Abstract
CD1d-restricted invariant NKT (iNKT) cells are a unique lineage of T lymphocytes that regulate both innate and adaptive immunity. The Mediator complex forms the bridge between transcriptional activators and the general transcription machinery. Med1/TRAP220 (also called DRIP205) is a key component of Mediator that interacts with ligand-bound hormone receptors, such as the vitamin D receptor. Here, we show that T-cell–specific Med1 deficiency results in a specific block in iNKT cell development but the development of conventional αβ T cells remains grossly normal. The defect is cell-intrinsic and depends neither on apoptosis, cell-cycle control, nor on CD1d expression of CD4+CD8+ double-positive thymocytes. Surprisingly, ectopic expression of a Vα14-Jα18 T-cell receptor transgene completely rescues the defect caused by Med1 deficiency. At the molecular level, thymic iNKT cells in Med1−/− animals display reduced levels of IL-2Rβ and T-bet expression and could not complete terminal maturation. Thus, Med1 is essential for a complete intrathymic development of iNKT cells.
Invariant NKT (iNKT) cells form a distinct lymphocyte lineage that regulates a broad range of immune responses. iNKT cells express a semi-invariant T-cell receptor (TCR), comprised of an invariant α-chain (Vα14-Jα18 in mice) and a restricted β-chain repertoire (Vβ8.2, Vβ2, or Vβ7 in mice), and recognize glycolipids presented by the nonclassical MHC molecule CD1d (1–4). Despite the limited antigen specificity resulting from their invariant TCR, iNKT cells can rapidly secrete large amounts of cytokines, such as IL-4 and IFN-γ, upon activation, thus contributing to the efficient licensing of other immune cell types to respond. Therefore, iNKT cells are key players of the immune response as they are considered to bridge innate and adaptive immunity (1, 5).
iNKT cells arise from CD4+CD8+ double-positive (DP) thymocytes and diverge from mainstream T-cell development upon expression of the invariant TCRα chain followed by the selection of CD1d-expressing DP thymocytes (6, 7). iNKT cell developmental stages could be identified using αGalCer-CD1d tetramers. The earliest detectable CD1d-tetramer+ subset has a CD24hi phenotype. Subsequently, the cells down-regulate CD24 expression and undergo a massive expansion to acquire a CD44hi effector/memory phenotype followed by terminal maturation, which is accompanied by the expression of NK cell-surface markers, such as NK1.1 (1–3).
Many transcription factors, signaling molecules, and cytokines have been shown to be crucial at multiple stages of iNKT cell development (3). The transcription factor RORγt is required for the productive rearrangement of the TCRα gene segment Vα14 to the distal Jα18, as it drives the expression of anti-apopototic factor Bcl-xL, allowing DP thymocytes to survive long enough for distal TCRα rearrangements to occur (6, 8). In contrast, transcription factors T-bet, vitamin D receptor (VDR) as well as IL-15 signaling are not implicated in this rearrangement process but control iNKT cell terminal maturation (9–11). Several other factors, such as GATA-3, Id2, and cytokine IL-7, are essential for the peripheral maintenance of iNKT cells (12–14).
The Mediator complex forms the bridge between transcriptional activators and the RNA polymerase II. Med1/TRAP220 (also termed DRIP205) is a key component of Mediator originally found to associate with nuclear hormone receptors, such as thyroid hormone-, estrogen-, and vitamin D-receptor (reviewed in ref. 15). It was shown that Med1 exists predominantly in a Mediator subpopulation enriched in RNA polymerase II (16). Previously, it has been suggested that Med1 not only interacts with hormone receptors but also with GATA factors (17). Consistent with an essential role in mouse development, ablation of Med1 leads to lethality at midgestation (11.5 d post coitum). Although Med1 is not required for cell division, it is critical for placental and cardiac development during embryogenesis (17–20). Using a conditional null mutation of Med1, we recently demonstrated a severe defect in erythropoiesis although the development of myeloid and conventional lymphoid lineages remain largely unaffected (21).
Here, we demonstrate that T-cell–specific ablation of Med1 specifically impairs the generation of iNKT cells. The block in iNKT cell development is cell-intrinsic and is not due to the influence of Med1 deficiency on DP thymocytes. Ectopic expression of a Vα14-Jα18 TCR transgene largely rescues the iNKT cell defect. At the molecular level, Med1-deficient thymic immature iNKT cells lack IL-2Rβ and T-bet expression, suggesting that Med1 plays an essential role for the activation of the genetic program involved in the terminal maturation stage of iNKT cell development.
Results
Generation of iNKT Cells Is Significantly Impaired in the Absence of Med1.
To investigate the role of Med1 in the development of T-cell lineages, we analyzed T-cell–specific Med1 conditional knockout mice mediated by the expression of Cre recombinase driven by the promoter of the mouse CD4 gene, hereafter referred to as Med1−/− mice. As Med1+/− mice were phenotypically indistinguishable to wild-type mice, they were used as littermate controls in all experiments. Quantitative RT-PCR and Western blotting analysis demonstrated that Med1 was efficiently deleted at the genomic DNA (gDNA) (Fig. S1A, Left), mRNA (Fig. S1A, Right), and protein (Fig. S1B) levels. As expected from our previous report (21), total thymocyte and splenocyte cellularity was normal (Fig. S2 A and B). We did not observe any gross alterations of the main thymic T-cell subsets, determined by a CD4/CD8α cell-surface staining (Fig. S2 C and D, Upper). However, in the periphery we detected a significant increase both in frequency and absolute cell number of CD4+ T cells concomitant with a decrease of CD8+ T cells (Fig. S2 C and D, Lower). Thus, conventional αβ T cells can be generated in the absence of Med1. The reason for the discrepancy observed in splenic CD4+/CD8+ T-cell ratio will be addressed in a further study.
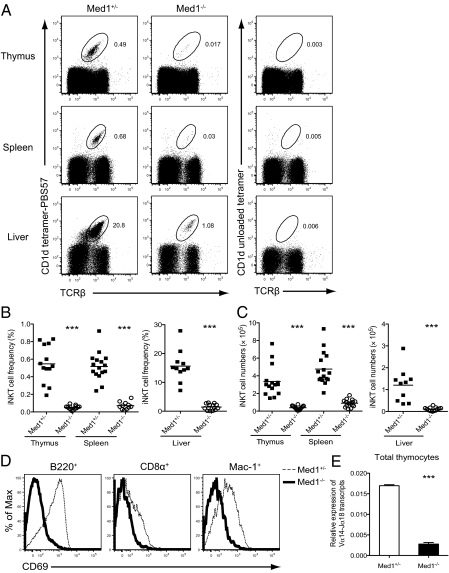
In contrast, the generation of iNKT cells, as stained by TCRβ antibody and CD1d-tetramer loaded with the α-GalCer analog PBS57 (22), was strongly affected in the thymus of Med1−/− mice [Fig. 1 A (Top), B, and C]. This block could not be compensated in the periphery, as seen by a profound decrease in iNKT cell frequencies and numbers in the spleen and liver [Fig. 1 A (Middle and Bottom), B, and C]. Alternative staining with antibodies against TCRβ and NK1.1 showed a similar pattern of reduction (Fig. S3).
Fig. 1.
Impairment of iNKT cell development in the absence of Med1. (A) Representative dot plots with the staining of anti-TCRβ antibody and PBS57-loaded CD1d tetramer (Left) or unloaded CD1d tetramer (Right) in the thymus, spleen, and liver from Med1+/− or Med1−/− mice. The percentages of CD1d-tetramer+ iNKT cells are indicated in each plot. (B and C) Statistical analysis of the percentage and absolute numbers of iNKT cells in the thymus, spleen, and liver mononuclear cells from Med1+/− or Med1−/− mice. Numbers represent the mean of at least 11 mice per group. (D) Activation of B cells (B220+), CD8α+ T cells, and macrophages (Mac-1+) in the spleens from Med1+/− or Med1−/− mice immunized intravenously with 2 μg α-GalCer. Up-regulation of CD69 was assessed 5 h after immunization. (E) Real-time PCR quantification of Vα14-Jα18 transcripts in total thymocytes. Level of Vα14-Jα18 transcripts was normalized to the quantity of Cα transcripts. Results are shown as mean ± SEM from two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired two-tailed t test).
A unique feature of iNKT cells is the rapid production of effector cytokines, including IFN-γ and IL-4, upon stimulation, resulting in the licensing and activation of other immune cells (23). In line with the massive reduction of iNKT cell numbers in Med1−/− mice, we could not detect any IFN-γ and IL-4 production in the serum of Med1−/− mice upon intravenous injection of α-GalCer (Fig. S4). Congruent with the defective cytokine production by Med1−/− iNKT cells, there was no measurable activation of B cells, T cells, and macrophages upon challenge with α-GalCer compared with Med1+/− cells, as assessed by the up-regulation of the early activation marker CD69 (Fig. 1D).
Using quantitative RT-PCR with primers specific for Vα14-Jα18 TCR gene rearrangement, we found a significant reduction in this transcript in total thymocytes from Med1−/− mice once normalized to Cα transcripts (Fig. 1E), consistent with the striking decrease of iNKT cells in the thymus.
Med1 Deficiency Has no Impact on iNKT Cell Precursor CD4+CD8+ Thymocytes.
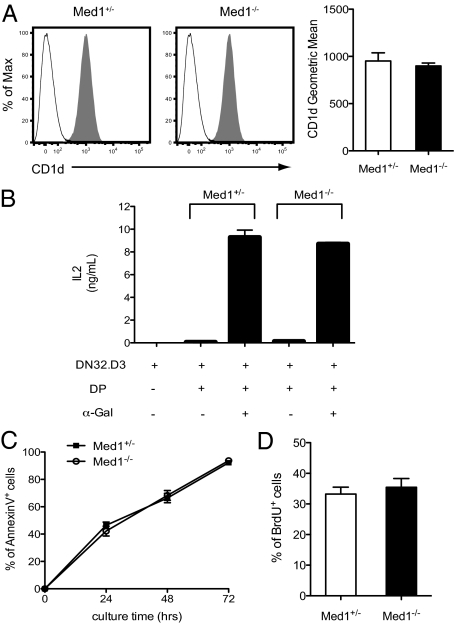
iNKT cells originate from CD4+CD8+ (DP) thymocytes (6, 7, 24) and are positively selected by CD1d-expressing cortical thymocytes (25). As CD1d is important for both positive selection and peripheral maturation of iNKT cells (26, 27), and mice lacking CD1d have severe defects in iNKT cell development (28, 29), we investigated whether the observed abrogation of iNKT cell development in Med1−/− mice was because of an impaired expression of CD1d. As shown in Fig. 2A, the expression level of CD1d was equivalent on Med1+/− and Med1−/− DP thymocytes. However, in addition to CD1d expression, efficient loading of glycolipid antigens through the endosomal-lysosomal pathway is also required for iNKT cell positive selection (1, 30). Using an NKT cell hybridoma DN32.D3, the ability of Med1−/− thymocytes to present lipid antigens was assayed. Thymocytes from Med1+/− and Med1−/− mice were incubated with DN32.D3 in the presence or absence of exogenous glycolipid α-GalCer, and NKT hybridoma activation was evaluated by IL-2 production. As shown in Fig. 2B, Med1−/− thymocytes were as efficient as Med1+/− thymocytes to induce IL-2 production by DN32.D3 cells in response to endogenous or exogenous lipid antigens.
Fig. 2.
Med1 deficiency has no influence on iNKT cell precursor CD4+CD8+ (DP) thymocytes. (A) Expression of CD1d on DP thymocytes from Med1+/− or Med1−/− mice. After gating on DP thymocytes, histograms correspond to CD1d staining (gray) and isotype control (white). Data are representative of three independent experiments. The geometric mean fluorescent intensity is shown on the right (mean ± SEM). (B) CD1d-dependent antigen presentation by thymocytes from Med1+/− and Med1−/− mice. The NKT cell hybridoma DN32.D3 was stimulated with total thymocytes from Med1+/− and Med1−/− mice with or without exogenous α-GalCer, as indicated. IL-2 production was measured by ELISA. Results are shown as mean ± SEM from two independent experiments. (C) Med1+/− or Med1−/− thymocytes were cultured in vitro for 24, 48, and 72 h and the percentage of Annexin V+ cells is shown after gating on the DP thymocyte population. (D) BrdU incorporation of Med1+/− or Med1−/− DP thymocytes after a 24-h in vivo pulse of BrdU. Results are shown as mean ± SEM from three independent experiments.
We subsequently assessed the survival and proliferation of DP thymocytes. As shown in Fig. 2C, thymocytes isolated from Med1+/− or Med1−/− mice displayed similar percentage of AnnexinV+ cells when cultured in vitro for 24, 48, and 72 h. In addition, the percentage of BrdU-labeled DP thymocytes in Med1−/− mice was comparable to that of the control mice (Fig. 2D). Hence, survival and proliferation of DP thymocytes are not affected by Med1 deficiency.
Furthermore, a microarray analysis on sorted Med1+/− and Med1−/− DP thymocytes revealed similar expression of genes related to iNKT cell development, which was further validated by quantitative RT-PCR (Fig. S5).
Defects in iNKT Cell Development Observed in Med1−/− Mice Is Cell-Intrinsic.
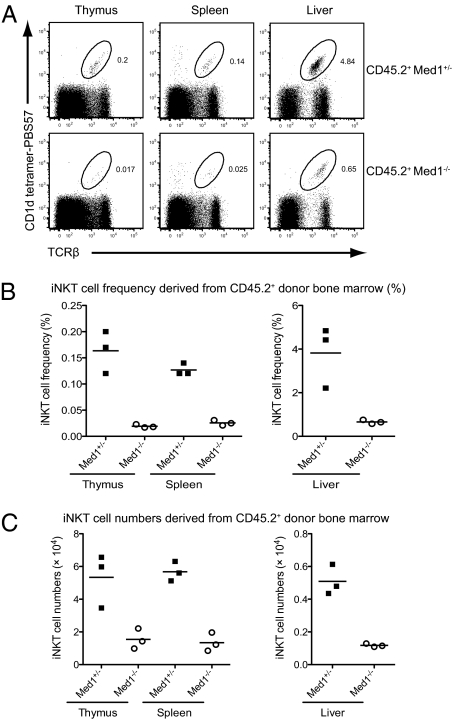
The findings above are suggestive of a cell-intrinsic defect of iNKT cell development in Med1−/− mice. Because DP thymocytes serve as both precursor and selecting cells, it remains possible that the defects of the signaling pathways elicited by homotypic interactions might account for the reduction of iNKT cells in Med1−/− mice. To address this issue, we generated bone-marrow chimeras by reconstituting lethally irradiated CD45.1+ congenic wild-type recipients with a 1:1 mixture of CD45.1+ congenic wild-type and either CD45.2+ Med1+/− or CD45.2+ Med1−/− bone-marrow cells. The mix of wild-type and Med1−/− donor cells ensured that wild-type cells would be present during the development of Med1−/− iNKT cells. Analysis of these chimeras revealed that Med1−/− donor bone-marrow cells reconstituted the iNKT cell compartment poorly (Fig. 3A) both in frequency and absolute numbers (Fig. 3 B and C), even in the presence of wild-type cells, which confirmed the cell-intrinsic defects of iNKT cell development in the absence of Med1.
Fig. 3.
Cell-intrinsic requirement for Med1 in iNKT cell development. Flow cytometric analysis of iNKT cells in irradiated CD45.1+ congenic recipient mice reconstituted with 1:1 mixture of CD45.1+ wild-type and CD45.2+Med1+/− or CD45.2+ Med1−/− bone-marrow cells. The mice were analyzed 10 to 12 wk after bone marrow reconstitution. (A) Representative flow cytometric plots of iNKT cells derived from CD45.2+Med1+/− (Upper) or CD45.2+Med1−/− (Lower) donors in the bone-marrow chimeras. iNKT cells in the thymus, spleen, and liver were identified by CD1d-tetramers. (B and C) The frequency and absolute numbers of iNKT cells derived from CD45.2+Med1+/− or CD45.2+Med1−/− donor cells in the bone-marrow chimeras. Three mice from each group were analyzed.
iNKT Cell Development Is Most Severely Affected at the Maturation Stage.
Immature iNKT cells undergo several well-defined developmental stages in the thymus. As depicted in Fig. S6A, the earliest detectable CD1d-tetramer+ cells are CD24hiCD44−NK1.1− (stage 0). Subsequently, the cells progress through three more developmental stages: CD24loCD44loNK1.1− (stage 1), CD24loCD44hiNK1.1− (stage 2), and CD24loCD44hiNK1.1+ (stage 3) (2, 3, 24, 31). The presence of a few residual iNKT cells in Med1−/− mice has allowed us to further assess the developmental status of Med1-deficient iNKT cells. First, to exclude that the residual iNKT cells from Med1−/− mice were because of the incomplete gene deletion mediated by Cre recombinase, thymic and splenic CD1d-tetramer+ cells were sorted from Med1+/− and Med1−/− mice and analyzed for Med1 deletion efficiency. Quantitative RT-PCR results at both the gDNA (Fig. S6B) and mRNA (Fig. S6C) levels demonstrated that Med1 deletion was efficient and complete. Then, thymocytes from 3- to 4-wk-old Med1+/− and Med1−/− mice were analyzed on the basis of stage-specific cell-surface marker expression. Analysis of CD1d-tetramer+ thymocytes showed that Med1−/− iNKT cells mostly up-regulate CD44 but failed to express NK1.1 to complete terminal maturation (Fig. 4A). Enumeration of iNKT cell stage intermediates revealed a twofold reduction at stage 0 and a three- to fourfold decrease at stages 1 and 2. The most severely affected stage was the transition from stage 2 to stage 3 (∼50-fold reduction at stage 3) (Fig. 4B), highlighting the profound block in iNKT cell terminal maturation upon Med1 ablation. These results indicate that although 50% of iNKT cells can overcome Med1 deletion at an early stage, they fail to go through the maturation stage because of the necessity for Med1.
Fig. 4.
Characterization of the developmental intermediates of iNKT cells in the absence of Med1. (A) The developmental stages of thymic iNKT cells were analyzed based on the expression of CD24, CD44, and NK1.1. The upper plots are gated on CD1d-tetramer+ cells and the lower plots are gated on CD24loCD1d-tetramer+ cells. (B) Absolute numbers of iNKT cells at different developmental stages were calculated. The fold-reduction at each stage is indicated. Results are shown as mean ± SEM; 11 mice of each genotype were analyzed.
In mice, iNKT cells can be further subdivided into CD4+ and double-negative (DN) subsets (3, 30). Thymic immature NK1.1−CD4+ iNKT cells could give rise to NK1.1+ NKT cells, including both CD4+ and DN cells, after intrathymic injection into Jα18−/− recipient thymi, suggesting a precursor/product relationship between CD4+ and DN iNKT subsets (24). Evaluation of the subset composition in Med1−/− animals showed that the CD4+ population was significantly reduced in the thymus and spleen (Fig. S7 A and B). Thus, in the absence of Med1, the CD4+ iNKT cell subset was most severely affected, especially in the spleen, where the absolute numbers of the DN cells was not altered at all (Fig. S7B). It has been shown that GATA-3 deficiency led to a selective impairment of the CD4+ subset of iNKT cells (14), and in PLZF-deficient mice, the ratio of CD4+ to DN subsets was skewed toward CD4+ iNKT cells (32). However, Med1−/− thymic iNKT cells express normal or slightly higher level of GATA-3, and the expression of PLZF, which is normally down-regulated upon maturation into stage 3, is increased, this is in line with the lack of mature stage 3 iNKT cells in Med1−/− mice (Fig. S7 C and D).
Vα14-Jα18 TCR Transgene Expression Bypasses iNKT Cell Defects in Med1−/− Mice.
The twofold reduction of iNKT cells at the developmental stage 0 in Med1−/− mice implied the presence of an earlier defect in iNKT cell development, which might be bypassed by the forced expression of an iNKT cell-specific Vα14-Jα18 TCR transgene. Therefore, we crossed Med1−/− mice with transgenic mice expressing a rearranged Vα14-Jα18 TCRα chain under the control of the CD4 promoter (Vα14+ Tg) and found that the relative proportion and absolute cell numbers of iNKT cells were largely restored in the thymus, spleen, and liver of Med1−/− mice by this transgene (Fig. 5 A and B). Further analysis of the thymic iNKT cell developmental stages showed no differences in the percentage and absolute cell numbers between Med1+/− and Med1−/− mice expressing the Vα14-Jα18 TCR transgene (Fig. 5 C and D). Of note, cell numbers of immature iNKT cells are much higher in Vα14-Jα18 TCR transgenic mice compared with the nontransgenic heterozygous mice. On the other hand, cells at the mature stage 3 are greatly underrepresented in the TCR-transgenic mice (Figs. 4B and 5D). Therefore, the transgenic mice might not be suitable to study defects at the final maturation step because the majority of iNKT cells genereated have an immature phenotype.
Fig. 5.
Transgenic expression of Vα14-Jα18 TCR restores iNKT cell development in Med1−/− mice. (A) iNKT cells identified by PBS57-loaded CD1d tetramer in the thymus, spleen, and liver from Vα14 Tg-Med1+/− or Vα14 Tg-Med1−/− mice. The percentages of CD1d-tetramer+ iNKT cells are indicated in each plot. (B) Statistical analysis of the percentage (Upper) and absolute numbers (Lower) of iNKT cells in the thymus, spleen, and liver from Vα14 Tg-Med1+/− or Vα14 Tg-Med1−/− mice. (C) Surface expression of CD24, CD44, and NK1.1 by thymic iNKT cells from Vα14 Tg-Med1+/− or Vα14 Tg-Med1−/− mice. The left plots are gated on total thymocytes and the right plots are gated on CD24loCD1d-tetramer+ cells. (D) Absolute cell numbers of iNKT cells at stages 0 to 3. Five mice from each genotype were analyzed. Data are representative of five independent experiments.
Thymic iNKT Cells in Med1−/− Mice Lack IL-2Rβ and T-bet Expression.
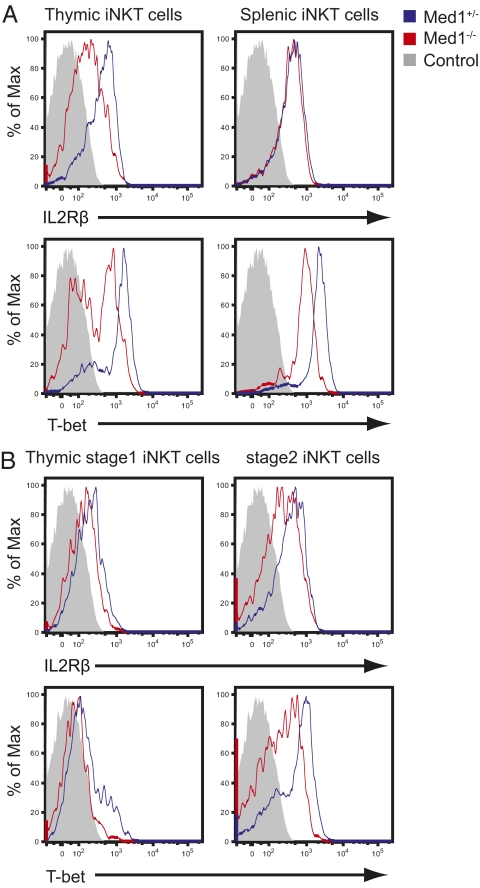
The developmental stage at which iNKT cell generation was most severely blocked was reminiscent of that of T-box transcription factor T-bet–deficient mice (10). It has been reported that T-bet controls not only iNKT cell maturation, but also migration, survival, and effector functions (33). IL-2Rβ (CD122) is a direct target gene of T-bet and is essential for iNKT cell maturation in an IL-15–dependent manner (34). These observations prompted us to examine whether the expression of IL-2Rβ and T-bet was affected in Med1−/− iNKT cells. As shown in Fig. 6A, gated on TCRβ+CD1d-tetramer+ iNKT cells, the expression of IL-2Rβ was drastically decreased in thymic Med1−/− iNKT cells (Fig. 6A, Upper Left), whereas it was maintained in splenic iNKT cells (Fig. 6A, Upper Right). T-bet expression was reduced in both thymic and splenic iNKT cells in Med1−/− mice (Fig. 6A, Lower). IL-2Rβ and T-bet expression are normally up-regulated upon iNKT cell maturation, with stage 3 cells showing the highest expression level (Fig. S8) (10, 12, 35). Thus, the overall decreased expression level of IL-2Rβ and T-bet might be the mere consequence of the paucity of mature iNKT cells in Med1−/− mice. Thus, we refined our analysis at the developmental stages 1 and 2 and found that Med1−/− immature stage 2 cells also displayed a reduced expression of IL-2Rβ and T-bet (Fig. 6B). Therefore, the lack of IL-2Rβ and T-bet is a potential molecular mechanism leading to the developmental block seen at the immature stage 2 in the thymus of Med1−/− animals.
Fig. 6.
Thymic and splenic iNKT cells in Med1−/− mice have reduced T-bet expression. (A) The expression of IL-2Rβ (Upper) and T-bet (Lower) in thymic and splenic CD1d-tetramer+ iNKT cells from Med1+/− or Med1−/− mice. (B) The expression of IL-2Rβ (Upper) and T-bet (Lower) in thymic iNKT cells at stage 1 and stage 2 (blue: Med1+/− mice; red: Med1−/− mice; gray solid: isotype control). Data are representative of five independent experiments.
Discussion
In the present study, we have shown that Med1 is essential for iNKT cell development but not the development of conventional αβ T cells. The specificity of the phenotype might be explained by the fact that Med1 is only found in a subset of Mediator complexes and is not required for the integrity of the complex per se (16). The defects observed at the early stages of iNKT cell development were bypassed by ectopic expression of Vα14-Jα18 TCR transgene, leading to a nearly complete restoration of iNKT cells (Fig. 5). One possible explanation for this finding is that in the transgenic mice, the majority of iNKT cells generated are immature and may not require factors, such as Med1, which is essential for terminal maturation. Hence, the transgenic mice might not be the best system to study the factors critical for the final maturation of iNKT cells. Alternatively, it is also possible that early expression of Vα14-Jα18 TCR transgene alters the normal developmental pathway of iNKT cells and bypasses the necessity for Med1, as has been discussed for Vα14+ Tg Fyn-deficient mice (23, 36).
The severe developmental block of the iNKT cell lineage before terminal maturation seen in Med1−/− animals resembles the one described for T-bet–deficient mice (10). The stepwise stage-dependent expression pattern of T-bet makes this transcription factor an attractive candidate for being a master regulator of iNKT cell terminal maturation. Mice deficient for costimulatory molecules (37), Tec kinase Itk and Rlk (38), and VDR (11) have been reported to exhibit iNKT cell defects accompanied by the diminished expression of T-bet or its direct target gene, IL-2Rβ. In Med1-deficient iNKT cells, T-bet and IL-2Rβ expression is markedly reduced (Fig. 6), most probably accounting for the block in the final maturation of thymic iNKT cells. The challenging question remaining is which transcription factor functions upstream of T-bet and requires Med1 as a cofactor. The most likely candidate is the VDR. It has been previously demonstrated that Med1 physically and functionally interacts with VDR (15). VDR is also required for iNKT cell development (11). Importantly, vitamin D deficiency during embryogenesis affects iNKT cell development (39). Alternatively, Med1 might interact with another iNKT cell master regulator RORγt. However, Med1 knockout mice neither display any defect in expression of Bcl-xL (Fig. S9A) nor do they have a defect in Vα14-Jα18 gene rearrangement (Fig. S9B), as described in RORγt-deficient mice. Still, it remains possible, that Med1 is crucial for the expression of only a subset of RORγt-regulated genes. Because RORγt target genes are currently unknown, this awaits further investigations.
In summary, our data reveal a surprisingly lineage-specific function for a generic cofactor, the Mediator subunit Med1, in iNKT cell development. Further studies are needed to elucidate the interplay between Med1 and VDR, and the potential VDR-dependent regulation of T-bet. Because vitamin D deficiency has been implicated in autoimmune disease (39), the VDR/Mediator interaction is of potential therapeutic interest.
Materials and Methods
Mice.
Med1 conditional knockout mice (40) and CD4-Cre mice (41) were described previously. B6.SJL mice expressing the CD45.1 alloantigen were purchased from The Jackson Laboratory. Vα14-Jα18 TCR transgenic mice were kindly provided by A.Bendelac (University of Chicago, Chicago, IL). Mice were bred and housed in specific pathogen free conditions in accordance with institutional, state, and federal guidelines on animal welfare.
Further materials and methods are provided in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. J. Reddy for providing us with Med1 floxed mice; Dr. A. Bendelac for Vα14-Jα18 TCR-transgenic mice and NKT cell hybridoma DN32.D3; Dr. C. Wilson for CD4-Cre transgenic mice; Drs. H. Luche, K. Beck, J. Swann, C. Waskow, and S. Martin for critical reading of the manuscript; and the National Institutes of Health Tetramer Facility for PBS57 loaded CD1d-tetramers. This work was supported by Deutsche Forschungsgemeinschaft Grants BOR-1639/4-1, the Emmy-Noether Fellowship BO1639/3, Collaborative Research Center Grant SFB592/C3, and funds from the Max Planck Society (to T.B.). X.Y. is part of the International Max-Planck-Research-School of Molecular and Cellular Biology. A.I. is supported by the Federal Ministry of Education and Research (BMBF 01 EO 0803).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109095108/-/DCSupplemental.
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.D'Cruz LM, Yang CY, Goldrath AW. Transcriptional regulation of NKT cell development and homeostasis. Curr Opin Immunol. 2010;22:199–205. doi: 10.1016/j.coi.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 4.Das R, Sant'Angelo DB, Nichols KE. Transcriptional control of invariant NKT cell development. Immunol Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 8.Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc Natl Acad Sci USA. 2005;102:5114–5119. doi: 10.1073/pnas.0408449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 11.Yu S, Cantorna MT. The vitamin D receptor is required for iNKT cell development. Proc Natl Acad Sci USA. 2008;105:5207–5212. doi: 10.1073/pnas.0711558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda JL, et al. Homeostasis of V alpha 14i NKT cells. Nat Immunol. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 13.Monticelli LA, et al. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc Natl Acad Sci USA. 2009;106:19461–19466. doi: 10.1073/pnas.0908249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim PJ, et al. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 15.Belakavadi M, Fondell JD. Role of the mediator complex in nuclear hormone receptor signaling. Rev Physiol Biochem Pharmacol. 2006;156:23–43. doi: 10.1007/s10254-005-0002-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, et al. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Crawford SE, et al. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 18.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 19.Landles C, et al. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol Endocrinol. 2003;17:2418–2435. doi: 10.1210/me.2003-0097. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y, et al. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J Biol Chem. 2000;275:14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- 21.Stumpf M, et al. Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proc Natl Acad Sci USA. 2010;107:21541–21546. doi: 10.1073/pnas.1005794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, et al. A modified alpha-galactosyl ceramide for staining and stimulating natural killer T cells. J Immunol Methods. 2006;312:34–39. doi: 10.1016/j.jim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Kronenberg M. Toward an understanding of NKT cell biology: Progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 24.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNab FW, et al. The influence of CD1d in postselection NKT cell maturation and homeostasis. J Immunol. 2005;175:3762–3768. doi: 10.4049/jimmunol.175.6.3762. [DOI] [PubMed] [Google Scholar]
- 27.Wei DG, et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005;202:239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 29.Mendiratta SK, et al. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–477. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 30.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 31.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 32.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda JL, et al. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood. 2006;107:2797–2805. doi: 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15 receptor beta-chain in natural killer 1.1+ T cell receptor-alpha beta+ cell development. J Immunol. 1997;159:5931–5935. [PubMed] [Google Scholar]
- 35.Matsuda JL, Gapin L. Developmental program of mouse Valpha14i NKT cells. Curr Opin Immunol. 2005;17:122–130. doi: 10.1016/j.coi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Gadue P, Yin L, Jain S, Stein PL. Restoration of NK T cell development in fyn-mutant mice by a TCR reveals a requirement for Fyn during early NK T cell ontogeny. J Immunol. 2004;172:6093–6100. doi: 10.4049/jimmunol.172.10.6093. [DOI] [PubMed] [Google Scholar]
- 37.Chung Y, et al. A critical role of costimulation during intrathymic development of invariant NK T cells. J Immunol. 2008;180:2276–2283. doi: 10.4049/jimmunol.180.4.2276. [DOI] [PubMed] [Google Scholar]
- 38.Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 39.Yu S, Cantorna MT. Epigenetic reduction in invariant NKT cells following in utero vitamin D deficiency in mice. J Immunol. 2011;186:1384–1390. doi: 10.4049/jimmunol.1002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia Y, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 41.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.