Abstract
Over half of all human cancers, of a wide variety of types, sustain mutations in the p53 tumor suppressor gene. Although p53 limits tumorigenesis through the induction of apoptosis or cell cycle arrest, its molecular mechanism of action in tumor suppression has been elusive. The best-characterized p53 activity in vitro is as a transcriptional activator, but the identification of numerous additional p53 biochemical activities in vitro has made it unclear which mechanism accounts for tumor suppression. Here, we assess the importance of transcriptional activation for p53 tumor suppression function in vivo in several tissues, using a knock-in mouse strain expressing a p53 mutant compromised for transcriptional activation, p5325,26. p5325,26 is severely impaired for the transactivation of numerous classical p53 target genes, including p21, Noxa, and Puma, but it retains the ability to activate a small subset of p53 target genes, including Bax. Surprisingly, p5325,26 can nonetheless suppress tumor growth in cancers derived from the epithelial, mesenchymal, central nervous system, and lymphoid lineages. Therefore, full transactivation of most p53 target genes is dispensable for p53 tumor suppressor function in a range of tissue types. In contrast, a transcriptional activation mutant that is completely defective for transactivation, p5325,26,53,54, fails to suppress tumor development. These findings demonstrate that transcriptional activation is indeed broadly critical for p53 tumor suppressor function, although this requirement reflects the limited transcriptional activity observed with p5325,26 rather than robust transactivation of a full complement of p53 target genes.
The p53 protein plays a critical role in suppressing tumorigenesis, as evidenced by its frequent mutation in human cancers and the fully penetrant cancer phenotype of p53 null mice (1, 2). p53 is thought to act as a tumor suppressor by inducing apoptosis or cellular senescence in response to cellular stresses such as DNA damage or oncogenic signals, with the relative importance of apoptosis or senescence as a tumor suppressor mechanism varying by tissue type (1, 3, 4). The molecular mechanism by which p53 blocks cancer development, however, has remained elusive.
The best-characterized molecular function of p53 is as a transcriptional activator (1), and in this capacity it induces the transcription of a plethora of target genes. However, whether transactivation is the p53 activity responsible for its tumor suppressor function has been controversial, as p53 possesses a variety of other biochemical activities. For example, p53 directly represses the transcription of numerous genes by binding to p53 response elements in the regulatory regions of these genes. Additionally, p53 induces apoptosis through mitochondrial membrane permeabilization via interactions with Bcl-2 family members (5).
To assess the contribution of transcriptional activation to p53 function, we previously generated a knock-in mouse strain expressing p5325,26, a p53 mutant bearing alterations in critical residues in the transactivation domain, from the endogenous p53 promoter (6). We showed that this mutant is severely compromised for the transactivation of the majority of known p53 target genes, including p21, Puma, and Noxa, but that it retains the ability to properly activate a small set of p53 target genes including Bax and other recently identified target genes (6, 7). Consistent with the critical role for well-established, classical p53 target genes, such as p21 or Puma, Noxa, and Perp in G1 arrest or apoptosis (8, 9), respectively, the p5325,26 protein is defective in inducing cell cycle arrest or apoptosis in response to acute DNA damage. In contrast, the p5325,26 mutant retains function in response to oncogenic signals, such as in engaging the p53 effector program of cellular senescence. Moreover, it is capable of serving as a tumor suppressor in a mouse model for nonsmall cell lung cancer (NSCLC), suggesting that efficient transactivation of most canonical target genes by p53 is dispensable for p53 tumor suppressor function in this epithelial lineage. Instead, tumor suppressor activity is associated with robust p53-mediated activation of a set of newly identified p53 target genes (7).
Given that p53 elicits different cellular effector responses, including cell cycle arrest, apoptosis, or senescence, to promote tumor suppression in diverse tissues and in response to different initiating oncogenic lesions (10–13), it is also possible that the biochemical mechanism underlying p53 action in tumor suppression might similarly vary according to context. For example, p53-mediated apoptosis limits development of Eμ-myc–driven B-cell lymphomas and large T-antigen–induced choroid plexus tumors (10, 13). In contrast, p53-dependent senescence is thought to restrict development of lung cancers caused by Ras pathway activation (11) or prostate cancers caused by Pten loss (12). Thus, here, using the p5325,26 knock-in mice, we specifically investigate the role of transcriptional activation by p53 in the suppression of tumors arising from diverse cell types of origin to assess whether p53 uses a common mechanism in tumor suppression in different tissues. We have extended our analysis based on epithelial tumors to encompass analysis of tissues representing the four major lineages that give rise to tumors: epithelial, mesenchymal, central nervous system, and lymphoid. We analyze the contribution of intact p53 transactivation function to the suppression of fibrosarcoma, medulloblastoma, and B-cell lymphoma development. Collectively, our studies reveal that full p53 transactivation potential is dispensable for tumor suppression in diverse tissue types, and that instead, robust transactivation of the small set of genes efficiently induced by p5325,26 and/or minimal activation of canonical p53 target genes accounts for tumor suppression. These observations provide important general insight into p53's mechanism of action in preventing cancer.
Results
p5325,26 Is an Effective Suppressor of Fibrosarcoma Growth.
We first sought to examine the importance of transcriptional activation for p53-mediated tumor suppression in mesenchymal cancers using a fibroblast transplant system in which mouse embryo fibroblasts (MEFs) transformed with the E1A and H-RasV12 oncogenes are injected into immunocompromised mice and tumor growth is assessed (14). In our knock-in mice, expression of the p5325,26 allele is regulated by a transcriptional stop element flanked by LoxP sites (Lox-Stop-Lox, LSL). Thus, homozygous p53LSL-25,26/LSL-25,26 MEFs derived from these mice can produce either no p53 protein or p5325,26, through infection with adenovirus-empty (Ad-empty) or adenovirus-Cre (Ad-Cre), respectively (Fig. 1 A and B). We introduced activated H-RasV12 and E1A into wild-type and p53LSL-25,26/LSL-25,26 MEFs through retroviral transduction, followed by infection with Ad-Cre or Ad-empty. We verified the severely compromised transcriptional activity of p5325,26 on several well-established p53 target genes (Fig. 1C (6). E1A-Ras MEFs expressing wild-type p53, no p53, or p5325,26 were s.c. injected into the flanks of Scid mice, and tumor growth was monitored over time. In this model, E1A-Ras MEFs lacking p53 rapidly formed large tumors, whereas cells expressing wild-type p53 formed small tumors that developed with longer latency (Fig. 2A). Similarly, cells expressing p5325,26 produced cancers dramatically smaller than those from p53-deficient cells, although these tumors were somewhat larger than those carrying wild-type p53. We hypothesized that the greater size of the p5325,26 tumors relative to wild-type tumors was not due to a difference in p53 activity, but rather to an outgrowth of the small percentage of p53-deficient cells retaining the LSL element in the mice injected with E1A-Ras p5325,26 cells (Fig. 1D). Indeed, although p5325,26 expression was detectable in over 96% of E1A-Ras MEFs before injection, immunohistochemical staining of the tumors derived from the p5325,26-expressing cells revealed that there was significant expansion of cells lacking p53 protein expression in every tumor (Fig. 2B). This kind of in vivo competition to assess the relative fitness of two types of cells—nonrecombinant p53-deficient versus recombinant p5325,26 mutant-expressing cells in this case—in cancer development is a highly informative and widely used approach (Fig. 1D). Our findings indicate that in this context, a strong selective advantage exists for cells lacking expression of p5325,26, reflective of the potent tumor suppressor activity of the p5325,26 protein (Fig. 1D). This finding is consistent with the observed tumor suppression potential of p5325,26 in a lung cancer model (7).
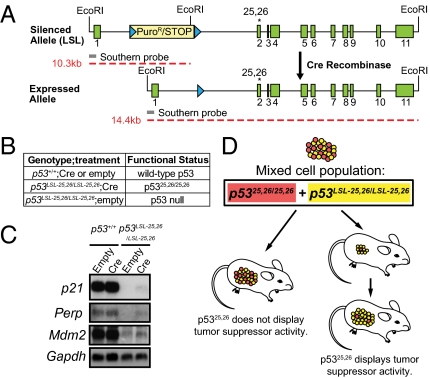
Fig. 1.
Experimental design for this study. (A) Schematic of the p5325,26 allele. The silenced LSL allele comprises exons 1–11 (green boxes) of p53 with the L25Q/W26S mutations in exon 2 (asterisk) and a transcriptional stop element/puromycin resistance cassette (yellow box) flanked by LoxP sites (blue triangles) inserted into intron 1. The transcription of mutant p53 is silenced until Cre mediates recombination between the LoxP sites and excision of the stop element. The location of the 5′ probe (gray bar) used for Southern blotting and the sizes of the EcoRI fragments (red dashed lines) generated from each allele are indicated. (B) Table summarizing conditional p53 protein expression strategies used throughout the manuscript. (C) Analysis of p53 target gene expression in E1A-Ras MEFs by Northern blotting. Gapdh serves as a loading control. Efficient adenoviral-Cre–mediated recombination of the LSL-25,26 allele was confirmed by immunofluorescence staining for p53, with over 94% of the cells expressing p5325,26. (D) An in vivo competition assay was used throughout the study to evaluate the tumor suppression potential of p5325,26. Red cells represent p5325,26 mutant-expressing tumor cells, and yellow ones represent tumor cells in which Cre failed to delete the LSL element, and therefore retain the p53LSL-25,26/LSL-25,26 status and are p53 null. If tumors form from p5325,26 mutant cells (Left), then the mutant is ineffective as a tumor suppressor. If tumors form due to the outgrowth of the p53 null cells (Right), then it suggests that the p5325,26 mutant is an effective tumor suppressor. In this case, tumor growth may appear retarded at early time points relative to p53 null tumors.
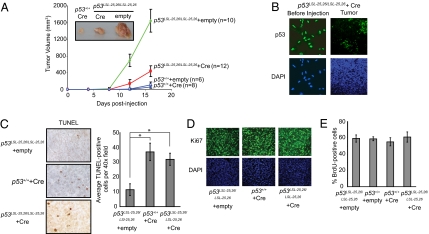
Fig. 2.
p5325,26 efficiently suppresses tumorigenesis in transplanted transformed MEF fibrosarcomas through the induction of apoptosis. (A) p5325,26 restricts tumor growth. In this and subsequent panels of this figure, MEFs expressed E1A, H-RasV12, and no p53 (p53LSL-25,26/LSL-25,26 + Ad-empty), p53wt (p53+/+ + Ad-empty or p53+/+ + Ad-Cre), or p5325,26 (p53LSL-25,26/LSL-25,26 + Ad-Cre). E1A-Ras MEFs of various p53 genotypes were injected into the flanks of Scid mice, and average tumor volume was monitored for 16 d. The number of tumors analyzed is indicated. Error bars show the SD. Photograph shows a representative example of tumors of each genotype at day 16. (B) A selective pressure exists for cells lacking p5325,26 expression during E1A-Ras tumor growth in vivo. (Left) Representative images showing that p5325,26 is expressed in greater than 96% of E1A-Ras MEFs before injection into mice, with DAPI as a nuclear stain. (Right) Representative example of p53 immunofluorescence staining of E1A-Ras p5325,26 tumors. DAPI stain shows that the observed field is uniformly filled with tumor cells. (C) TUNEL staining of histological sections from tumors. (Left) Representative example showing TUNEL+ cells staining brown, with hematoxylin as a counterstain. (Right) Quantification of TUNEL staining. The number of TUNEL+ cells in three 40× fields for each of three to five tumors of each genotype was averaged and graphed ±SEM. *P < 0.03 (Student's t test). (D) p5325,26 does not affect proliferation in vivo. E1A-Ras MEF tumors expressing no p53, p53wt, or p5325,26 were stained for Ki67, with DAPI as a nuclear stain. Shown are representative pictures from multiple experiments. (E) p5325,26 does not inhibit proliferation in vitro. E1A-Ras MEFs expressing no p53, p53wt, or p5325,26 were pulsed for 4 h with BrdU and immunostained for p53 and BrdU. Graph shows the average BrdU-labeling index in three independent experiments, ±SD. Efficient Cre-mediated recombination of the LSL-25,26 allele was confirmed by immunofluorescence staining for p53, with over 94% of the cells expressing p5325,26. For C and D, due to the outgrowth of p53-deficient cells in the p5325,26 tumors, we costained tumors for p53 and either TUNEL or Ki67 and analyzed only regions with homogenous p5325,26 expression.
As apoptosis is thought to be the mechanism through which p53 limits tumor growth in this fibrosarcoma model (14), we evaluated the capacity of p5325,26 to drive apoptosis in this setting. As reported, tumors expressing wild-type p53 displayed a significantly higher apoptotic index than those lacking p53 protein expression (Fig. 2C). In the regions of the tumors expressing p5325,26, the apoptotic index mirrored that seen in wild-type p53-expressing tumors. In contrast, analysis of Ki67 expression in tumors, as well as BrdU pulse labeling of E1A-Ras MEFs in vitro, showed no obvious difference in the number of cycling cells between the three genotypes (Fig. 2 D and E). Together, these data indicate that p5325,26, like wild-type p53, triggers apoptosis to suppress tumor growth in this setting. Thus, full p53 transactivation potential is dispensable for the induction of apoptosis in response to hyperproliferative signals and for the consequent suppression of tumor growth in this fibrosarcoma model.
p5325,26 Suppresses Medulloblastoma Formation in Vivo.
To examine the requirement for p53 transactivation in tumor suppression in CNS tumors, we used an autochthonous tumor model for medulloblastoma in which p53 loss greatly affects the penetrance of the cancer phenotype. As with humans carrying one inactivated allele of the Sonic hedgehog receptor-encoding gene Patched1 (Ptch), Ptch+/− mice are predisposed to developing tumors in the cerebellum (15). Loss of p53 synergizes with Ptch heterozygosity, resulting in a marked increase in the rate and frequency of medulloblastoma formation, with the penetrance increasing from ∼15% to >95% (16).
To determine the importance of p53 transactivation for tumor suppression in this context, we examined the consequence of p5325,26 expression in Ptch+/− mice. For these experiments, we used mice expressing Cre under the control of the Math1 promoter, which specifically drives Cre expression in cerebellar granule neuron precursor cells (GNPs), the cell of origin for medulloblastoma (17). We generated cohorts of Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre and Ptch+/−; p53+/+; Math1-Cre mice as well as Ptch+/−; p53LSL-25,26/LSL-25,26 mice that served as p53 null controls. As an additional wild-type control, we generated Ptch+/−; p53LSL-wt/LSL-wt; Math1-Cre mice, and as another p53 null control, we generated Ptch+/−; p53LSL-wt/LSL-wt mice. The efficacy of p53 expression was demonstrated by immunostaining for p53 in P9 cerebellum, which revealed that ∼98% of GNPs express p5325,26 (Fig. 3A). All cohorts were aged and assessed for tumor burden upon morbidity.
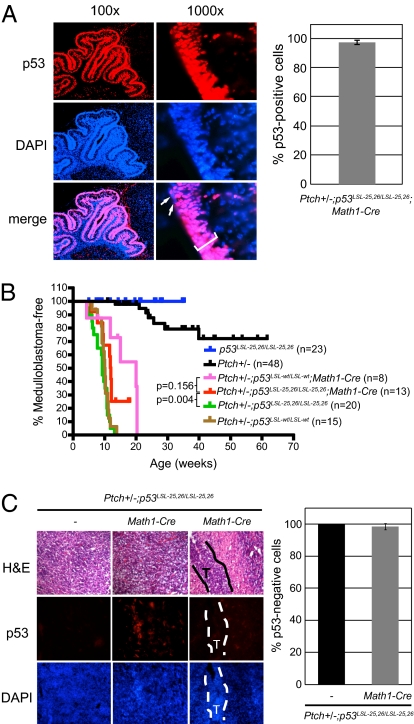
Fig. 3.
Expression of p5325,26 suppresses medulloblastoma in Ptch+/− mice. (A) p5325,26 is efficiently expressed in granule neuron precursor cells. (Left) Sections of Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre P9 cerebella were stained for p53 (Top) and DAPI (Middle). Merged image (Bottom) reveals that only a small percentage of cells are p53 deficient (arrows). (Right) Quantification of the percentage of p53+ cells in the external granular layer (bracket) of the cerebellum is shown. Error bars represent ±SD. (B) Kaplan–Meier analysis showing medulloblastoma incidence for Ptch+/− (p53wt; black), Ptch+/−; p53LSL-25,26/LSL-25,26 (p53null; green), Ptch+/−; p53LSL-wt/LSL-wt (p53null; brown), Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre (p5325,26; red), Ptch+/−; p53LSL-wt/LSL-wt; Math1-Cre (p53wt; pink), and p53LSL-25,26/LSL-25,26 (p53null; blue) mice. (C) Tumors are mainly composed of p53null cells. (Top) Representative H&E-stained sections of advanced tumors from Ptch+/−; p53LSL-25,26/LSL-25,26 (p53null) and Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre (p5325,26) mice and an intermediate tumor (T; outlined region) from a Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre mouse. Sections from corresponding samples were stained for p53 (Middle) and counterstained with DAPI (Bottom). Graph shows the percentage of p53− cells relative to DAPI-stained cells in tumors. Three to four samples per genotype were examined and error bars represent ±SD.
Consistent with previous reports, p53 deficiency in Ptch+/− mice resulted in 100% incidence of medulloblastoma by 12 wk of age, whereas only 15% of Ptch+/−; p53+/+ mice developed medulloblastomas, and did so with longer latency (Fig. 3B). p53 loss alone did not result in medulloblastomas. Interestingly, we observed a significant delay in the onset of medulloblastoma in Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre animals relative to Ptch+/−; p53LSL-25,26/LSL-25,26 mice (P = 0.0042, log rank test). The Ptch+/−; p53LSL-wt/LSL-wt; Math1-Cre mice displayed a similar decrease in medulloblastoma incidence relative to Ptch+/−; p53LSL-wt/LSL-wt animals (P = 0.0009, log rank test), and the medulloblastoma incidence was not statistically significantly different from that of the Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre animals (P = 0.156, log rank test). However, the incidence of medulloblastoma in Ptch+/−; p53LSL-wt/LSL-wt; Math1-Cre mice was greater than the 15% seen in the Ptch+/−; p53+/+ animals, suggesting that the medulloblastomas developing in Lox-Stop-Lox mice might result from an outgrowth of p53 null cells (Fig. 1D). To determine whether this was the case, cerebellar tumor sections from Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre mice were stained for p53 expression. Indeed, the majority of cells in the Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre tumors lacked p53 expression, with the exception of a few small patches of p53-expressing cells in two of the tumors (Fig. 3C). These findings suggest that tumorigenesis in Ptch+/−; p53LSL-25,26/LSL-25,26; Math1-Cre mice is largely due to a selective advantage for p53 null cells over p5325,26-expressing cells, consistent with our observations from the E1A-Ras MEF tumor experiments, and these data highlight the potency of p5325,26 tumor suppressor activity. Thus, full p53 transcriptional activation potential is not required for tumor suppression in the cerebellum.
p5325,26 Suppresses B-Cell Lymphoma Development in Vivo.
To assess the role of transcriptional activation for p53-mediated tumor suppression in the lymphoid lineage, we used Eμ-myc transgenic mice, which provide a well-characterized model for human non-Hodgkin's B-cell lymphoma in which p53 is critical for suppressing lymphoma development (18, 19). Specifically, transgenic Eμ-Myc mice heterozygous for a p53 null allele develop fully penetrant, aggressive lymphomas by 1–2 mo, accompanied by Loss of Heterozygosity (LOH) for p53, demonstrating that loss of p53 is a key step for lymphomagenesis. For our study, it was not possible to obtain Eμ-Myc; p53LSL-25,26/LSL-25,26 mouse cohorts through breeding because the Eμ-Myc; p53LSL-25,26/+ mice do not survive long enough to breed, just like Eμ-Myc; p53+/− mice. We therefore took advantage of the fact that lymphomas developing in Eμ-Myc mice heterozygous for a p53 null allele (i.e., Eμ-myc; p53LSL-25,26/+) display LOH, leading to the loss of the wild-type p53 allele (Fig. 4A (18). Lymphomas could then be derived from Eμ-myc; p53LSL-25,26/+ mice, manipulated in vitro to induce p5325,26 expression and transplanted into a number of recipient mice to examine lymphoma growth. We generated cohorts of Eμ-myc; p53LSL-25,26/+ mice expressing a Rosa26-CreER transgene (20) and allowed them to develop lymphomas. Lymphoma cells were isolated and cultured in vitro, and the Eμ-myc; Rosa26-CreER; p53LSL-25,26/LSL-25,26 status of the cells expected upon LOH was confirmed by Southern blotting (Fig. 4B and Fig. S1). We then optimized the dose and time of 4-hydroxytamoxifen (4-OHT) treatment to achieve the highest level of recombination of the LSL element possible (Fig. 4B), but we could never achieve better than 50% recombination in Eμ-myc; Rosa26-CreER; p53LSL-25,26/LSL-25,26 cells. Therefore, as in the cases of the fibrosarcoma and medulloblastoma models, we aimed to assess the relative fitness of nonrecombinant p53-deficient and recombinant p5325,26-expressing cells (Fig. 1D). The 4-OHT–treated lymphoma cells were introduced into a cohort of isogenic wild-type recipient mice, and the recipient mice were monitored for lymphoma development. Visible lymphomas formed after 2–3 wk in the vast majority of the recipients. Interestingly, despite the injected cells showing ∼50% recombination and expressing abundant p5325,26 protein before injection, the reconstituted lymphomas resulting from them did not carry the recombined p53LSL-25,26 allele or express p5325,26 protein (Fig. 4 C–E). The clear selective advantage for the functionally p53 null lymphoma cells relative to lymphoma cells expressing p5325,26 suggests that the p5325,26 mutant displays tumor suppressor activity. Together, our observations from multiple tumor studies suggest that full p53 transactivation function is unnecessary for p53 tumor suppressor function in a wide range of tissues.
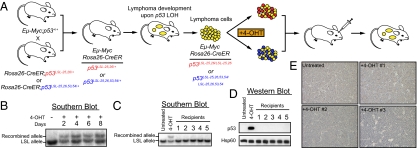
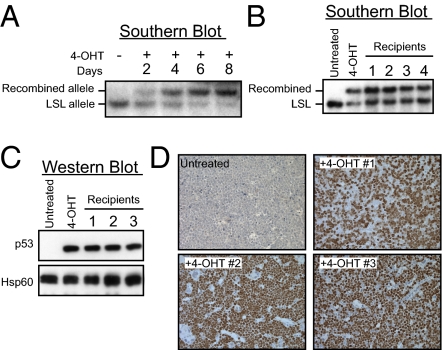
Fig. 4.
p5325,26 displays tumor suppressor activity in Eμ-Myc–driven B-cell lymphomas. (A) Schematic of the Eμ-Myc study design. Eμ-Myc mice were crossed to Rosa26-CreER; p53LSL-25,26/+ and Rosa26-CreER; p53LSL-25,26,53,54/+ mice. The resulting Eμ-Myc; Rosa26-CreER; p53LSL-25,26/+ and Eμ-Myc; Rosa26-CreER; p53LS-25,26,53,54/+ mice developed aggressive lymphomas with short latency (tumors in yellow), and the p53 locus underwent LOH, resulting in Eμ-Myc; Rosa26-CreER; p53LSL-25,26/LSL-25,26 and Eμ-Myc; Rosa26-CreER; p53LSL-25,26,53,54/LSL-25,26,53,54 tumors. Lymphoma cells were cultured and treated with 4-OHT to activate Cre and induce recombination of the LSL element and expression of the mutant p53 proteins (p5325,26 in red and p5325,26,53,54 in blue). Tumor cells were retroorbitally injected into syngeneic recipient mice and lymphoma growth was monitored. (B) Time course of recombination after 4-OHT treatment. Eμ-Myc; Rosa26-CreER; p53LSL-25,26/LSL-25,26 lymphoma cells were treated with 1 μM 4-OHT and harvested at different time points for Southern blot analysis using a probe that can differentiate the recombined (Upper band) and nonrecombined (Lower band) LSL-p53 allele (Fig. 1A). The first lane displays DNA from untreated lymphoma cells. (C) Representative Southern blot analysis of DNA from lymphomas that developed in recipient mice. Southern blotting was performed to check the ratio of the recombined and nonrecombined alleles. The first two lanes show untreated cells and 4-OHT–treated cells used for injection. (D) Representative Western blot analysis on p5325,26 expression in the reconstituted lymphomas. The first two lanes show untreated cells and 4-OHT–treated cells used for injection. Hsp60 serves as a loading control. (E) Representative p53 immunohistochemistry (IHC) in the reconstituted lymphomas. The tumor in the upper left corner was reconstituted from untreated Eμ-Myc;Rosa26-CreER;p53LSL-25,26/LSL-25,26 lymphoma cells and tumors 1–3 were from 4-OHT-treated cells and correspond to the same tumors in the Southern blot in (C) and Western blot in (D).
p5325,26,53,54 Fails to Suppress B-Cell Lymphoma Development in Vivo.
The ability of p5325,26 to suppress the development of multiple tumor types suggests that the limited transactivation capacity of this mutant may account for tumor suppression. Indeed, analysis of another knock-in mouse strain that we generated, in which mutations were introduced into both transactivation domains (p5325,26,53,54), thereby completely inactivating p53 transactivation function, showed that transcriptional activation is critical for tumor suppression in a NSCLC cancer model (7). To assess this possibility in another tumor type, we generated Eμ-myc; Rosa26-CreER; p53LSL-25,26,53,54/+ mice, from which we derived lymphoma cells as described above. In these lymphomas, the wild-type p53 allele was lost through LOH (Fig. 5A). The cultured tumor cells were treated with 4-OHT to induce recombination of the LSL element and expression of p5325,26,53,54. Akin to the situation with the Eμ-myc; Rosa26-CreER; p53LSL-25,26/LSL-25,26 lymphoma cells, recombination was incomplete (Fig. 5A), and we again sought to examine the competitive growth of this mixed population of tumor cells in vivo. Lymphoma cell transplantation into isogenic recipient mice was performed, and tumor development was monitored.
Fig. 5.
Suppression of B-cell lymphoma growth by p53 requires transcriptional activation. (A) Time course of recombination after 4-OHT treatment. Eμ-Myc;Rosa26-CreER;p53LSL-25,26,53,54/LSL-25,26,53,54 lymphoma cells were treated with 4-OHT and harvested at different time points for Southern blot analysis, as in Fig. 4. (B) Representative Southern blot analysis of DNA from lymphomas that developed in recipient mice to check the ratio of the recombined and nonrecombined alleles. The first two lanes show untreated cells and 4-OHT-treated cells used for injection. (C) Representative Western blot analysis on p5325,26,53,54 expression in the reconstituted lymphomas. The first two lanes show untreated cells and 4-OHT-treated cells used for injection. Hsp60 serves as a loading control. (D) Representative p53 IHC in the reconstituted lymphomas. The tumor in the Upper Left corner was reconstituted from untreated Eμ-Myc; Rosa26-CreER; p53LSL-25,26,53,54/LSL-25,26,53,54 lymphoma cells and tumors 1–3 were from 4-OHT–treated cells and correspond to the same tumors in the Southern blot in B and Western blot in C.
In contrast to the lymphomas derived from Eμ-myc; Rosa26-CreER; p53LSL-25,26/LSL-25,26 cells, lymphomas resulting from the Eμ-Myc; Rosa26-CreER; p53LSL-25,26,53,54/LSL-25,26,53,54 cells typically retained the recombined p5325,26,53,54-expressing allele at approximately the same ratio as the injected cells (Fig. 5B). Furthermore, strong p5325,26,53,54 protein expression was detected in the lymphomas by Western blot and immunohistochemical analysis (Fig. 5 C and D). Such a scenario, in which tumors that develop stain prominently for mutant p53, suggests that the p5325,26,53,54 mutant is ineffective in tumor suppression, as there is no selective advantage for p53 null cell outgrowth. The inability of the transactivation-dead p5325,26,53,54 mutant to suppress tumor growth indicates that the transactivation function of p53 is indeed critical for lymphoma suppression in vivo. However, the tumor suppressor activity of the p5325,26 transactivation hypomorph suggests that only limited p53 transactivation potential is required for lymphoma suppression.
Discussion
Here, we set out to determine whether p53 uses similar or different molecular mechanisms for tumor suppression in distinct tumor types. Specifically, we investigated the requirement for transcriptional activation for p53-mediated suppression of tumors derived from different lineages, through analysis of knock-in mice expressing a p53 transcriptional activation mutant, p5325,26. This mutant is severely impaired in its ability to activate the majority of canonical p53 target genes, but it does retain the ability to properly induce a small subset of p53 target genes, such as Bax and recently described targets, including Phlda3, Abhd4, and Sidt2 (6, 7). Interestingly, our studies of the p5325,26 mutant reveal that it retains tumor suppressor activity in all tissues examined, indicating that full transactivation potential is not required for p53 to elicit a tumor suppressor response in a variety of different contexts.
In our tumor studies, we used a Cre-regulated conditional system in which some transformed cells expressed p5325,26, whereas others that failed to delete the stop element retained a p53 null status. Thus, our tumorigenesis assays were based on in vivo competition between p5325,26 mutant cells and p53 null cells, a powerful approach to analyze the relative fitness of two populations of cells. In such experiments, the relative abundance of cells of each genotype is evaluated in the final tumors and compared with the starting population. If a certain population of tumor cells exhibits a selective growth advantage over the other, it will dominate within the final tumor mass. This strategy has been applied successfully to measure tumorigenic phenotypes previously (10). In our study, the selective proliferative advantage of p53 null cells over p5325,26-expressing cells in multiple tumor types, including fibrosarcomas, medulloblastomas, and lymphomas, has demonstrated that the p5325,26 mutant displays potent tumor suppressor activity in cancers of different origins. It will be interesting in the future to determine whether this tumor suppressor activity is strictly cell autonomous or may reflect non–cell-autonomous effects as well.
Our findings contrast with another recent study, also using a conditional system to drive p5325,26 expression in thymocytes, which concluded that p5325,26 cannot suppress spontaneous T-cell lymphomagenesis (21). In accord with this study, we find that spontaneous tumors, including thymic lymphomas, can develop efficiently in tamoxifen-treated, aging p53LSL-25,26/LSL-25,26; Rosa26-CreER mice (Fig. S2A). However, as in the studies described here, we find that upon close inspection, tumors derive from the fraction of cells that fail to delete the LSL element, which are therefore p53 null, as in all of the scenarios presented here (Fig. S2B). Thus, our findings collectively indicate a strong selection pressure against p5325,26 expression, underscoring its potent tumor suppressor activity. Importantly, in the development of both spontaneous tumors and Eμ-Myc–driven B-cell lymphomas, we observe no selection pressure against p5325,26,53,54 expression in tumors (Fig. 5D and Fig. S2B).
The importance of p53 transcriptional activation for tumor suppression has been suggested by two major lines of evidence. First, p53 mutations found in tumors typically lie within the DNA binding domain, suggesting that inactivation of DNA binding is critical for tumor development (4). This idea is consistent with an important role for transactivation, which relies on sequence-specific DNA binding through the DNA binding domain. Second, studies of specific p53 target genes, through either RNAi-based knockdown approaches or the generation of knockout mice, have implicated these genes as important for p53 effector functions, including apoptosis and senescence (9, 22–24). Although these target gene studies have been of great importance in defining the pathways p53 uses to initiate these responses, no target gene knockout has ever produced the dramatic tumor predisposition observed in p53-deficient mice. Previous explanations have posited that the combined actions of proteins encoded by a host of p53 target genes mediate p53's tumor suppressor function, and thus the knockout of any individual target gene has minimal, if any, consequence for p53 tumor suppressor activity. Our study helps to address this issue by using p5325,26, which is severely compromised for activation of myriad p53 target genes, allowing us to effectively phenocopy the knockdown of numerous p53 targets in one mouse. Despite the inability of p5325,26 to efficiently induce most p53 target genes, it still remains a potent tumor suppressor, supporting the idea that strong transactivation of the majority of established p53-inducible genes is not essential for p53 tumor suppressor function. In contrast, the ability of p53 to induce either cell cycle arrest or apoptosis in vivo in response to acute DNA damage signals does depend on full p53 transcriptional activation potential (7).
Our studies do nonetheless suggest that some flavor of transcriptional activation is critical for p53 tumor suppressor function, as the p5325,26,53,54 mutant, which has lost all transactivation potential, fails to display tumor suppressor activity in NSCLC and both B-cell and T-cell lymphomas (ref. 7 and this study). The transcriptional activation requirement for tumor suppression may be through low-level transactivation of classical p53 targets such as p21 or Puma or through full transactivation of the small subset of p53 targets properly induced by p5325,26, such as Bax and other recently described targets (7). The latter possibility is supported by the finding that Bax loss compromises Myc-induced apoptosis and alleviates the selection pressure for p53 mutations during Eμ-Myc–driven lymphomagenesis (25) and the observed tumor suppressor activity of several of these recently described targets, such as Phlda3, Abhd4, and Sidt2 (7). It may also be that combined low-level activation of canonical p53 target genes and strong induction of recently discovered p53 target genes, accounts for p53 tumor suppressor activity, a possibility supported by the observed tumor suppressor activity of p21 and Puma in certain settings (26, 27). Future studies will further elaborate the transcriptional networks fundamental for tumor suppression in different settings.
Our findings have significant implications for improving cancer therapy, especially by suggesting strategies to restore critical p53 functions in tumor suppression. Current p53-oriented therapeutic strategies are focused on ectopically expressing wild-type p53 in p53 null tumors or restoring wild-type p53 function in tumors in which p53 is incapacitated by alterations in other pathway components (28). Whereas the first strategy is limited by such factors as efficiency of delivery, the second approach relies on the presence of wild-type p53, which is mutated in over half of human cancers. Defining components of the p53 transcriptional network critical for tumor suppression greatly expands our options in target selection for therapeutic intervention, thereby helping to overcome obstacles associated with strategies relying on p53 as the sole target.
Materials and Methods
Immunostaining.
p53 immunofluorescence (CM5, 1:150, Novocastra) and TUNEL staining were performed as described (6, 7). IHC was performed using standard methods. Unmasking for p53 was performed by incubating 10 min in 0.01 M citrate buffer pH 6.0 in a pressure cooker and for Ki67 by incubating 15 min in unmasking solution (Vector Labs) in a boiling water bath. p53 was detected with CM5 (1:250) and Ki67 with anti-Ki67 (1:100; BD Pharmingen).
Fibrosarcoma and Medulloblastoma Tumorigenesis Assays.
MEFs were isolated, cultured, and infected with adenoviruses or retroviruses as described (6). For allograft assays, 1 × 106 E1A-Ras MEFs were injected into the flanks of Scid mice, and tumor growth was monitored by three orthogonal measurements using digital calipers. For medulloblastoma experiments, Kaplan–Meier survival curves and associated statistics were generated using GraphPad Prism4.0 software. Tumors were processed for histological examination using standard procedures.
Lymphoma Reconstitution Assays.
Eμ-Myc transgenic mice on a C57BL/6 background were crossed to Rosa26-CreER transgenic; p53LSL-25,26/+ or p53LSL-25,26,53,54/+ mice of a mixed C57BL/6 and 129S/v background to generate the Eμ-Myc; Rosa26-CreER; p53LSL-25,26/+ and Eμ-Myc; Rosa26-CreER; p53LSL-25,26,53,54/+ mice. The lymphoma reconstitution assay was adapted from a previous study (18) and is described in the SI Materials and Methods. All work was performed in accordance with the Stanford University Administrative Panel for Laboratory Animal Care.
Supplementary Material
Acknowledgments
We thank C. Miething, P. Viatour, S. Jacobs, and J. Jones for technical assistance; D. Rowitch for the Math1-Cre mice; and S. Artandi, J. Sage, A. Brunet, and A. Sweet-Cordero for critical reading of the manuscript. This work was supported by Stanford Graduate Fellowships and National Science Foundation Graduate Research Fellowships (to T.M.J and C.A.B.), a National Institutes of Health (NIH) National Research Service Award Fellowship (National Cancer Institute 5F32CA117775-03) (to E.Y.L.), Damon Runyon Cancer Research Foundation, American Cancer Society, Leukemia and Lymphoma Society, NIH grants (to L.D.A.), and a National Institutes of Health Grant (RO1CA088060, to M.P.S.). M.P.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111245108/-/DCSupplemental.
References
- 1.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Kenzelmann Broz D, Attardi LD. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis. 2010;31:1311–1318. doi: 10.1093/carcin/bgp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson TM, Hammond EM, Giaccia A, Attardi LD. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet. 2005;37:145–152. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- 7.Brady CA, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brugarolas J, et al. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 9.Ihrie RA, Attardi LD. Perp-etrating p53-dependent apoptosis. Cell Cycle. 2004;3:267–269. [PubMed] [Google Scholar]
- 10.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 11.Collado M, et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Symonds H, et al. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- 14.Lowe SW, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich LV, Milenković L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 16.Wetmore C, Eberhart DE, Curran T. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res. 2001;61:513–516. [PubMed] [Google Scholar]
- 17.Matei V, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 21.Gaidarenko O, Xu Y. Transcription activity is required for p53-dependent tumor suppression. Oncogene. 2009;28:4397–4401. doi: 10.1038/onc.2009.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Stanchina E, et al. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 23.Villunger A, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 24.Jeffers JR, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 25.Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol. 2001;21:7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci USA. 2006;103:19842–19847. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemann MT, et al. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci USA. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.