Abstract
An essential regulator of gene transcription, nuclear receptor liver receptor homologue 1 (LRH-1) controls cell differentiation in the developing pancreas and maintains cholesterol homeostasis in adults. Recent genome-wide association studies linked mutations in the LRH-1 gene and its up-stream regulatory regions to development of pancreatic cancer. In this work, we show that LRH-1 transcription is activated up to 30-fold in human pancreatic cancer cells compared to normal pancreatic ductal epithelium. This activation correlates with markedly increased LRH-1 protein expression in human pancreatic ductal adenocarcinomas in vivo. Selective blocking of LRH-1 by receptor specific siRNA significantly inhibits pancreatic cancer cell proliferation in vitro. The inhibition is tracked in part to the attenuation of the receptor’s transcriptional targets controlling cell growth, proliferation, and differentiation. Previously, LRH-1 was shown to contribute to formation of intestinal tumors. This study demonstrates the critical involvement of LRH-1 in development and progression of pancreatic cancer, suggesting the LRH-1 receptor as a plausible therapeutic target for treatment of pancreatic ductal adenocarcinomas.
Keywords: protein target, gene regulation
With mortality rate nearing its incidence, pancreatic ductal adenocarcinoma (PDAC) presents a challenge for modern oncology. Current chemotherapy drugs approved for pancreatic cancer are not organ specific and are modestly effective. Thus, there is a need for improved therapeutic options and effective pancreatic cancer drugs. Recent studies reveal that signaling pathways are similar in pancreatic development and malignant growth in the adult pancreas (1, 2). One of the common driving factors in pancreatic embryo- and oncogenesis is the nuclear receptor liver receptor homologue 1 (LRH-1, NR5A2; ref. 3). In adults, this protein is expressed primarily in liver, intestine, and pancreas, where it regulates expression of proteins maintaining cholesterol homeostasis (3, 4). LRH-1 is also expressed in the ovary and breast adipose tissue where it controls biosynthesis of steroids (5, 6). LRH-1 is vital in early development; it maintains undifferentiated ES cells by controlling expression of two transcription factors, Oct4 and Nanog (7–9). Recent studies demonstrated that LRH-1 can substitute for Oct4 in derivation of induced pluripotent stem (iPS) cells (9). LRH-1 is classed as an orphan nuclear receptor because its activating hormones have not been identified. Crystallographic and biochemical studies presented compelling evidence that LRH-1 could bind ligands (10–13) and suggested phosphatidyl inositols as potential hormone candidates for this receptor (10). LRH-1 is also regulated via posttranslational modifications, phosphorylation, and sumoylation (14, 15). Known transcriptional regulators of LRH-1 include steroid receptor coactivators (SRCs), the CREB-binding protein (CBP), and the peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) as well as the corepressors silencing mediator for retinoic acid and thyroid hormone receptor (SMRT), short heterodimer partner (SHP), the prospero homeobox protein 1 (Prox1), and the orphan nuclear receptor dosage sensitive sex reversal, adrenal hypoplasia critical region on chromosome X gene 1 (Dax-1) (3, 16, 17). No synthetic antagonists of LRH-1 are available to date. LRH-1 is linked to multiple developmental pathways including Wnt/β-catenin (8, 18), Hedgehog signaling (19), and regulatory cascades controlled by the pancreas-specific transcription factor pancreatic and duodenal homeobox 1 (PDX-1) (20). In pancreatic cancer, these pathways are reactivated and shown to be essential for development of pancreatic tumors (1, 2, 21, 22). Recent genome-wide association studies link mutations in the LRH-1 gene and its up-stream regulatory regions to development of PDAC (23).
In this study, we demonstrate that transcription of LRH-1 gene is misregulated, and its expression is activated in human PDACs compared to normal adult pancreas. We show that the aberrant expression of LRH-1 in pancreatic cancer cells can be attenuated by receptor specific siRNA. This suppression results in significant inhibition of cancer cell proliferation, suggesting that LRH-1 has a major role in development of pancreatic cancer. This finding complements the previous report implicating LRH-1 in formation of intestinal tumors (24).
Results
Identification of LRH-1 in Pancreatic Cancer Cells Cultured in Vitro.
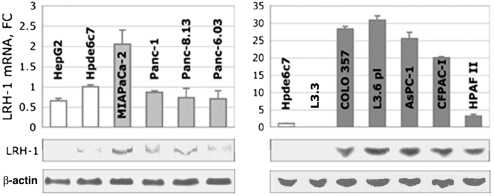
Using quantitative PCR (qPCR) and Western blot analyses, we showed that LRH-1 receptor is present in nine well-characterized pancreatic cancer cell lines derived either from the primary PDAC tumors or metastatic sites (Fig. 1). The receptor levels in most tested primary tumor cells were comparable to those found in nonneoplastic pancreatic ductal epithelium (Hpde6c7, Fig. 1, Left). In contrast, in most metastatic pancreatic cancer cells, the levels of the receptor transcripts were significantly increased (25–30-fold change in AsPC-1, COLO 357, and its derivative cell line L3.6 pl; Fig. 1, Right). In contrast to COLO 357 and L3.6 pl, in cells L3.3 (another derivative of COLO 357) the LRH-1 transcription appeared to be disabled. Western blot analysis confirmed the presence of LRH-1 in primary cancer cells and revealed its significantly higher expression in cells originated from metastatic tumors (shown at the bottom of Fig. 1). No LRH-1 protein was detected in L3.3 cells; because of the apparent absence of the receptor in these cells, we used them as a negative control in our further experiments. In four cancer cell lines, the activated transcription of LRH-1 gene correlated with reappearance of PDX-1, a known transcriptional regulator of the receptor (20). However, no clear reciprocity was revealed between these two proteins (Fig. S1 A and B). Notably, we observed significantly decreased levels of Prox1, an established corepressor of LRH-1 (16), in eight out of 10 cancer cell lines (Fig. S1C). We also found the transcripts for SHP, another corepressor of LRH-1 (3, 11), diminished in seven cancer cell lines tested (Fig. S1D).
Fig. 1.
Expression of LRH-1 in pancreatic cancer cells. Levels of LRH-1 mRNA in pancreatic cancer cells originated from primary and metastatic PDACs (Left, Right, in light and dark gray) are shown relative to control (nonneoplastic pancreatic ductal epithelial cells Hpde6c7, in white); note 10-fold scale change in the right panel. (Left) For comparison, the levels of LRH-1 mRNA in hepatocarcinoma cells HepG2 are shown in white. Standard deviations are drawn as black lines. Western blots at the bottom of the panels show normalized expression of LRH-1 protein in the corresponding cells (imaged by enhanced chemiluminescence, 1-min exposure). The expression levels of LRH-1 are normalized to those of β-actin (detected after 10-s exposure).
Expression of LRH-1 in Human PDAC Tumors.
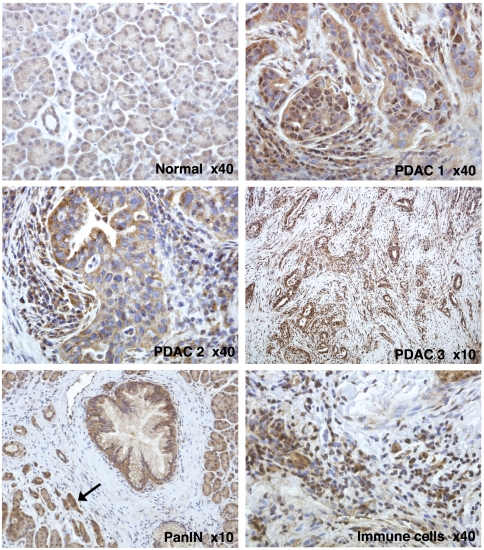
We analyzed the expression of the receptor in human resection specimens obtained from normal and cancerous pancreas. Twelve independent cases of PDAC and four samples of normal pancreas were studied using LRH-1 specific immunohistochemistry (IHC) staining with two receptor-specific antibodies [HPA005455 by Sigma-Aldrich and LS-A2447 by LifeSpan Biosciences; the specificity of antibodies was confirmed using positive (liver) and negative (heart muscle tissue] controls (Fig. S2). The IHC experiments revealed clear differences between the levels of LRH-1 in normal and cancerous pancreas (Fig. 2 and Fig. S2; for statistical analysis of IHC data, see Table S1). In normal exocrine pancreas, the LRH-1 protein was present at relatively low levels. Both acini and ducts showed weak LRH-1 staining in the cytoplasm and weak-to-moderate punctate staining in the nuclei. In contrast, in all tested PDACs, the presence of LRH-1 was markedly increased, with heightened levels of the protein detected either in the nuclei or the cytoplasm of cancerous cells. In some neoplastic cells, the receptor appeared to localize predominantly at the cytoplasm. Whereas the fibroblasts in the stroma surrounding the neoplastic lesions did not show significant staining, many infiltrating immune cells were positively stained for LRH-1. An increased presence of LRH-1 was also detected in the acinar cells affected by pancreatitis as well as in pancreatic intraepithelial neoplasia (PanIN) lesions (Fig. 2 and Fig. S2, indicated).
Fig. 2.
Differential expression of LRH-1 in normal and cancerous pancreas. Shown are IHC images of human normal (Normal) and neoplastic pancreas (PDAC 1–3). The samples were treated with primary antibody HPA005455 (Sigma-Aldrich), stained with ImmPACT diaminobenzidine (Vector Labs) and counterstained with hematoxylin; the original magnification (× 40 or × 10) is specified. Weak cytoplasmic and weak-to-moderate punctate nuclear staining is observed for all components of normal exocrine pancreas. In neoplastic cells, elevated levels of LRH-1 are observed either in the nuclei (PDAC 1) or in the cytoplasm (PDAC 2) or both. Whereas the fibroblasts surrounding neoplastic lesions do not show significant LRH-1 specific staining (PDAC 3), many infiltrating immune cells (indicated) are positively stained for LRH-1. Heightened levels of LRH-1 are also observed in PanIN lesions (indicated) and in the acinar and ductal cells affected by pancreatitis (indicated by arrow).
Blocking LRH-1 by Specific siRNA.
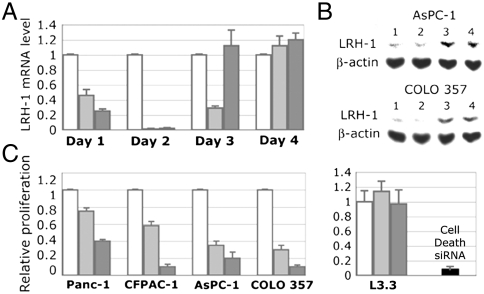
We analyzed whether the LRH-1 transcripts and protein levels in pancreatic cancer cells are diminished after treatments with receptor specific siRNA. Two different siRNAs corresponding to the receptor N-terminal region and its ligand-binding domain [nucleotides 411–431 (siRNA-1) and 1504–1524 (siRNA-2); ref. 25] were used for these experiments. Quantitative PCR analysis showed nearly complete inhibition of LRH-1 gene transcription on the second day after the treatments (Fig. 3A; the transcript levels returned to the original by day 4 likely due to siRNA degradation). In cells treated with control siRNA, the transcription of LRH-1 was not affected. Complementing qPCR data, Western blot analysis showed that expression of LRH-1 protein in cells treated with specific siRNAs was substantially reduced; treatment with control siRNA had no effect on LRH-1 expression (Fig. 3B).
Fig. 3.
Blocking LRH-1 function by siRNA specific to the receptor. (A) Inhibition of LRH-1 transcription in AsPC-1 cells. Cells were analyzed by qPCR for relative levels of LRH-1 mRNA at days 1–4 following transfections. Control in white corresponds to cells treated with irrelevant siRNA (see Materials and Methods); light- and dark-gray bars indicate mRNA in cells transfected with two LRH-1 specific siRNAs. (B) Western blot analyses of AsPC-1 and COLO 357 cells transfected with anti-LRH-1 siRNAs. Lanes: 1 and 2, cells treated with two LRH-1 specific siRNA; 3, cells transfected with control siRNA; 4, nontransfected cells. Data are shown for day 2 following the transfections. Expression of β-actin was used as an internal control. (C) Effect of LRH-1 knockdown on cancer cell proliferation. Proliferation of four pancreatic cancer cells expressing LRH-1 (indicated in the left panel) was compared following their transfections with two LRH-1 specific siRNAs (in light and dark gray). Data are shown for day 4 relative to the control (white). Proliferation of cells L3.3 that do not express LRH-1 was not affected by the anti-LRH-1 siRNAs (shown in the right panel relative to the control). The efficiency of transfections (> 90%) was optimized by monitoring proliferation of cells treated with AllStars Hs Cell Death Control siRNA (in black).
Inhibition of Pancreatic Cancer Cell Proliferation by Anti-LRH-1 siRNA.
Using siRNA as an efficient tool for silencing the receptor, we analyzed the role of LRH-1 in proliferation of pancreatic cancer cells. These experiments revealed that specific blocking of LRH-1 results in significant inhibition of cancer cell proliferation; inhibition was observed in four different cell lines expressing the receptor (Fig. 3C, Left). Notably, proliferation of the same cells treated with control siRNA was not affected. Furthermore, proliferation of pancreatic cancer cells L3.3, which do not express LRH-1 (Fig. 1), was not affected either (Fig. 3C, Right).
Transcription of LRH-1 Regulated Gene Targets.
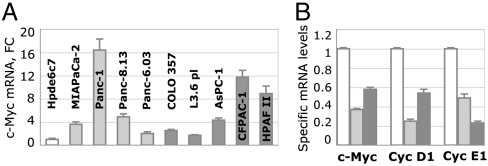
We tested possible mechanisms of LRH-1 involvement in pancreatic cancer by evaluating the levels of mRNA for its established targets, including cyclins D1, E1 (Cyc D1, Cyc E1), and C-Myc genes (9, 18), which are essential for cell growth and proliferation (26–29). Quantitative PCR data revealed that C-Myc gene is up-regulated in all tested pancreatic cancer cells (Fig. 4A). Quantitative PCR analyses also showed that transcription of C-Myc was compromised in pancreatic cancer cells treated with LRH-1 specific siRNAs (Fig. 4B). Specific inhibition of LRH-1 also resulted in diminished transcription of cyclins D1 and E1 (Fig. 4B). Treatment of cells with control siRNA had no effect on transcription of any of the tested targets. All measurements were performed between 24 and 48 h after the transfections, when the levels of LRH-1 were minimal (Fig. 3). Complementing these analyses, we evaluated the levels of other known gene targets of LRH-1 that control cell differentiation (Oct4 and Nanog; refs. 7–9). Although the levels of Oct4 in pancreatic cancer cells were comparable to that observed in nonneoplastic ductal epithelium (Fig. S3A), Nanog gene was found to be activated in five cancer cell lines tested (Fig. S3B).
Fig. 4.
Effects of blocking LRH-1 on transcription of genes controlling cell proliferation. (A) Quantitative PCR data showing levels of C-Myc mRNA. Levels of C-Myc in primary and metastatic cells are shown as light- and dark-gray bars, relative to that found in nonneoplastic cells Hpde6c7. Standard deviations are drawn as black lines. (B) Inhibition of C-Myc, Cyc D1, and Cyc E1 gene transcription in AsPc-1 cells treated with anti-LRH-1 siRNA. Cells were analyzed by qPCR for the relative levels of mRNA corresponding to C-Myc, Cyc D1, and Cyc E1 (indicated). Controls in white correspond to cells treated with irrelevant siRNA; light- and dark-gray bars indicate the levels of corresponding mRNA in cells transfected with two LRH-1 specific siRNAs.
Cell Cycle Arrest Following Inhibition of LRH-1 by Specific siRNA.
We analyzed the cell cycle of AsPC-1 cells treated with either irrelevant or LRH-1 specific siRNAs using propidium iodide (PI) DNA staining followed by fluorescence-activated cell sorting. These studies allowed assessment of normal and apoptotic nuclei as well as distribution of cell populations in G0/G1, S, and G2/M phases of the cell cycle. Our data revealed that G0/G1 population in specifically treated cells was increased, whereas the populations of cells progressing through S phase and those entering G2 phase were decreased (Fig. S4). Based on these observations, we conclude that specific blocking of LRH-1 in pancreatic cancer cells induces a cell cycle arrest in G0/G1 phase by slowing the G1/S transition. No increase in the apoptotic cell population was observed for cells transfected with either control or specific siRNAs compared to untreated AsPC-1 cells.
Discussion
In this study, we provide evidence of the up-regulated transcription of LRH-1 gene in PDAC cells compared to nonneoplastic pancreatic ductal epithelium. Up-regulation is modest in cancer cells derived from primary tumors; however, in cells originated from metastatic sites, the observed levels of LRH-1 transcripts exceed those seen in nonneoplastic epithelium 20–30 times (Fig. 1). Activation of LRH-1 does not always correlate with the reappearance of PDX-1, a known up-stream activator of LRH-1 (20) (Fig. S1 A and B), suggesting that other regulatory mechanisms, including possible genomic alterations influencing expression and activity of the receptor, might be involved in the observed up-regulation of LRH-1 in pancreatic cancer cells. In agreement with this idea, genome-wide association study identified five SNPs associated with pancreatic cancer that mapped to LRH-1 gene and its up-stream regulatory region (23). No evidence of alterations in the LRH-1 gene copy number was found in independent comparative genomic hybridization (CGH) studies using pancreatic cancer cells cultured in vitro (30). Our data showed decreased levels of Prox1 and SHP, two established corepressors of LRH-1 (3, 11, 16), in the majority of tested pancreatic cancer cell lines (Fig. S1 C and D), suggesting that changes in regulation of transcriptional activity of LRH-1 might be associated with pancreatic oncogenesis.
Consistent with activation of LRH-1 in pancreatic cancer cells in vitro, we observe increased levels of LRH-1 in human PDAC samples (Fig. 2, Fig. S2, and Table S1). In accordance with the receptor’s established function in normal exocrine pancreas (4), relatively low levels of LRH-1 are found in the nuclei and cytoplasm of normal acini and ducts. In contrast, in PDAC tumors, the presence of LRH-1 is markedly increased, with heightened levels of receptor found either in the nuclei or cytoplasm of the neoplastic cells. No evidence of LRH-1 gene amplification was found in independent CGH studies on human PDAC tumors (31, 32) to account for the observed receptor overexpression. Interestingly, in some neoplastic lesions, the LRH-1 protein is localized predominantly at the cytoplasm of cancerous cells (Fig. 2 and Fig. S2). Previously, similar cytoplasmic localization was reported for overexpressed LRH-1 receptor in colon and breast cancer tumors (24, 25). The implications of these observations are not understood. It is hypothesized that localization of nuclear receptors in the cytoplasm of cancer cells might be linked to nongenomic signaling essential for tumor survival and growth (33, 34). Such cytoplasmic accumulation could also be related to the receptor-mediated transcription and nuclear-cytoplasmic shuttling of microRNAs in neoplastic cells; these processes are often impaired during tumorigenesis (35, 36).
Our data show that activated expression of LRH-1 is associated with cancer growth and is evident in samples of invasive pancreatic cancer (Fig. 2, Fig. S2, and Table S1). These observations suggest that the increased activity of LRH-1 in cancer cells might correlate with their aggressive growth and exaggerated proliferative properties. The latter hypothesis is supported by the established role of LRH-1 in cell proliferation. The receptor is known to induce cell proliferation through the induction of cyclins D1 and E1, an effect thought to be potentiated by interactions of LRH-1 with β-catenin, a key component of the Wnt signaling pathway (8, 18). In addition to cyclins D1 and E1, expression of Myc genes, which are required for cell proliferation and are also regulated via Wnt/β-catenin signaling, is shown to be controlled by LRH-1 (9, 18). Consistent with these data, we observe decreased levels of C-Myc as well as Cyc D1 and Cyc E1 as a result of specific blocking of LRH-1 (Fig. 4). Importantly, the diminished levels of LRH-1 and its transcriptional targets correlate with significant inhibition of pancreatic cancer cell proliferation (Fig. 3C). Analysis of underlying mechanisms of LRH-1 mediated control over cell proliferation revealed that blocking the receptor affects the cell cycle by slowing the G1/S transition (Fig. S4). This finding is congruent with the described role of LRH-1 as a regulator of cyclins D1 and E1, which induce progression through the cell cycle and stimulate the G1/S transition (18).
Because LRH-1 is critical for maintenance of pluripotent stem cells (7, 8) and is shown to mediate reprogramming of somatic cells (9), we examined levels of most common markers associated with dedifferentiation and pluripotency, including transcription factors Oct4 and Nanog, the established targets of LRH-1 (7–9). Although we did not find significant differences in Oct4 transcription between normal and neoplastic cells (Fig. S3A), we found dramatic amplification of Nanog transcripts in a subset of cancer cells (Fig. S3B); the reappearance of Nanog in these cells might be indicative of their dedifferentiated state. The receptor-mediated activation of genes controlling cell growth, proliferation, and differentiation (Fig. 5) suggest that aberrant expression of LRH-1 might be linked to proliferation of dedifferentiated pancreatic cancer cells, which are associated with aggressive pancreatic tumors (37). Recent studies demonstrated the inherent plasticity of pancreatic cells that can be reprogrammed to the transient de-differentiated progenitor-like states, which can give rise to PDAC (38–40). Because of the established roles of LRH-1 in pancreatic and stem cell differentiation (7–9, 20), we hypothesize that this receptor might contribute to the reprogramming events in the adult pancreas that lead to its transformation. This hypothesis implies that LRH-1 might be involved in maintenance of pancreatic cancer stem cells (37) that are thought to be the major contributor to the relapse of pancreatic cancer and its metastasis.
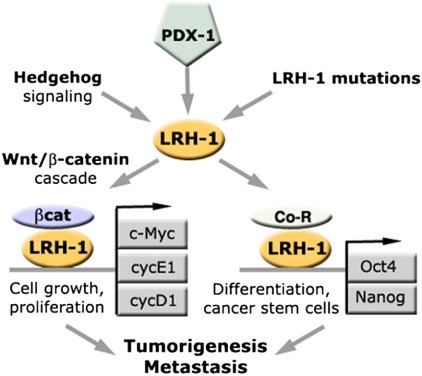
Fig. 5.
Possible mechanisms of involvement of nuclear receptor LRH-1 in development of pancreatic cancer. Indicated are major transcriptional targets of LRH-1 and regulatory pathways mediated by the receptor. The expression of LRH-1 (yellow symbol) is controlled by the pancreas-specific master transcription factor PDX-1 (indicated in green) and via hedgehog signaling pathway that are both reactivated in pancreatic cancer. Activating mutations in the LRH-1 gene and its regulatory regions might contribute to the receptor up-regulation. The activated receptor functions synergistically with β-catenin (shown in purple), activating multiple target genes regulating cell growth and proliferation (Cyc D1, Cyc E1, and C-Myc, indicated in gray) as well as cell differentiation (Oct4, Nanog, indicated in gray). The cumulative effect of the aberrant, LRH-1 mediated activation of these multiple targets in adult pancreas results in tumorigenesis and metastasis.
While investigating the diverse and intersecting mechanisms of the receptor involvement in pancreatic oncogenesis, we noted that tumor-associated infiltrating immune cells, including lymphocytes and neutrophils, also express high levels of LRH-1 (Fig. 2 and Fig. S2). Similar expression of LRH-1 was previously detected in lymphocytes infiltrating breast carcinomas (25). These observations are captivating as accumulating data indicate that infiltration of solid tumors by immune cells might enhance the metastatic potential of cancer cells by facilitating their invasion into surrounding tissues (41). Notably, human neutrophils produce high levels of interleukin 1 receptor antagonist (IL-1RA), an established transcriptional target of LRH-1 (42, 43). This protein modulates the inflammatory responses during carcinogenesis, and its overexpression correlates with poor prognosis in patients with diverse malignancies (44). LRH-1 also regulates the extraadrenal biosynthesis of glucocorticoids, which modulate local inflammatory responses (45) and prolong the survival of neutrophils by delaying their apoptosis (46). These findings point to another possible role for LRH-1 in pancreatic tumorigenesis, through its impact on inflammatory pathways. Previously, LRH-1 was shown to contribute to intestinal tumor formation through effects on cell cycle and inflammation (24). In agreement with these data, we observe heightened expression of LRH-1 in pancreatic tissue affected by pancreatitis and in PanIN lesions (Fig. 2 and Fig. S2).
In summary, our work reveals that nuclear receptor LRH-1 is up-regulated and overexpressed in neoplastic pancreas compared to normal pancreatic tissues. We show that specific blocking of LRH-1 by siRNA inhibits pancreatic cancer cell proliferation in vitro; the inhibition is tracked to the attenuation of the receptor’s transcriptional targets controlling cell growth, proliferation, and differentiation. Our data, combined with recent results of genome-wide association study (23), demonstrate that LRH-1 receptor has a major role in development of pancreatic cancer. Our study also suggests that LRH-1 might be a plausible therapeutic target for treatment of PDAC. Although many other regulatory molecules have been identified as the important players in pancreatic tumorigenesis, critical factors contributing to the LRH-1 appeal as a drug target include the receptor’s well-defined ligand-binding pocket, which could be targeted by specific synthetic modulators, and the proven efficiency of nuclear receptor inhibitors for treatment of different types of malignancies, including breast and prostate cancer.
Materials and Methods
Cells and Culture Conditions.
Pancreatic cancer cells and immortalized pancreatic ductal epithelium cells were kindly provided by M. McMahon [University of California, San Francisco (UCSF)]. Cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2, in DMEM supplemented with 10% FBS, 1% l-glutamine, and antibiotics; cells were harvested and passaged when they reached 80% confluence.
Immunohistochemistry.
IHC experiments were done on 5-μm formalin-fixed paraffin-embedded sections of normal pancreas (four cases) and pancreatic ductal adenocarcinoma samples (12 cases; obtained from UCSF Cancer Center Tissue Core and from LifeSpan Biosciences, Inc.). The tissue sections were mounted on slides, treated with xylene and rehydrated using alcohol–water mixtures before heat treatment in 10 mM citric acid (pH 6.0) for antigen retrieval. The endogenous peroxidase activity was quenched using 0.3% H2O2 solution. Following blocking with 10% serum, sections were incubated at +4 °C overnight with primary human LRH-1 (hLRH-1) antibodies [LS-A2447 (LifeSpan Biosciences) or HPA005455 (Sigma-Aldrich), 1∶200 dilution], then rinsed and incubated with biotinylated anti-rabbit IgG (Vector Laboratories, 1∶200 dilution) for 30 min at room temperature. After rinsing, the samples were treated with streptavidin and biotinylated horseradish peroxidase (Vectastain Elite kit, Vector Labs), stained with ImmPACT diaminobenzidine (Vector Labs) and counterstained with hematoxylin. Images were analyzed using confocal microscopy (Nikon Eclipse Ti, magnification × 10 or × 40). The specificity of staining was confirmed using positive (liver) and negative (heart muscle tissue) controls.
siRNA Blocking.
Pancreatic cancer cells AsPC-1 and COLO 357 were transfected with 10 nM of either of the two anti-LRH-1 siRNAs [corresponding to nucleotides 411–431 (siRNA-1) and 1504–1524 (siRNA-2) of hLRH-1] or control siRNA using HiPerfect reagent (Qiagen) according to manufacturer’s protocol. Either siRNA targeting GFP or ALLStars siRNA labeled with Alexa Fluor 488 (Qiagen) were used for controls, depending on the specifics of the experiments. The transfection efficiencies (75–90%) were evaluated by flow cytometry (BD LSR II flow cytometer, Becton Dickinson) and confirmed using AllStars Hs Cell Death Control siRNA (Qiagen).
RNA Purification, cDNA Synthesis, and qPCR Analysis.
For each sample, total RNA was isolated using TRIzol (Invitrogen) according to manufacturer’s protocol. Complementary DNA was synthesized from 1.5 μg of total RNA, at 42 °C for 60 min, using the SuperScript-II First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative PCR amplification of mRNA for LRH-1 and other targets was performed in triplicates using the Mx3005P Real-Time PCR System and the SYBR Green I dye (Stratagene) for detection. No Template Control and No Reverse Transcription Control were included in each run. The amplification curves were analyzed with the Mx3005P software using the comparative cycle threshold method. All target mRNAs levels were normalized relative to that of RS9 (internal control).
Western Blot Analysis.
Cell lysate samples were separated by SDS PAGE and transferred to nitrocellulose membranes. Western blot analyses were performed using goat anti-LRH-1 antibody and donkey antigoat horseradish peroxidase conjugated IGs (sc-5995 and sc-2033, Santa Cruz Biotechnology). The membranes were developed using enhanced chemiluminescence according to a standard protocol. The expression levels of LRH-1 were normalized to that of β-actin (detected using goat anti-β-actin antibody; sc-1616, Santa Cruz Biotechnology).
Cell Growth and Proliferation.
Cultured cells were pelleted by centrifugation at 900 × g and resuspended in phenol red free medium supplemented with 10% charcoal-stripped FBS (Hyclone) and 2% l-glutamine. Cells were plated in replicates of four (104 cells/mL) in 96-well microtiter plates. At 24 h after seeding, cells were transfected with either anti-LRH-1 or control siRNAs (10 nM) using HiPerfect reagent (Qiagen) according to manufacturer’s protocol. At 96 h after the transfections, cell proliferation in each well was quantified using CellTiter-Glo assay (Promega).
Cell Cycle Analysis.
Untreated cells AsPc-1 and cells treated with anti-LRH-1 or control siRNAs were fixed with ethanol, stained with propidium iodide (Sigma), and analyzed using BD LSR II flow cytometer [Becton Dickinson; 488-nm excitation, 562–588 nm emission (R-phycoerythrin (PE) fluorescence); single cells have been gated using PE-Area/PE-Width plot]. The cell cycle profiles were analyzed with FlowJo V.8.8.6 (Flow Cytometry Analysis Software), using Dean–Jett–Fox algorithm.
Supplementary Material
Acknowledgments.
The authors thank Drs. S. Gysin and K. Shumate (Helen Diller Family Comprehensive Cancer Center, UCSF) for reagents and experimental suggestions, Drs. M. McMahon, M. Tempero (Cancer Center, UCSF), and M. Hebrok (Diabetes Center, UCSF) for helpful discussions, and R. Uthayaruban for technical assistance. This work was supported by National Institute of Health Grants R01 DK078075 and R21 CA140751 (to R.J.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112047108/-/DCSupplemental.
References
- 1.Thayer SP, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pasca di Magliano M, et al. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS One. 2007;2:e1155. doi: 10.1371/journal.pone.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fayard E, Auwerx J, Schoonjans K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Fayard E, Schoonjans K, Annicotte J-S, Auwerx J. Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J Biol Chem. 2003;278:35725–35731. doi: 10.1074/jbc.M302370200. [DOI] [PubMed] [Google Scholar]
- 5.Kim JW, Peng N, Rainey WE, Carr BR, Attia GR. Liver receptor homolog-1 regulates the expression of steroidogenic acute regulatory protein in human granulosa cells. J Clin Endocrinol Metab. 2004;89:3042–3047. doi: 10.1210/jc.2003-031599. [DOI] [PubMed] [Google Scholar]
- 6.Clyne CD, et al. Regulation of aromatase expression by the nuclear receptor LRH-1 in adipose tissue. Mol Cell Endocrinol. 2004;215:39–44. doi: 10.1016/j.mce.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Gu P, et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner RT, Xu X, Yi F, Merrill BJ, Cooney AJ. Canonical Wnt/β-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells. 2010;28:1794–1804. doi: 10.1002/stem.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heng JC, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Krylova IN, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 11.Ortlund EA, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- 12.Whitby RJ, et al. Small molecule agonists of the orphan nuclear receptors steroidogenic factor-1 (SF-1, NR5A1) and liver receptor homologue-1 (LRH-1, NR5A2) J Med Chem. 2011;54:2266–2281. doi: 10.1021/jm1014296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JM, et al. A nuclear receptor dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474:506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MB, et al. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol Cell Biol. 2005;25:1879–1890. doi: 10.1128/MCB.25.5.1879-1890.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YK, Choi YH, Chua S, Park YJ, Moore DD. Phosphorylation of the hinge domain of the nuclear hormone receptor LRH-1 stimulates transactivation. J Biol Chem. 2006;281:7850–7855. doi: 10.1074/jbc.M509115200. [DOI] [PubMed] [Google Scholar]
- 16.Qin J, et al. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and supresses the transcription of the cholesterol 7-a-hydroxylase gene. Mol Endocrinol. 2004;18:2424–2439. doi: 10.1210/me.2004-0009. [DOI] [PubMed] [Google Scholar]
- 17.Sablin EP, et al. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci USA. 2008;105:18390–18395. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botrugno OA, et al. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell. 2004;15:499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Sheela SG, Lee WC, Lin WW, Chung BC. Zebrafish ftz-f1a (nuclear receptor 5a2) functions in skeletal muscle organization. Dev Biol. 2005;286:377–390. doi: 10.1016/j.ydbio.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 20.Annicotte JS, et al. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol Cell Biol. 2003;23:6713–6724. doi: 10.1128/MCB.23.19.6713-6724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quint K, et al. The expression pattern of PDX-1, SHH, patched and Gli-1 is associated with pathological and clinical features in human pancreatic cancer. Pancreatology. 2009;9:116–126. doi: 10.1159/000178882. [DOI] [PubMed] [Google Scholar]
- 22.Heiser PW, et al. Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterololy. 2008;135:1288–1300. doi: 10.1053/j.gastro.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson G, et al. A genome-wide association study identified pancreatic cancer susceptibility loci on chromosomes 13q22 1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoonjans K, et al. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc Natl Acad Sci USA. 2005;102:2058–2062. doi: 10.1073/pnas.0409756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Annicotte JS, et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24:8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caldon CE, Musgrove EA. Distinct and redundant functions of cyclin E1 and E2 in development and cancer. Cell Div. 2010;5:2. doi: 10.1186/1747-1028-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacy DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 29.Ekholm SV, Reed SI. Regulation of cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 30.Gysin S, Rickert P, Kastury K, McMahon M. Analysis of genomic DNA alterations and mRNA expression patterns in a panel of human pancreatic cancer cell lines. Genes Chromosomes Cancer. 2005;44:37–51. doi: 10.1002/gcc.20216. [DOI] [PubMed] [Google Scholar]
- 31.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada T, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004;89:6130–6138. doi: 10.1210/jc.2004-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakravarty D, et al. Extranuclear functions of ER impact invasive migration and metastasis by breast cancer cells. Cancer Res. 2010;70:4092–4101. doi: 10.1158/0008-5472.CAN-09-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamagata K, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transfomation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 37.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 38.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris JP, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris J, Hebrok M. It’s free for all—insulin-positive cells join the group of potential progenitors for pancreatic ductal adenocarcinoma. Cancer Cell. 2009;16:359–361. doi: 10.1016/j.ccr.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McColl SR, Paquin R, Menard C, Beaulieu AD. Human neutrophils produced high levels of the interleukin 1 receptor antagonist in response to granulocyte/macrophage colony-stimulating factor and tumor necrosis factor alpha. J Exp Med. 1992;176:593–598. doi: 10.1084/jem.176.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venteclef N, Delerive P. Interleukin-1 receptor antagonist induction as an additional mechanism for liver receptor hololog-1 to negatively regulate the hepatic acute phase response. J Biol Chem. 2007;282:4393–4399. doi: 10.1074/jbc.M608993200. [DOI] [PubMed] [Google Scholar]
- 44.Mustea A, et al. Decreased IL-1 RA concentration in ascites is associated with a significant improvement in overall survival in ovarian cancer. Cytokine. 2008;42:77–84. doi: 10.1016/j.cyto.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Mueller M, et al. The nuclear receptor LRH-1 critically regulates extra-adrenal glucocorticoid synthesis in the intestine. J Exp Med. 2006;203:2057–2062. doi: 10.1084/jem.20060357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox G. Glucocorticoid treatment inhibits apoptosis in human neutrophils. Separation of survival and activation outcomes. J Immunol. 1995;154:4719–4725. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.