Abstract
Chromatin folding in eukaryotes fits the genome into the limited volume of the cell nucleus. Formation of higher-order chromatin structures attenuates DNA accessibility, thus contributing to the control of essential genome functions such as transcription, DNA replication, and repair. The 30-nm fiber is thought to be the first hierarchical level of chromatin folding, but the nucleosome arrangement in the compact 30-nm fiber was previously unknown. We used cryoelectron tomography of vitreous sections to determine the structure of the compact, native 30-nm fiber of avian erythrocyte nuclei. The predominant geometry of the 30-nm fiber revealed by subtomogram averaging is a left-handed two-start helix with approximately 6.5 nucleosomes per 11 nm, in which the nucleosomes are juxtaposed face-to-face but are shifted off their superhelical axes with an axial translation of approximately 3.4 nm and an azimuthal rotation of approximately 54°. The nucleosomes produce a checkerboard pattern when observed in the direction perpendicular to the fiber axis but are not interdigitated. The nucleosome packing within the fibers shows larger center-to-center internucleosomal distances than previously anticipated, thus excluding the possibility of core-to-core interactions, explaining how transcription and regulation factors can access nucleosomes.
The basic repeating unit of chromatin is the nucleosome, the structure of which is known at atomic resolution (1). In vitro, in the presence of linker histones and a moderate concentration of monovalent cations, nucleosome arrays adopt an irregular zigzag topology (2, 3). Further increase of monovalent salt and/or the presence of divalent cations leads to more compact structures, referred to as 30-nm chromatin fibers, in which nucleosomes are assumed to be helically packed with a density in the range of 6–11 nucleosomes/turn (4, 5). The biological relevance of this compaction is supported by direct visibility of 30-nm fibers in situ by electron microscopy (EM) in some cell types of evolutionary distant species (6–8). Despite this fundamental knowledge, the resolution of the electron micrographs was not sufficient to visualize the organization of the individual nucleosomes within the compact 30-nm fiber. Thus the nucleosome arrangement within the fiber, and accordingly the interpretation of basic chromatin functions, was based on indirect methods, leading to various models, which have hitherto been disputed (9).
Two main types of model have been proposed: The first model is a one-start helix in which consecutive nucleosomes are arranged as a solenoid (4, 10), and the second model is a twisted two-start helix formed by a zigzag ribbon of nucleosomes (11). The most recent evidence for the one-start model arises from measurements of two-dimensional (2D) electron microscopic images of nucleosome arrays reconstituted in vitro, which suggest an interdigitated solenoid with the linker DNA bent in the interior of the fiber (12). The two-start model was recently supported by the crystallographic study of the tetranucleosome (13) as well as by cross-linking experiments (14).
Cryoelectron tomography has the unique ability to visualize molecular assemblies in an unaltered frozen hydrated state at close-to-native conditions. In conventional electron microscopy of plastic sections, the sample is chemically fixed, dehydrated, embedded in resin, and stained. These steps, especially the process of dehydration, results in structural rearrangement that is particularly prominent for polyelectrolites such as DNA molecules (15). Another disadvantage of conventional microscopy is that the surrounding stain is viewed and not the biological structure itself. In cryoelectron tomography, the samples are high-pressure frozen, and the preserved frozen-hydrated sample is then sectioned at cryotemperatures without dehydration. Because the samples are unstained, the intensities seen in images of the vitrified samples represent the true densities of the macromolecules. Here we use cryoelectron tomography of vitreous sections to directly visualize the nucleosome arrangement within the compacted 30-nm fibers of mature chicken erythrocyte nuclei, without the use of fixatives, dehydration, or any cross-linking agents used in conventional electron microscopy and other techniques.
Results
Cryo-EM of Chicken Erythrocyte Nuclei.
Mature avian erythrocyte nuclei are well known to contain 30-nm chromatin fibers when isolated at an ionic strength close to the physiological, for example in buffer containing 2 mM MgCl2 and 75 mM monovalent cations (16). We isolated chicken erythrocyte nuclei in such buffer and vitrified them in buffer containing 20 mM NaCl with 20% 40 kDa dextran. The osmotic effect of this particular dextran maintains fiber compaction that is equivalent to that of 2 mM MgCl2 ionic strength (17) (Fig. S1). The nuclei were intact and maintained their shape through the isolation and vitrification procedures. Chicken erythrocyte nuclei are advantageous for the data analysis because (i) they have low amounts of nonhistone chromosomal protein and thus contain no other protein complexes that might affect the image processing (18), and (ii) the interfiber distance is large enough such that the fibers can be extracted separately. Chicken erythrocyte chromatin contains the full complement of linker histone H1 (0.4 H1 per nucleosome) and H5 (0.9 per nucleosome) (19) and is representative of transcriptionally inactive chromatin (20). In this study we quantitatively analyze vitreous sections of these nuclei, which show distinct 30-nm fibers in both the 2D projections and the three-dimensional (3D) reconstructions (Fig. 1 A–F).
Fig. 1.
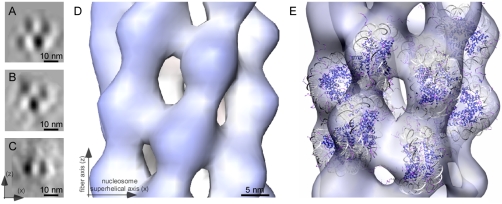
Cryo-EM images of chromatin in chicken erythrocytes. (A) A 2D overview image of an approximately 50-nm-thick cryosection of a chicken erythrocyte nucleus. Cross-sections of fibers (top views) have higher contrast compared to the longitudinally sectioned fibers and are thus much more obvious, but all possible orientations are visible. Some longitudinally sectioned fibers are highlighted by red boxes; some cross-sectioned fibers are indicated by red circles. The box length is 200 nm, and the width is 50 nm. Inset: A 1D rotationally averaged power spectrum of the nucleus shows a strong peak in the 30–40-nm range. (B) A 10-nm-thick slice through a 3D reconstruction of a chicken erythrocyte nucleus. The nucleus contains compact yet distinct and well-separated 30-nm fibers in which individual nucleosomes are visible. A longitudinal and a cross-sectioned fiber are indicated by a red box and a red circle, respectively. (C and D) Cross-sections of individual fibers; the slices are 1.2 nm thick. (E and F) Longitudinal sections of individual fibers; the slices are 1.2 nm thick. The size and shape of the high-density positions correspond well to individual nucleosomes.
Subtomogram Averaging of Native 30-nm Fibers from Chicken Erythrocytes.
Subtomogram averaging involves aligning many similar 3D images (the so-called subtomograms) to a reference template, which iteratively improves the signal-to-noise ratio of the data until convergence (21). Even though the excellent quality of the reconstruction allows us to directly visualize nucleosomes in the reconstruction (Fig. 1 C–F), an averaging procedure is performed in order to improve the signal-to-noise ratio and to average out the missing wedge. We detected the fibers in tomographic reconstructions by cross-correlation against a filtered binary cylinder, 30 nm in diameter and 60 nm in length, which was rotated and translated in space (22).
The length of the template cylinder was chosen to be 60 nm in order to avoid selecting top views as side views. Because the cylinder does not contain any structurally relevant information, any high-resolution features that arise after alignment must come from the data itself, thus completely avoiding any template bias. We thus automatically extracted approximately 1,000 subtomograms of 30-nm fibers and applied a subtomogram alignment and averaging procedure (see SI Methods for detailed protocol). This involved aligning each extracted 3D subtomogram to an initial binary cylinder, where the missing wedge was considered and compensated for in both the alignment and averaging algorithms (21), until the structure reached convergence. We initially classified the subtomograms, in order to remove false positives, as well as fibers with incorrect fiber-axis orientations (21, 23). The subtomogram average with a full length of 60 nm had poor resolution resulting from structural heterogeneity in the fibers over a long distance, but when masked over the central region of approximately 40 nm along the fiber axis, the resolution improved. This indicates that the helical order in the fiber is evidently short-range (not more than 3 nucleosome gyres). The subtomograms were realigned using this mask with an angular restriction that prevented rotating the fibers more than 30 ° from their originally estimated fiber axis.
The resulting structure in the subtomogram average is a fiber with a diameter of approximately 32 nm and a length of approximately 60 nm (Fig. S2), containing discrete elliptical densities, the size of which correspond to individual nucleosomes. The spatial positions and orientations of the nucleosomes can be clearly recognized (Fig. 2). A cross-section through the subtomogram average shows six radially organized nucleosomes spaced equidistantly on the plane (Fig. 2 E and F). There are 6.5 nucleosomes per 11 nm, resulting in a helical pitch of approximately 22.8 nm. Nucleosomes display a checkerboard pattern when the fiber is viewed from the side (Figs. 2C and 5A), indicating that nucleosomes are juxtaposed above and between those of the previous gyre, without interdigitation. A plot of the Euler angle distribution (Fig. S3) shows that there is a complete coverage of angles, resulting in an isotropic point-spread function. Because the orientations of the fibers are uniformly distributed and the missing wedge was compensated for, the subtomogram average does not have any missing information in Fourier space. The resolution of the subtomogram average assessed by the Fourier shell correlation at 0.5 cutoff is 4.3 nm (Fig. S4).
Fig. 2.
Subtomogram average of chicken erythrocyte fibers. (A) A depiction of the locations and mutual positions of the slices and isosurface visualized in B–H. (B) Computational projection image parallel to the fiber axis through the subtomogram average. Six radially oriented elliptical densities are visible that surround a central density. The size of one of the radially placed densities corresponds to the dimensions of an individual nucleosome. (C) A computational projection image calculated perpendicular to the fiber axis through the periphery of the fiber shows a checkerboard pattern of nucleosomes. (D) 0.6 nm thick longitudinal slice showing the off-superhelical-axis nucleosome juxtapositioning present in the fiber. (E and F) Two 0.6-nm-thick slices, 10 nm apart, showing radially arranged nucleosomes. (G) Isosurface of the subtomogram average shown in perspective view. Two gyres of the fiber are visualized. The fiber axis and nucleosome spacing are indicated. (H) Isosurface of the subtomogram average contoured at two threshold levels to indicate the center of the nucleosome positions at the higher threshold and the appearance of the 30-nm fiber at the lower threshold. The isosurface at the higher threshold shows distinct densities in elliptical form, depicting the center of the positions of the individual nucleosomes. The isosurface at lower threshold has been selected in order to faithfully represent a complete nucleosome and is shown as transparent, pseudocolored according to the cylindrical radius. Red indicates positions at the outer radius of the fiber, while green-blue positions are toward the center of the fiber. Positions of the nucleosomes are well defined.
Fig. 5.
Nucleosome juxtapositioning and the idealized fiber after symmetry imposition. (A and B) Two classes from the mononucleosome classification procedure depicting a checkerboard pattern, which is completely compatible with the side views of the subtomogram average when viewed in a direction perpendicular to the fiber axis. The classes have a slightly different organization of nucleosomes, but both show nucleosomes that are juxtaposed face-to-face but shifted off their superhelical axes. The superhelical axis is parallel to the x axis. (C) A representative class from the mononucleosome classification procedure depicting a face-to-face on-axis juxtaposition of nucleosomes, which is different from that seen in the subtomogram average, which can be attributed to local distortions of the helical structure. (D) Isosurface of the idealized fiber after symmetrization based on the determined helical parameters. (E) Individual nucleosomes are fitted within the idealized fiber (visualized in the transparent white-blue isosurface). The fiber axis and nucleosome superhelical axis are indicated by arrows.
Determination of the Symmetry Parameters.
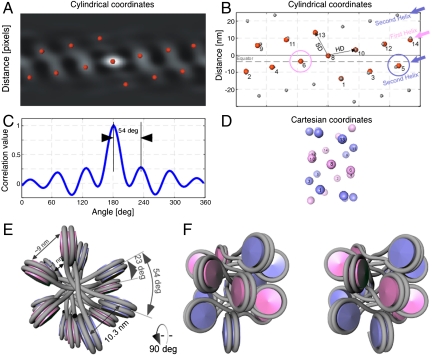
We did not impose symmetry during the iterative alignment procedure, thus presumption of helical symmetry was completely avoided. When the autocorrelation function of the subtomogram average is plotted in cylindrical coordinates, it shows a remarkable periodicity at a radius of 13 nm (Fig. 3A). In cylindrical coordinates, the autocorrelation peaks may be considered as the lattice points of a helical net, which may be indexed by direction vectors (Fig. 3B). The direction vector to the highest correlation peak (HD) and the vector of second highest correlation peak (SD) are shown in Fig. 3B. HD shows the direction of the basic helix (indicated by the pink arrow), from which the nucleosome repeat distance can be calculated. SD indicates the spacing between helix gyres (the second gyre is indicated by the blue arrow) (Fig. 3B). Remarkably, the autocorrelation function shows distinct lattice points up to the third order, capturing all nucleosomes within two gyres showing that the subtomogram average has approximately 6.5-fold symmetry (Fig. 3C). Both the first and the second helix intersect a horizontal line (the equator), evidencing that a two-start helix is the smallest possible startedness.
Fig. 3.
The fiber shows a two-start organization. (A) The autocorrelation function of the subtomogram average in cylindrical coordinates at a radius of 13 nm. The red spheres indicating the positions of the local maxima show a remarkable periodicity and are used to define the lattice points. (B) An idealized lattice, shown in gray spheres, was calculated based on the positions of the local maxima. In the idealized lattice the first helix is indicated by a pink arrow, and the second helix is indicated by a blue arrow. The positions of the nucleosomes visible in the real data are indexed by small numbers. Both helices are shown to intersect the equator at the positions indicated by the pink and the blue circle respectively. This shows that the fiber has a clear two-start geometry. (C) A plot through the autocorrelation function at the position of the basic helix shows that the subtomogram average has approximately 6.5-fold symmetry with a rotation per subunit of approximately54°. (D) Cartesian space depiction of the idealized lattice in B, facilitating indexing of the nucleosomes on the lattice. The numbering of the nucleosomes is identical to B. The two-start helix is visible in Cartesian coordinates. (E) Top view of the positions of the nucleosomes placed at the positions of the correlation peaks shown in B, with the linker DNA modeled in gray, which is assumed to cross the fiber-axis. The azimuthal rotation and center-to-center nucleosome distance are indicated. (F) Side view of the depiction in E (related by a 90° rotation) shown in crossed-eyes stereo view.
By deducing the symmetry parameters, we thus determined that the subtomogram average is a left-handed helix with an axial translation (Δz) per subunit of 3.4 nm (± 0.48 nm) and an azimuthal rotation (Δϕ) per subunit of -54° (± 0.4°), which relates each nucleosome to its neighbor with a center-to-center distance within a gyre of 10.3 nm (± 0.12 nm) (Fig. 3 and Movie S1). This results in approximately 6.7 nucleosomes per turn (Fig. 3 E and F) or approximately 6.5 nucleosomes per 11 nm. This number indicates that the fibers are fully compact, because a compaction rate of 6–7 nucleosomes per 11 nm turn corresponds to the saturation point of compaction in isolated chicken erythrocyte chromatin fibers, according to measurements provided by previous studies (5, 24). We have verified the reproducibility of the final subtomogram average using three individual datasets from different tomograms with independent initialization parameters and the results were very similar. Among the processed datasets, the resulting subtomograms showed an approximately 6.5-fold structure with Δz per subunit of approximately 3.6 nm (± 0.6 nm) and an azimuthal rotation (Δϕ) per subunit of approximately -54° (± 3°) (tilt-series alignment shown in Table S1).
Classification: Variation and Flexibility within the Fibers.
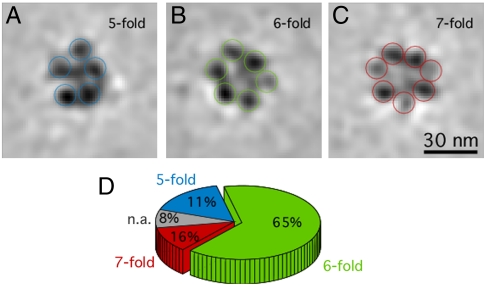
Our average describes the most statistically significant symmetry that is characteristic of the dataset, but after classification of the subtomograms (21), we see that there is some level of variation and flexibility within the fibers. To identify the structural variability we classified the final set of aligned particles using principal components analysis (PCA) using masks of 30 nm in length. This classification procedure showed that variance existed within the symmetries of the extracted fibers. Because we ignored variations smaller than our resolution limit, we could ascertain that we identified true structural variation because the densities we classified were in the size order of a nucleosome. Most of the class averages (65%) display 6 nucleosomes per cross-section, but others show 5 (11%) or 7 (16%), demonstrating the level of variability that may be attributed to slight distortions or bending within the individual fibers (Fig. 4). However, we never obtain classes with more than 7 nucleosomes per turn—which contradicts the extremely high packing ratio of 11 nucleosomes per 11 nm obtained by measurements of in vitro reconstituted arrays (12). It is evident that in nature a structural variability may exist within the range of a given set of parameters. This may of course be a different case for reconstituted fibers, where structural homogeneity is aimed for even if this may not be the true reflection of the structure in nature.
Fig. 4.
Structural variability within the dataset. A characteristic subset of the resulting subtomogram class averages from the PCA classification is shown; the slices are 1.2 nm thick. Most of the class averages display 6 nucleosomes per cross-section (B), but others show 5 (A) or 7 nucleosomes per cross-section (C), which demonstrates the level of variability with our data. The nucleosomes have varying densities because they are computationally sectioned at different heights. (D) Pie chart depicting the percentages of particles contributing to each class.
Characteristics of the Native 30-nm Fiber.
The autocorrelation function in cylindrical coordinates unambiguously shows that the fiber has a two-start geometry formed by two opposing helices, which is in agreement with the two-start model (Fig. 3 B and D). The symmetry cannot be accounted for by a one-start helix, because the rise per nucleosome precludes the possibility of completing the number of nucleosomes per 11 nm within the determined pitch. A far smaller axial translation (Δz) per subunit would be required (< 1.7 nm) in order for this requirement to be met. By fitting nucleosomes into the density using an algorithm to take into account the symmetry parameters, it is possible to determine that the two-start geometry of the fiber is formed by two opposing helices where the second helix starts at 203° with respect to the first.
Due to the resolution of the subtomogram average, we are unable to unambiguously resolve DNA connectivity between the nucleosomes. However, we consistently see densities crossing the fiber axis in the subtomogram average and even in individual subtomograms at favorable projection angles (Fig. 1 C and D). Computational projections down the fiber axis reveal a density in the fiber cavity, which cannot be accounted for by H1/H5 or histone tail domains due to their small size and location (Fig. 2B) (SI Methods). Because there have been no previous reports of any such density being visualized in real data, we would suggest that this provides additional evidence that DNA indeed crosses the fiber.
Next, we docked nucleosomes into individually segmented nucleosome densities of the subtomogram average by means of cross-correlation-based techniques (22). Although the accuracy is restricted by the resolution, the docking is unambiguous (Fig. S5). Nucleosome fitting within the density of a single nucleosome shows that the angle between the nucleosome superhelical axis and the fiber axis is approximately 90°, with a variance of approximately 20° (Movie S2). This angle agrees with previous values determined from measurements of the photochemical dichroism for chicken erythrocyte chromatin (10) and has a reasonable agreement with the angle between the superhelical axes of the nucleosome stacks in the idealized fiber model generated from the tetranucleosome X-ray structure, where the angle was increased to 70° in order to relieve steric interference between tetranucleosomes (13). Nucleosomes in our fiber are mostly arranged face-to-face but shifted off their superhelical axes. We assume that the nucleosomes are numbered according to a linear array dependent on the DNA connectivity, where nucleosome n2 is attached to n1 and n3. In the fiber, nucleosomes are arranged such that nucleosome n8 is asymmetrically positioned above nucleosomes n1 and n6, without interdigitation. While nucleosomes n6 and n8 are on the same gyre, nucleosome n1 resides on the subsequent gyre (Fig. 3 D and F and Movie S2).
Differential Face-to-Face Juxtapositioning.
In order to visualize local areas of order that might not have been captured by the cylindrical subtomogram averaging process, we used an additional method to determine the local arrangement of nucleosomes surrounding another nucleosome in question. We thus automatically extracted subtomograms around dense positions with the approximate volume of a single nucleosome and aligned them against a mononucleosome template (25) (SI Methods). The majority of resulting classes (Fig. 5 A–C) show an ordered nucleosome organization, arranged face-to-face, and exhibit a checkerboard pattern that is completely compatible with the side views of the subtomogram average when viewed in a direction perpendicular to the fiber axis. The idealized symmetrized model of our fiber calculated from the helical parameters derived from the subtomogram average distinctly shows this geometry (Fig. 5 D and E; details of the symmetrization procedure are provided in SI Text). Within the classes, the nucleosomes have the propensity to orient face-to-face shifted off their superhelical axes (Fig. 5 A and B). However, in some cases, classes also reveal nucleosomes juxtaposed with their superhelical axes aligned, which is different from that seen in the subtomogram average (Fig. 5C) and can be attributed to local distortions of the helical structure. We thus find that the nucleosomes can align face-to-face with a relative shift in the juxtaposition of approximately 4 nm. These variations in the face-to-face juxtapositioning of nucleosomes may reflect structural variability at the local scale resulting from bending, twisting, or intercalation of the fibers.
In most crystal structures, the center-to-center distance between two similarly oriented nucleosomes is approximately 5.7 nm, which leaves a small distance of approximately 4 Å between the octomer faces (1). Nucleosomes in our fiber, however, are related with a center-to-center distance within a gyre of approximately 10.3 nm (Fig. 3E and Fig. S6). This value varies with the distance from the fiber axis because adjacent nucleosomes are at an angle to each other (Movie S3). Thus, due to the geometry of our fiber, the nucleosomal DNA at the center of the fiber is almost in contact (approximately 2 nm distance), but the nucleosomal DNA on the periphery of the fiber is highly accessible, leaving an internucleosomal gap of approximately 10 nm (Fig. S6).
Discussion
Using cryoelectron tomography, we show that the most predominant form of chromatin in chicken erythrocyte nuclei is a 30-nm fiber arranged in a two-start helix with approximately 6.7 nucleosomes per turn. This geometry is similar to the structure of the tetranucleosome (13) and the two-start cross-linker model derived from evidence based on chromatin studies using reconstituted chromatin fibers (14). In our data, instances of electron densities crossing the fiber axis and the presence of a central density in the fiber cavity provide direct support for the cross-linker organization. Thus, a transition from the irregular zigzag ribbon observed at low salt to the compact 30-nm fiber can occur without major topological changes, thereby preserving the two-start geometry. However, the important difference between these compaction levels is that while the irregular zigzag fiber shows few or no face-to-face nucleosome interactions (2), the face-to-face stacking motif represents an essential feature of the 30-nm fiber observed directly in nuclei.
The higher-order structural arrangement of nucleosomes is also revealed by subtomogram classification, giving rise to a checkerboard pattern that reflects the side views of the 30-nm fiber (Fig. 5 A–C). The face-to-face juxtaposition of nucleosomes may occur with superhelical axes aligned or with a shift in the position of the superhelical axes with respect to one another. The helical rise per nucleosome in our subtomogram average is similar to the nucleosome rise in the idealized crossed-linker model (13). However, the larger azimuthal rotation in our fiber results in a more homogeneous nucleosome distribution compared to the idealized cross-linker model (13) where stacked nucleosomes form two dense gyres that are widely separated. In this aspect, our model is closer to the solenoid model (12), where the gyres are more evenly spaced—however, without interdigitation of the nucleosomes. We suggest that the uniform, nearly hexagonal, packing of nucleosomes (Fig. 3B) is a more energetically favorable solution to minimize repulsion from noncompensated negative charges of nucleosomal DNA.
Face-to-face nucleosome binding interactions have previously been demonstrated in various crystal structures (1, 13, 25). However, we find that nucleosomes in situ interact at a center-to-center distance of approximately 10.3 nm, leaving a large distance of at least 3 nm between the two octomer cores (Fig. 3E and Fig. S6), which is much larger than that reported in crystal structures. This binding distance excludes core-to-core interactions (13, 26) but allows interactions mediated by histone N-terminal tails in a tail-to-core interaction. The large internucleosomal spacing suggests high accessibility of the octamer face and tails to functional interactions in compact chromatin in situ, which is consistent with the high dynamic permeability observed by light microscopy methods (27). The rather open structure suggests that mechanisms other than the mechanical barrier of DNA accessibility posed by tight nucleosome packing are involved in transcriptional silencing.
In contrast to artificial arrays with a defined nucleosome repeat length and uniform histone composition, native chromatin has inherent local variations in nucleosome repeat length and modifications and variants of the core and linker histones. We demonstrate that despite this variability, native chromatin shows remarkable features of structural regularity within compact 30-nm fibers. Our study provides a 3D view for the native chromatin fiber organization in the compact state. It also provides structural interpretation for previous studies of gene activity control using the chicken erythroid differentiation system (28). The type of histone modifications as well as the absence of HP1 in transcriptionally silent chicken erythrocyte chromatin indicate that it differs from constitutive heterochromatin (29), and we therefore speculate that its 30-nm fiber organization may be more representative of facultative heterochromatin. This hypothesis is in agreement with recent studies by Kizilyaprak et al. (6) on the mouse rod photoreceptor cell, where the heterochromatin areas enriched with facultative heterochromatin show a 30-nm fiber organization; whereas the clustered constitutive chromatin appears without order, which is compatible with concept of a chromatin melt (17). Other evidence is provided by studies of polytene chromosomes in Cheronomus tentans, where termination of transcription resulted in expanded “puffed” chromatin fibers rearranging into 30-nm fibers (7).
Extrapolation of the features of the chicken erythrocyte 30-nm fiber to facultative heterochromatin in situ provides structural clues for the reactivation of repressed genes. The internucleosomal gaps, as seen in our 30-nm fiber, may provide an initial “pore” for pioneer transcription factors, like FoxA, which interacts with H3 and H4 histones (30) to access their binding sites in compact chromatin. Indeed, completely inactive chicken erythrocyte genomes can also be reactivated once placed in an active cytoplasmic environment such as heterokaryons (31). Because the internucleosomal gap at the periphery of our fiber is approximately 10 nm, large molecules such as chromatin factor RCC1 can bind directly to the octamer face (32) without the necessity of fiber decompaction. Further cryoelectron tomographic analysis applied to these and other systems has the potential to reveal essential information of chromatin fiber arrangement in different functional states.
Methods
Chicken erythrocyte nuclei were isolated from a chicken erythrocyte suspension (Charles River Laboratories) essentially as previously described (8, 16). The nuclei were high-pressure frozen and then cryosectioning was performed using a 35° diamond knife, with the nominal cutting feed set to 80–100 nm. Tilt-series were collected on a Tecnai G2 Polara microscope (FEI), recorded on a 2k × 2k pixel CCD camera at a defocus level between -8 μm and -6.5 μm, with a pixel size at the specimen level of 0.6 nm. The total electron dose was approximately 90 e-/Å2 per tilt-series. Tilt-series were aligned using the fiducialless alignment software Alignator (33). Subtomograms of fibers were extracted, iteratively aligned, and averaged using algorithms to take the missing wedge into account, until the final structure reached convergence (Fig. S7). All processing was performed using subtomograms with a voxel size of 1.2 nm, and carried out with custom scripts written in MATLAB (The Mathworks), which are available at http://www.biophys.uni-frankfurt.de/frangakis. For volume visualization and isosurface rendering the Amira software package (Mercury Computer Systems, http://www.amiravis.com) (34) and the Chimera package were used (35). Details of protocols, sample preparation, electron microscopy techniques, and image-processing methods are available in SI Text.
Supplementary Material
Acknowledgments.
We would like to thank Helen Saibil, Christoph Mueller, and Trevor Sewell for carefully reading the manuscript; the Frankfurter Cluster for macromolecular complexes; and the European Research Council for the starting grant to A.S.F.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron density map of the 30-nm fiber has been deposited in the EBI Macromolecular Structure Database with accession nos. EMD-1781 and EMD-1782.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108268108/-/DCSupplemental.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol. 1994;125:1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bednar J, Horowitz RA, Dubochet J, Woodcock CL. Chromatin conformation and salt-induced compaction: Three-dimensional structural information from cryoelectron microscopy. J Cell Biol. 1995;131:1365–1376. doi: 10.1083/jcb.131.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerchman SE, Ramakrishnan V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc Natl Acad Sci USA. 1987;84:7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kizilyaprak C, Spehner D, Devys D, Schultz P. In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS One. 2010;5:e11039. doi: 10.1371/journal.pone.0011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson K, Bjoerkroth B, Daneholt B. Packing of a specific gene into higher order structures following repression of RNA synthesis. J Cell Biol. 1984;98:1296–1303. doi: 10.1083/jcb.98.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodcock CL. Chromatin fibers observed in-situ in frozen-hydrated sections—native fiber diameter is not correlated with nucleosome repeat length. J Cell Biol. 1994;125:11–19. doi: 10.1083/jcb.125.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremethick DJ. Higher-order structures of chromatin: The elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 10.McGhee JD, Nickol JM, Felsenfeld G, Rau DC. Higher order structure of chromatin: Orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983;33:831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- 11.Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: Evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: Evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 14.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 15.Kellenberger E. The potential of cryofixation and freeze substitution: Observations and theoretical considerations. J Microsc. 1991;161:183–203. doi: 10.1111/j.1365-2818.1991.tb03083.x. [DOI] [PubMed] [Google Scholar]
- 16.Langmore JP, Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 17.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dingman CW, Sporn MB. Studies on chromatin. I. Isolation and characterization of nuclear complexes of deoxyribonucleic acid, ribonucleic acid, and protein from embryonic and adult tissues of the chicken. J Biol Chem. 1964;239:3483–3492. [PubMed] [Google Scholar]
- 19.Bates DL, Thomas JO. Histones H1 and H5: One or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron IL, Prescott DM. RNA and protein metabolism in the maturation of the nucleated chicken erythrocyte. Exp Cell Res. 1963;30:609–612. doi: 10.1016/0014-4827(63)90344-6. [DOI] [PubMed] [Google Scholar]
- 21.Forster F, Pruggnaller S, Seybert A, Frangakis AS. Classification of cryo-electron sub-tomograms using constrained correlation. J Struct Biol. 2008;161:276–286. doi: 10.1016/j.jsb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Frangakis AS, et al. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc Natl Acad Sci USA. 2002;99:14153–14158. doi: 10.1073/pnas.172520299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z, Frangakis AS. Classification of electron sub-tomograms with neural networks and its application to template-matching. J Struct Biol. 2011;174:494–504. doi: 10.1016/j.jsb.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Ghirlando R, Felsenfeld G. Hydrodynamic studies on defined heterochromatin fragments support a 30-nm fiber having six nucleosomes per turn. J Mol Biol. 2008;376:1417–1425. doi: 10.1016/j.jmb.2007.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frouws TD, Patterton HG, Sewell BT. Histone octamer helical tubes suggest that an internucleosomal four-helix bundle stabilizes the chromatin fiber. Biophys J. 2009;96:3363–3371. doi: 10.1016/j.bpj.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verschure PJ, et al. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 2003;4:861–866. doi: 10.1038/sj.embor.embor922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood WI, Felsenfeld G. Chromatin structure of the chicken beta-globin gene region. Sensitivity to DNase I micrococcal nuclease, and DNase II. J Biol Chem. 1982;257:7730–7736. [PubMed] [Google Scholar]
- 29.Gilbert N, et al. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 2003;22:5540–5550. doi: 10.1093/emboj/cdg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 31.Linder S, Zuckerman SH, Ringertz NR. Pattern of chick gene activation in chick erythrocyte heterokaryons. J Cell Biol. 1982;95:885–892. doi: 10.1083/jcb.95.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castano-Diez D, Scheffer M, Al-Amoudi A, Frangakis AS. Alignator: a GPU powered software package for robust fiducial-less alignment of cryo tilt-series. J Struct Biol. 2010;170:117–126. doi: 10.1016/j.jsb.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Pruggnaller S, Mayr M, Frangakis AS. A visualization and segmentation toolbox for electron microscopy. J Struct Biol. 2008;164:161–165. doi: 10.1016/j.jsb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.