 protein was much higher in non-cancer tissues than that in the cancer tissues. According to our data, ADAM23 was more downregulated in NSCLC with lymph node metastasis (6.9%, 4/28) when compared to non-lymph nodes metastasis (66.7%, 16/24), and its expression level decreased with the development of pathologic TNM stage (66.7%, 8/12; 60.0%, 9/15; 16.7%, 2/12 and 7.7%, 1/13). Moreover, tumours with poor differentiation displayed the lower expression of ADAM23 (21.7%, 5/23 vs. 51.7%, 15/29). A statistically significant correlation was found between the positive rate of ADAM23 and tumour stages, lymph nodes metastasis, together with histological differentiation (P<0.05), suggesting that ADAM23 may play an important role in the invasion and metastasis of NSCLC. However, the staining of this marker and parameters such as age, gender and pathology typing proved to have no significant. It was worth noting that ADAM23 may take part in the development and progression of NSCLC. This is consistent with a previously published study (Wang et al. 2008). However, how ADAM23 performs its functions in lung carcinogenesis remains to be determined.
To investigate the relationship between the expression level of ADAM23 protein and methylation status of its promoter region of ADAM23 gene, we carried out MSP and found that the rates of ADAM23 gene promoter methylation were 40.4% in cancer tissues and 7.6% in adjacent normal tissues together with 0% in benign pulmonary lesions (0/8; P<0.000). In 32 cancer tissues of ADAM23-negative expression, the rate of the ADAM23 gene methylation accounts for 50.3% (17/32). The relationship between the expression level of ADAM23 and its promoter methylation was negatively associated (r = −0.328, P = 0.017). Thus, we could conclude that ADAM23 acts as a functional tumour suppressor gene in NSCLC, and epigenetic inactivation of ADAM23 by CpG island methylation may be an important event contributing to lung carcinogenesis and tumour progression, although the detailed mechanism underlying remains largely unknown.
Furthermore, the results of treating methylated cell lines with or without 5-aza-2′-deoxycytidine (5-Aza-2′-dC) confirmed that methylation was responsible for the gene downregulation. Thus, we proposed that hypermethylation of ADAM23 promoter CpG island reduces the expression level of the ADAM23 gene and that methylation seems to occur in a tumour-specific manner. Some samples showed methylation both in tumours and its corresponding normal tissues while other samples showed that methylation and unmethylation co-existed in the same tumour tissues (data not shown). Unmethylation was observed in all tumour tissues. That might be the presence of contaminating normal tissue or infiltrating lymphocytes. Additionally, there was a group of NSCLC in which the ADAM23 gene silenced displaying neither methylation nor unmethylation. Therefore, we believed that there are other reasons apart from methylation which silence ADAM23 gene, such as histone deacetylation, the loss of homozygosity or the contamination of the tumour tissues.
The specific mechanism of the downregulation of ADAM23 in NSCLC will be investigated in the future. To the best of our knowledge, a body of increasing evidence indicates that the methylation status of CpG sites is a promising molecular marker for human cancer, for changing in CpG methylation is associated with multistage carcinogenesis from precancerous conditions to malignant tumour progression (Mulero-Navarro & Esteller 2008; Toyota et al. 2009), and methylation of genes appears to follow a chronological order during carcinogenesis in lung cancer (Liu et al. 2010). Therefore, tumour-related genes, including ADAM23, by aberrant methylation of CpG islands are likely to be established during an early stage in the multistep process of lung carcinogenesis (Anglim et al. 2008). Although no statistically significant correlation was found between clinicopathological features and the degree of methylation in this study, some interesting features may deserve future investigation, such as methylation frequency. As a consequence, the DNA methylation correlation with TSGs's expression is a suitable marker not only for the tumour characteristics but also for the estimation of carcinogenic risk and early diagnosis.
ADAM23 is also involved in mediating cell-cell adhesion and cell-matrix interactions. It was recently shown to interact specifically with integrin of αvβ3 in an RGD-independent manner during invasion and metastasis (Verbisck et al. 2009). It has been demonstrated that ADAM23 negatively modulates integrin αvβ3 activation during metastasis (Liu et al. 2010). In our experiment, we also found that the positive rates between ADAM23 and αvβ3 were associated (χ2 = 9.026, r = −0.417, P<0.05) by analysing immunohistochemistry and the result was further confirmed in cells with or without treating with 5-aza-2′-dC (data not shown). So we proposed that the loss of ADAM23 expression might have enhanced cancer cell adhesion and invasion via modulation of integrin αvβ3 in NSCLC.
In conclusion, our findings displayed that the expression level of ADAM23 is likely to be involved in the progression of NSCLC and its downregulation probably correlated with promoter methylation. These results may proof of potential diagnostic and prognostic values in NSCLC. However, the exact role of ADAM23 in carcinogenesis remains largely unknown.
Acknowledgments
This work was supported by Nature Science Foundation of Hunan Province (06JJ2098) China and Graduate School of Central South University. We are grateful to Prof. Jifang Wen and Dr Hui Lv for their helpful comments. We thank Dr Guoqing Pan for technical help.
References
- Anglim PP, Alonzo TA, Laird-Offringa IA. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: an update. Mol. Cancer. 2008;23:7–81. doi: 10.1186/1476-4598-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky SA. Silencing of genes by promoter hypermethylation: key event in rodent and human lung cancer. Carcinogenesis. 2005;26:1481–1487. doi: 10.1093/carcin/bgi020. [DOI] [PubMed] [Google Scholar]
- Choi JS, Kim KH, Jeon YK. Promoter hypermethylation of the ADAM23 gene in colorectal cancer cell lines and cancer tissues. Cancer. 2009;124:1258–1262. doi: 10.1002/ijc.24023. [DOI] [PubMed] [Google Scholar]
- Costa FF, Verbisck NV, Salim AC, et al. Epigenetic silencing of the adhesion molecule ADAM23 is highly frequent in breast tumors. Oncogene. 2004;23:1481–1488. doi: 10.1038/sj.onc.1207263. [DOI] [PubMed] [Google Scholar]
- Costa FF, Colin C, Shinjo SM, et al. ADAM23 methylation and expression analysis in brain tumors. Neurosci. Lett. 2005;380:260–264. doi: 10.1016/j.neulet.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Emi M, Katagiri T, Harada Y, et al. A novel metalloprotease/disintegrin -like gene at 17q21.3 is somatically rearranged in two primary breast cancers. Nat. Genet. 1993;5:151–157. doi: 10.1038/ng1093-151. [DOI] [PubMed] [Google Scholar]
- Frigola J, Sole X, Paz MF, et al. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum. Mol. Genet. 2005;14:319–326. doi: 10.1093/hmg/ddi028. [DOI] [PubMed] [Google Scholar]
- Hagihara A, Miyamoto K, Furuta J, et al. Identification of 27 5′ CpG islands aberrantly methylated and 13 genes silenced in human pancreatic cancers. Oncogene. 2004;23:8705–8710. doi: 10.1038/sj.onc.1207783. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. CA Cancer J. Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Karim RZ, Gerega SK, Yang YH, et al. Proteins from the Wnt pathway are involved in the pathogenesis and progression of mammary phyllodes tumours. Clin. Pathol. 2009;62:1016–1020. doi: 10.1136/jcp.2009.066977. [DOI] [PubMed] [Google Scholar]
- Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WB, Liu JY, Ao L, et al. Epigenetic silencing of cell cycle regulatory genes during 3-methylcholanthrene and diethylnitrosamine-induced multistep rat lung cancer. Mol. Carcinog. 2010;49:556–565. doi: 10.1002/mc.20621. [DOI] [PubMed] [Google Scholar]
- Longo-Sorbello GS, Chen B, Budak-Alpdogan T, Bertino JR. Role of pemetrexed in non-small cell lung cancer. Cancer Invest. 2007;25:59–66. doi: 10.1080/07357900601130748. [DOI] [PubMed] [Google Scholar]
- Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2007;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: the time is now. Crit. Rev. Oncol. Hematol. 2008;68:1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Neal CL, Yao J, Yang W, et al. 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 2009;69:3425–3432. doi: 10.1158/0008-5472.CAN-08-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi SA, Bashir MU, Yaqinuddin A. Utility of DNA methylation markers for diagnosing cancer. Int. J. Surg. 2010;8:194–198. doi: 10.1016/j.ijsu.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Sagane K, Ohya Y, Hasegawa Y, et al. Metalloproteinase-like, cysteine-rich proteins MDC2 and MDC3: novel human cellular disintegrins highly expressed in the brain. Biochem. J. 1998;334:93–98. doi: 10.1042/bj3340093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Yoshino I. Aberrant methylation in non-small cell lung cancer. Surg. Today. 2010;40:602–607. doi: 10.1007/s00595-009-4094-6. [DOI] [PubMed] [Google Scholar]
- Takada H, Imoto I, Tsuda H, et al. ADAM23, a possible tumor suppressor gene, is frequently silenced in gastric cancers by homozygous deletion or aberrant promoter hypermethylation. Oncogene. 2005;24:8051–8060. doi: 10.1038/sj.onc.1208952. [DOI] [PubMed] [Google Scholar]
- Takanami I, Abiko T, Koizumi S. Expression of maspin in non-small-cell lung cancer: correlation with clinical features. Clin. Lung Cancer. 2008;9:361–366. doi: 10.3816/CLC.2008.n.052. [DOI] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Yamashita T, et al. Cancer epigenomics: implications of DNA methylation in personalized cancer therapy. Cancer Sci. 2009;100:787–791. doi: 10.1111/j.1349-7006.2009.01095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Muller-Hermelink HK, et al. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon: IARC Press; 2004. [Google Scholar]
- Verbisck NV, Costa ET, Costa FF, et al. ADAM23 negatively modulates alpha(v)beta(3) integrin activation during metastasis. Cancer Res. 2009;69:5546–5552. doi: 10.1158/0008-5472.CAN-08-2976. [DOI] [PubMed] [Google Scholar]
- Wang FJ, Zhang Q, Yu HL, et al. Expression of ADAM23 gene and its significance in human colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2008;16:3406–3409. [Google Scholar]
- Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of tumour suppressor and tumour-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–592. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


