Abstract
To catalog protein-altering mutations that may drive the development of prostate cancers and their progression to metastatic disease systematically, we performed whole-exome sequencing of 23 prostate cancers derived from 16 different lethal metastatic tumors and three high-grade primary carcinomas. All tumors were propagated in mice as xenografts, designated the LuCaP series, to model phenotypic variation, such as responses to cancer-directed therapeutics. Although corresponding normal tissue was not available for most tumors, we were able to take advantage of increasingly deep catalogs of human genetic variation to remove most germline variants. On average, each tumor genome contained ∼200 novel nonsynonymous variants, of which the vast majority was specific to individual carcinomas. A subset of genes was recurrently altered across tumors derived from different individuals, including TP53, DLK2, GPC6, and SDF4. Unexpectedly, three prostate cancer genomes exhibited substantially higher mutation frequencies, with 2,000–4,000 novel coding variants per exome. A comparison of castration-resistant and castration-sensitive pairs of tumor lines derived from the same prostate cancer highlights mutations in the Wnt pathway as potentially contributing to the development of castration resistance. Collectively, our results indicate that point mutations arising in coding regions of advanced prostate cancers are common but, with notable exceptions, very few genes are mutated in a substantial fraction of tumors. We also report a previously undescribed subtype of prostate cancers exhibiting “hypermutated” genomes, with potential implications for resistance to cancer therapeutics. Our results also suggest that increasingly deep catalogs of human germline variation may challenge the necessity of sequencing matched tumor-normal pairs.
Prostate carcinoma is a disease that commonly affects men, with incidence rates dramatically rising with advancing age (1). The vast majority of these malignancies behave in an indolent fashion, but a subset is highly aggressive and resistant to conventional cancer therapeutics. Although recent studies have detailed the landscape of genomic alterations in localized prostate cancers, including a report describing the whole-genome sequencing of seven primary tumors (1–4), the genetic composition of lethal and advanced disease is poorly defined. Previous work demonstrates the importance of chromosomal rearrangements that include TMPRSS2-ERG gene fusion as a frequent attribute of prostate cancer genomes, with clear implications for tumor biology (5–7). However, considerably less is known about the contribution of somatic point mutations to the pathogenesis of prostate cancer (3, 4, 8), including those specific somatic mutations that may drive metastatic progression or the development of resistance to specific therapeutics, such as those targeting the androgen receptor (AR) program (2–4). In this study, we describe the application of whole-exome sequencing (9) to determine the mutational landscape of 23 prostate cancers representing aggressive and lethal disease, including both metastases and primary carcinomas. All tumors were propagated in immunocompromised mice as tumor xenografts (10) to model the heterogeneity in tumor growth, response to treatment, and lethality that exists in prostate cancer. Furthermore, these tumor xenografts have the advantage of little to no human stromal contamination and provide the means to test the consequences of mutations functionally. Although corresponding normal tissue was not sequenced for most samples, we find that comparisons with increasingly deep catalogs of segregating germline variants based on unrelated individuals provide an effective filter, challenging the necessity of sequencing matched tumor-normal pairs. We identify a number of genes in which nonsynonymous alterations (somatic mutations or very rare germline mutations) are recurrently observed, including variants in TP53, DLK2, GPC6, and SDF4. Surprisingly, we also identify 3 aggressive prostate cancers that exhibit a “hypermutated” phenotype (i.e., a gross excess of point mutations relative to the other tumors sequenced here as well as those prostate cancers that have been evaluated to date). Finally, a comparison of castration-resistant (CR) and castration-sensitive (CS) matched tumor pairs derived from the same site of origin highlights mutations in the Wnt pathway as potentially contributing to the development of resistance to therapeutic targeting of AR signaling.
Results
Landscape of Prostate Cancer Mutations.
We performed whole-exome sequencing of 23 prostate cancers derived from 16 different lethal metastatic tumors and three high-grade primary carcinomas using solution-based hybrid capture (Nimblegen; Roche) followed by massively parallel sequencing (Illumina). Samples were designated as LuCaP 23.1 through LuCaP 147 in the order in which they were initially established as xenografts in mice (SI Appendix, Table S1). Three tumors representing CR variants of the original cancers (LuCaP 23.1AI, LuCaP 35V, and LuCaP 96AI) were also analyzed. Eight samples were captured against regions defined by the National Center for Biotechnology Information Consensus Coding Sequence Database (CCDS, 26.6 Mb), whereas the remaining 15 samples were captured using a more inclusive definition of the exome (RefSeq, 36.6 Mb) (SI Appendix, Table S2).
To filter contamination by mouse genomic DNA, sequence reads were independently mapped to both the mouse (mm9) and human (hg18) genome sequences, and only sequences that mapped exclusively to the latter were considered further. In each xenograft, 4–19% of total reads were discarded because of mapping to the mouse genome. After also removing duplicates, we achieved an average of ∼100-fold coverage of the 26.6-Mb target in samples captured using the CCDS target definition and an average of ∼140-fold coverage of the 36.6-Mb target in samples captured using the RefSeq definition. Samples had 90–95% of their respective target definitions covered to sufficient depth to enable high-quality base calling (SI Appendix, Figs. S1–S3 and Table S3). Across 23 tumors, we identified a nonredundant set of ∼80,000 single-nucleotide variants occurring within coding regions.
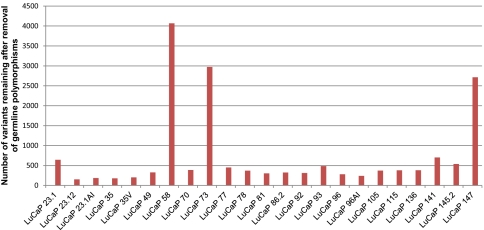
Most tumor sequencing analyses use matched tumor-normal pairs to distinguish somatic mutations present in the tumor from variants present in the germline of a given individual, with few exceptions (11). However, the fact that the overwhelming majority of germline variation in an individual human genome is “common,” coupled with the availability of increasingly deep catalogs of germline variation segregating in the human population, challenges the assumption that this is essential. Because corresponding normal tissue was not available for many of these tumor samples, we used the approach of sequencing tumor tissue only, removing from consideration all variants that were also observed in the pilot dataset of the 1,000 Genomes Project (12, 13), as well as variants present in any of ∼2,000 additional exomes sequenced at the University of Washington. After filtering, 3 tumors (LuCaP 58, LuCaP 73, and LuCaP 147) were observed to contain a very large number of single-nucleotide variants relative to all other tumors: 4,067, 2,972, and 2,714, respectively (Fig. 1). We refer to these xenografts as “hypermutated” and discuss their features below. Excepting these 3 tumors, the applied filters reduced the number of coding variants under consideration from ∼13,500 to ∼350 per tumor (Fig. 1 and SI Appendix, Tables S1 and S4). Of the 14,705 novel variants observed across the 23 tumors, 13,827 variants were called as heterozygous and 878 were called as homozygous, and 8,617 variants were predicted to cause amino acid changes (nonsynonymous), including 8,176 missense, 346 nonsense, and 95 splice site variants (SI Appendix, Table S5). These novel single-nucleotide variants (nov-SNVs) likely comprise a mixture of (i) somatic mutations that were present in the original tumor, (ii) somatic mutations occurring after tumor propagation and evolution in the mouse hosts, (iii) germline variants that were present in the individual of origin but are very rare in the population (i.e., “private” germline variation), and (iv) false-positive variant calls.
Fig. 1.
Subset of xenografts exhibits a high number of mutations. After filtering to remove common germline polymorphisms, three xenografts (LuCaP 73, LuCaP 147, and LuCaP 58) exhibit a hypermutated phenotype, with several thousand nov-SNVs each. This contrasts with the other 20 xenografts, which have 362 ± 147 coding alterations remaining after filtering.
We next sought to assess the efficiency of filtering against databases of germline variation in enriching for somatic variants. For three tumors, LuCaP 92, LuCaP 145.2, and LuCaP 147, normal tissue and tumor tissue were also collected directly from patients before propagation as xenografts. For two xenografts, LuCaP 145.2 and LuCaP 147, the fresh tumors were neighboring metastases from the same patient, whereas only the fresh tumor for LuCaP 92 was the exact precursor lesion from which the xenograft was derived. However, based on the observations of Liu et al. (14), metastases from a given patient are likely to be closely related. We sequenced the exomes of both normal and tumor tissues to determine true somatic mutations. For this analysis, we required that each base be covered by at least 24-fold in xenograft, tumor, and normal tissue and used less stringent requirements to call a variant within the normal tissue to reduce the number of false-positive somatic calls. In two of these three tumors (LuCaP 92 and LuCaP 145.2), filtering against germline databases reduced the number of variants under consideration from ∼21,000 to ∼400 (SI Appendix, Table S1). such that 0.2% of all SNVs but ∼33% of nov-SNVs (Table 1) represented true somatic mutations (i.e., a ∼150-fold enrichment). Of note, ∼11% of apparently true somatic mutations were removed by filtering against our databases of germline variation. These could either represent false-negative variant calls within normal tissue or true recurrence of a somatic mutation in the same position as found in the germline database. The third tumor, LuCaP 147, clearly contained a high number of somatic mutations and represents a tumor class we term “hypermutated” (discussed below).
Table 1.
Efficiency of germline filtering in identifying somatic mutations
| Sample ID | No. coding variants | No. xenograft nov-SNVs | No. true somatic mutations | No. true somatic mutations observed within set of xenograft nov-SNVs |
| LuCaP 92 | 17,092 | 193 | 56 | 51 |
| LuCaP 145.2* | 18,455 | 281 | 122 | 106 |
| LuCaP 147* | 22,458 | 2,122 | 2,045 | 1,823 |
We sequenced the exomes of normal and metastatic cancer tissue corresponding to three xenografts (LuCaP 92, LuCap 145.2, and LuCaP 147), and, for this analysis, considered only those positions called at high confidence across all three tissues. The first two columns represent the number of coding variants and nov-SNVs (variants observed in xenograft exome that remained after filtering) occurring at coordinates that could be confidently base-called in all three samples. The next two columns describe the number of true somatic mutations (defined by comparison of the exomes of normal and metastatic cancer tissue) within the set of all variants and the set of nov-SNVs. For example, filtering reduced the number of variants in LuCaP 92 from 17,092 to 193 while preserving 51 of 56 somatic mutations (sensitivity of 91%).
*Original tumor sample could not be identified, so a neighboring metastasis was used.
Recurrent Nonsynonymous Genomic Sequence Alterations in Prostate Cancers.
We examined the set of novel nonsynonymous single-nucleotide variants (nov-nsSNVs) to identify those genes that may be recurrently affected by protein-altering point mutations across different tumors. To reduce spurious findings attributable to inconsequential passenger mutations, we excluded the 3 hypermutated tumors from this analysis. We also manually examined read pileups for variants in genes with potential recurrence attributable to base-calling artifacts caused by either insertions/deletions or poorly mapping reads. Across 16 tumors from unrelated individuals, 131 genes had nov-nsSNVs in two or more exomes and 23 genes had nov-nsSNVs in three or more exomes (SI Appendix, Table S6).
A subset of the novel variants is likely attributable to instances where very rare germline variants (i.e., not seen in several thousand other chromosomes) occur in the same gene, because we cannot distinguish these from somatic mutations. We therefore excluded from consideration the 1% of genes with the highest rate of very rare germline variants (i.e., singletons), based on an analysis of control exomes (because some genes are much more likely to contain very rare germline variants than other genes) (15, 16). This reduced the number of candidates to 104 genes with nov-nsSNVs in 2 or more exomes and 12 genes with nov-nsSNVs in 3 or more exomes. To segregate candidate genes further, with the goal of identifying those with recurrent somatic mutations, we estimated the probability of recurrently observing germline nov-nsSNVs in each candidate gene by iterative sampling from 1,865 other exomes sequenced at the University of Washington. We excluded from consideration genes for which the probability of observing the genes recurrently mutated attributable to germline variation was greater than 0.001. This reduced the number of candidates to 20 genes with nov-nsSNVs in 2 or more exomes and 10 genes with nov-nsSNVs in 3 or more exomes (Table 2). Notably, whereas we began with 4 genes with nov-nsSNVs in 4 or more exomes (MUC16, SYNE1, UBR4, and TP53), only 1 of these (TP53) remained in our final candidate list, where it is the most significant (Table 2).
Table 2.
Genes with recurrent novel nonsynonymous alterations
| No. samples seen out of 16 | Gene ID | Gene name | Estimated P value of being germline | Individual mutations seen in specific LuCaP samples |
| 5 | TP53 | Tumor protein p53 (Li-Fraumeni syndrome) | <0.00005 | 73(ARG306GLN), 136(ARG280stop), 23.1AI(CYS238TYR), 92(GLU198stop)*, 73(ARG175CYS), 70(TYR163HIS), 77(PRO278SER) |
| 3 | SDF4 | Stromal cell-derived factor 4 | <0.00005 | 105(ASP276ASN), 78(GLY76SER), 115(ALA9SER) |
| 3 | PDZRN3 | PDZ domain containing RING finger 3 | <0.00005 | 23.1AI(ARG727CYS), 105(GLY570SER), 73(ARG463CYS), 92(ILE331LEU) |
| 3 | DLK2 | Delta-like 2 homolog | 0.00005 | 70(ARG371HIS), 145.2(SER361ARG)†, 96AI(HIS280GLN) |
| 3 | FSIP2 | Fibrous sheath interacting protein 2 | 0.00005 | 81(LYS22ASN), 92(THR698ILE)*, 136(GLN1526HIS) |
| 3 | NRCAM | Neuronal cell adhesion molecule | 0.00015 | 115(MET1094ILE), 86.2(LYS645GLU), 145.2(SER329CYS)* |
| 3 | MGAT4B | Mannosyl glycosyltransferase | 0.0002 | 105(ALA504THR), 23.1AI(ARG168CYS), 136(VAL150MET) |
| 3 | PCDH11X | Protocadherin 11 X-linked | 0.0003 | 145.2(VAL38PHE)†, 58(MET867VAL), 105(VAL1007ILE), 49(THR1296ASN) |
| 3 | GLI1 | Glioma-associated oncogene 1 | 0.0003 | 86.2(ARG20TRP), 78(ARG81GLN), 96AI(PRO210THR) |
| 3 | KDM4B | Lysine-specific demethylase 4B | 0.00035 | 73(ALA265VAL), 105(ARG534TRP), 35V(ALA555VAL), 73(ALA827VAL), 86.2(SER1036CYS) |
| 2 | DKK1 | dickkopf homolog 1 (Xenopus laevis) | <0.00005 | 92(GLU151GLN)*, 93(SER244TYR) |
| 2 | RAB32 | Member RAS oncogene family | 0.00005 | 93(VAL66ILE), 141(SER109stop) |
| 2 | PLA2G16 | Phospholipase A2, group XVI | 0.00015 | 115(SER85LEU), 35V(PRO19HIS) |
| 2 | TFG | TRK-fused gene | 0.00015 | 23.1AI(ASN134HIS), 141(GLN318stop), 147(TYR319stop) |
| 2 | TBX20 | T-box 20 | 0.0002 | 77(ARG437HIS), 96AI(ALA52SER) |
| 2 | ZNF473 | Zinc finger protein 473 | 0.00025 | 105(VAL465ILE), 115(GLY652ARG) |
| 2 | SF3A1 | Splicing factor 3a, subunit 1 | 0.0006 | 70(PRO558LEU), 96AI(VAL479ILE) |
| 2 | NMI | N-myc (and STAT) interactor | 0.00075 | 141(ILE302ARG), 86.2(GLN101ARG) |
| 2 | IKZF4 | IKAROS family zinc finger 4 (Eos) | 0.0008 | 93(ASP106ASN), 81(ASP498ASN) |
| 2 | BDH1 | 3-Hydroxybutyrate dehydrogenase | 0.00095 | 73(VAL190ILE), 23.1AI(THR176MET), 147(VAL142ILE), 115(HIS74TYR), 147(ALA50VAL) |
This analysis excludes LuCaP 73, LuCaP 147, and LuCaP 58 as well as the castration resistant lines LuCaP 35V, LuCaP 96AI, and LuCaP 23.1AI. P values were estimated by randomly sampling from 1,865 other exomes sequenced at the University of Washington to estimate the probability of recurrently observing nov-nsSNVs in a given candidate gene. These are the 20 genes with the best estimated P values; a full list of 131 candidates is provided in SI Appendix, Table S6.
*This nov-nsSNV was determined to be a somatic mutation within this xenograft.
†This nov-nsSNV was determined to be a rare germline mutation within this xenograft.
To estimate the “background” rate for calling genes as recurrently mutated via this approach, we analyzed 16 germline exomes from normal individuals that were captured using equivalent methods and applied the same filters. With the caveat that the overall number of coding alterations was lower in this set (an average of ∼250 instead of ∼350 novel variants per individual tumor), we identified 58 genes with nov-nsSNVs in 2 or more exomes with no P value cutoff. Using the same threshold criteria (i.e., removing the top 1% of genes with the highest rate of germline variants and a P value threshold of 0.001) reduced the number of genes with nov-nsSNVs in 2 or more exomes to 4 genes.
To segregate candidate genes further, we annotated positions with their conservation as measured with the Genomic Evolutionary Rate Profiling (GERP) score; variants at highly conserved positions would be predicted to be functionally significant (17). This allowed us to identify a subset of “best candidates” that includes several previously determined to be mutated in advanced prostate cancer (e.g., TP53) and others with described roles in tumorigenesis but not previously implicated in prostate cancer, including DLK2 and SDF4 (Discussion and Table 2). Determining which of these genes may be true driver mutations in prostate cancer will require the interrogation of larger cohorts, as well as functional characterization.
Mutations Associated with CR Prostate Cancer.
Castration, or androgen deprivation therapy, is a commonly used treatment for advanced disseminated prostate cancer. Although effective initially, resistance inevitably develops, leading to a disease state called castration-resistant prostate cancer (CRPC) with high rates of cancer-specific mortality (2, 13). Our study included three tumors with CS and CR derivatives: LuCaP 96/LuCaP 96AI, LuCaP 23.1/LuCaP 23.1AI, and LuCaP 35/LuCaP 35V (13) (SI Appendix, Fig. S4). A comparison of exomes from each CR xenograft with those of its CS counterpart identified ∼12–50 genes with nonsynonymous mutations that were present uniquely in the CR xenografts (SI Appendix, Table S7). There were no genes recurrently mutated exclusively in CR tumors. To look for enrichment of mutations in genes encoding proteins comprising specific biochemical pathways in CRPC, we examined 880 gene sets using the MSigDB pathways database (http://www.broadinstitute.org/gsea/msigdb/). We found a significant enrichment for genes participating in Wnt signaling in CR tumors: of 86 mutations unique to CRPCs, each tumor had at least 1 mutation in a member of the Wnt pathway (q < 0.01) (18). These included FZD6 (in LuCaP 23.1AI), GSK3B (in LuCaP 96AI), and WNT6 (in LuCaP 35V) (SI Appendix, Table S7).
Prostate Cancers with Hypermutated Genomes.
The genomes of three prostate cancers, LuCaP 58, LuCaP 73, and LuCaP 147, possessed a strikingly high number of nov-nsSNVs, nearly 10-fold more than other tumors (P = 0.0097) (Fig. 1). There were no distinctive features to suggest why these tumors should have more variants. Each tumor originated as a high-grade Gleason 9 cancer; all were from individuals of Caucasian ancestry; and one represented a primary neoplasm, one a lymph node metastasis, and one a metastasis to the liver. The hypermutated phenotype also does not appear to be solely determined by the length of time a tumor was passaged in animals, because LuCaP 147 was started nearly 10 y after most other xenografts in this panel. Further, tumors with hypermutated genomes did not exhibit substantially different patterns of structural changes compared with nonhypermutated tumors. As ascertained by array comparative genomic hybridization (array CGH), LuCaP 58, LuCaP 73, and LuCaP 147 had 1,582, 1,577, and 1,295 copy number variation (CNV) calls, respectively, compared with 1,470, 1,769, and 2,129 CNVs in nonhypermutated LuCaP 70, LuCaP 92, and LuCaP 145.2 tumors (SI Appendix, Table S8).
We hypothesized that the large number of nov-SNVs observed in three prostate cancers may be attributable to a “mutator phenotype” that either developed during the initial stages of tumorigenesis as a consequence of therapeutic pressures and subsequent clonal selection or evolved while being passaged in the mouse hosts. To determine if these results reflect truly elevated numbers of somatic mutations within human tumors and are not a result of passage within mice, we sequenced the exomes of paired normal and directly resected nonxenografted tumor samples corresponding to one hypermutated xenograft line (LuCaP 147) and two nonhypermutated xenograft lines (LuCaP 92 and LuCaP 145.2) (SI Appendix, Table S9). Of 2,122 nov-SNVs in LuCaP 147 able to be called across all three samples (xenograft, derivative tumor, and normal tissue) 1,464 were somatic and present in metastasis tissue (Tables 1 and 3). In contrast, the other two nonxenografted tumors (corresponding to LuCaP 92 and LuCaP 145.2) had 31 and 57 somatic mutations, respectively. Furthermore, because we sequenced a neighboring metastasis rather than the exact metastasis from which LuCaP 147 was derived, this result indicates that at least these ∼1,400 somatic mutations were shared between these two metastases. The vast majority of the ∼600 somatic mutations observed in the LuCaP 147 xenograft but not observed in the metastasis likely occurred during passage within mice, or else were mutations specific to the metastasis from which LuCaP 147 was derived. The pattern of somatic mutations within the metastasis corresponding to hypermutated LuCaP 147 appears to be heavily dominated by transition mutations, with G→A and C→T transitions accounting for greater than 70% of mutations observed (SI Appendix, Fig. S5).
Table 3.
Hypermutation phenotype arose before xenografting within LuCaP 147
| Sample ID | No. somatic mutations unique to metastasis | No. somatic mutations shared by metastasis and xenograft | No. somatic mutations unique to xenograft |
| LuCaP 92 | 8 | 31 | 25 |
| LuCaP 145.2* | 35 | 57 | 65 |
| LuCaP 147* | 91 | 1,464 | 581 |
After sequencing metastases and normal tissue corresponding to three xenografts, we calculated the number of somatic mutations shared by xenografts and a corresponding metastasis. In this table, somatic mutations are classified according to their presence in the metastasis and xenograft (in metastasis only, in both metastasis and xenograft, and in xenograft only). A total of 1,464 of ∼2,045 nov-SNVs within LuCaP 147 were also present within a different lung metastasis from the same individual. However, in all xenografts, a substantial number of mutations (25 within LuCaP 92 and 65 within LuCaP 145.2) appear to have developed after xenografting.
*Original tumor sample could not be identified, so a neighboring metastasis was used. These numbers therefore represent the minimal overlap between a xenograft and the metastasis from which it was derived.
Discussion
In this study, we performed a genome-wide analysis of protein-coding variation to identify sequence alterations in highly aggressive lethal prostate cancers. Despite having only limited access to matched normal tissue for comparisons, we were able to exploit increasingly deep catalogs of segregating germline variation to highlight genes that may be recurrently mutated in prostate cancer. This strategy may be highly relevant for the genomic analysis of carcinomas or tumor-derived cell lines for which corresponding benign tissue is not available.
Overall, we identified 131 genes that had nov-nsSNVs in two or more tumors. Additional analysis based on the likelihood of observing rare germline variation highlighted 20 genes as candidates for recurrent somatic alteration, with the known cancer gene TP53 emerging as the top candidate from the analysis. We acknowledge that the genetic alterations observed in xenograft lines may not reflect changes originally present in a tumor or may be a result of previously unseen germline variation, and it will be important to validate these candidates by establishing their prevalence in larger numbers of tumors for which matched normal tissue is available. However, these data provide an intriguing set of candidates for follow-up analysis. Several of these are discussed in further detail below.
We identified nov-nsSNVs in TP53 in 5 of the 16 independent tumors used to evaluate recurrence as well as in 1 of the hypermutated tumors. These variants included two positions that were called as homozygous (likely attributable to loss of heterozygosity) and are predicted to cause premature termination of the protein (Table 2). Hypermutated LuCaP 73 possessed two nov-nsSNVs in TP53 after filtering, including one in a mutational hotspot (175 ARG→CYS). LuCaP 77 possessed a homozygous nov-nsSNV (278 PRO→SER) that is also present in Single Nucleotide Polymorphism Database (dbSNP 131). This SNV was previously described in a case of familial cancer syndrome (Li-Fraumeni syndrome) and would have been removed from the analysis if we had filtered against dbSNP entries (19). Three tumors harbored nov-nsSNVs within the gene encoding DLK2, a protein that shares similarity with the Delta transcription factor and has recently been shown to be involved in NOTCH1 signaling during development (20). Two DLK2 nov-nsSNVs are in close proximity (at positions 361 and 371) in what is predicted to be a cytoplasmic domain and are in residues that are highly conserved evolutionarily (GERP score above 4.5). Three tumor genomes encoded variants in stromal-derived factor (SDF4), a 363-aa calcium-binding protein whose function is poorly understood (21). Two of the residues affected by nov-nsSNVs are highly conserved evolutionarily, with a GERP score above 4. Recent work has correlated low levels of SDF4 expression with a poor prognosis in metastatic breast cancer (22).
Recently, whole-genome sequencing of localized primary prostate cancers identified 165 genes that harbored somatic nonsynonymous mutations (1). Of these, PCDH15, LAMC1, and GPC6 also had nov-nsSNVs in two or more advanced prostate cancers characterized in the present study. Both PCDH15 and LAMC1 are large (>1,500 aa) and complex extracellular proteins that have a higher prior probability for somatic mutation or rare germline variants. GPC6 encodes a smaller protein (∼350 aa) and contains nov-nsSNVs at positions that are highly conserved (GERP score above 5) in 2 of 16 nonhypermutated tumors as well as in 1 hypermutated tumor. GPC6 encodes a glypican, a class of cell surface coreceptors for proteases implicated in cell growth and division (23–25).
Unexpectedly, we identified three tumors (representing 15% of those analyzed) with very high numbers of nov-SNVs. We confirmed that this hypermutator phenotype arose before passage in mice for at least one of these tumors (LuCaP 147), for which a nonxenografted tumor was available for comparison. These mutation frequencies far exceed those found in primary prostate cancers, as well as in most neoplasms arising in the breast, pancreas, and brain, where comprehensive exome or genome sequencing studies have been performed (26–28). However, cancers in the colon with mismatch repair gene defects (29) and those that arise in the lung and skin, where environmental genotoxins like tobacco or UV sun exposure are implicated in disease etiology, have numbers of mutations that approach those present in these hypermutated prostate cancers (30, 31). The pattern of mutation observed in whole-genome data argues against tobacco exposure within the metastasis corresponding to LuCaP 147, because the characteristic predominance of G→T transversion mutations caused by polycyclic aromatic hydrocarbons was lacking (30). Several nov-nsSNVs in the hypermutated tumors affect genes previously implicated in prostate cancer. For example, nov-nsSNVs in AR were observed in two of the hypermutated tumors, LuCaP 147 and LuCaP 73, including one well-characterized gain-of-function mutation (877 THR→ALA) (32). However, the very large number of nov-nsSNVs within these tumors renders it difficult to distinguish disease-relevant mutations from likely passenger events.
One potential explanation for the large number of mutations seen in these samples is acquisition of a mutator phenotype, in which alterations in DNA polymerase or DNA repair genes result in an accelerated rate of mutations (33, 34). In support of this, LuCaP 58 possessed three candidate mutations in MSH6, a gene known to promote mismatch repair and microsatellite stability, including a particular substitution, 1284 THR→MET, observed in individuals with Lynch syndrome (35). This gene was previously seen to be mutated in prostate cancer, where it was associated with an increase in overall mutation rate, although with a more limited assessment of genomic sequence (1.3 Mb) (4). Tumors with microsatellite instability are known to possess more mutations than other cancers; a recent analysis of colorectal cancer genomes detected approximately eightfold more nonsynonymous variation in a tumor that displayed microsatellite instability, consistent with the number of mutations seen here (29). We did not find nov-nsSNVs within DNA mismatch repair genes within the other two hypermutated prostate tumors (LuCaP 73 and LuCap 147); thus, a plausible explanation for the elevated mutation frequencies in these cancers remains to be established.
One limitation of this study is the use of tumor xenografts that may not precisely reflect the status of the tumor genome sampled directly from the patient. For those xenografts for which a corresponding nonxenograft tumor was available, the xenograft harbored approximately twofold more mutations (Table 3). This finding likely reflects continued tumor evolution and genotoxic stress over numerous population doublings or further selective pressure to adapt to a murine host. However, these xenografts are able to recapitulate many aspects of prostate cancer in vivo (36, 37). Thus, defining the genetic landscapes of these tumors allows one to use the xenografts as a means to test the consequences of mutation functionally and to evaluate therapeutics directed against pathways that are disrupted by specific genetic lesions.
In summary, by sequencing the exomes of 23 tumors representing a spectrum of aggressive advanced prostate cancers, we identified a large number of previously unrecognized gene coding variants with the potential to influence tumor behavior. Our results also indicate that, with notable exceptions, very few genes are mutated in a substantial fraction of tumors. Furthermore, although the overall mutation frequencies approximate those found in other cancers of epithelial origin, we also identified a distinct subset of tumors that exhibit a hypermutated genome. It will be important to determine the mechanism(s) responsible for the enhanced point mutation rates in these malignancies, particularly if further studies demonstrate enhanced resistance to cancer therapeutics.
Materials and Methods
Xenograft Tissues.
The LuCaP series of prostate cancer xenografts used in this study was obtained from the University of Washington Prostate Cancer Biorepository and developed by one of the authors (R.L.V.) within the Department of Urology (38). DNA was isolated from frozen tissue blocks using the QIAGEN DNeasy Blood and Tissue kit.
Exome Capture and Massively Parallel Sequencing.
The Nimblegen EZ SeqCap kit (Roche) was used as previously described to capture exons (39). Shotgun libraries were constructed by shearing DNA and ligating sequencing adaptors. Libraries were hybridized to either the EZSeqCap V1 or V2 solution-based probe, amplified, and sequenced on either the Illumina GAIIx or HiSeq platform (SI Appendix, Table S2). V1 probes (used in 8 samples) targeted 26.6 Mb corresponding to the CCDS definitions of exons, whereas V2 probes (used in 15 samples) targeted 36.6 Mb corresponding to the RefSeq gene database.
Read Mapping and Base Calling.
We dealt with the possibility of mouse gDNA contamination by mapping sequence reads to both the human (hg18) and mouse (mm9) genome sequences using a Burrows–Wheeler transform (40). Reads that mapped to the mouse genome were excluded from further analysis. Mapping statistics and calculations of mapping complexity are shown in SI Appendix, Figs. S1–S3. Sequence variant calls were performed by SAMtools (41) after removing potential PCR duplicates and were filtered to consider only positions with more than eightfold coverage and a Phred-like consensus quality of at least 30 (9).
Identification of Genes with Sequence Variation.
To eliminate common germline polymorphisms from consideration, variants that had the same position as variants present in pilot data from the 1,000 Genomes Project or in ∼2,000 exomes corresponding to normal (nontumor, nonxenografted) tissues sequenced at the University of Washington were removed from consideration (Fig. 1). Genotypes were annotated using the SeattleSeq server (http://gvs.gs.washington.edu/SeattleSeqAnnotation/), and only nonsynonymous variants (missense/nonsense/splice-site mutations) were considered in identifying genes with recurrent mutations. The subset of genes that were recurrently mutated was validated manually using the Integrated Genomics Viewer (IGV) to identify and remove false-positive calls attributable to the presence of an insertion/deletion or incorrect mapping read (12). To estimate the significance of the three hypermutator xenografts, the numbers of nov-SNVs present in LuCaPs 58, 73, and 147 were compared against other xenografts using a one-sided t test assuming unequal variance.
Estimation of Significance in Genes with Recurrent Nov-nsSNVs.
To distinguish genes that are observed to be recurrently mutated as a result of sampling germline variation among individuals from genes that are recurrently mutated as a result of somatic mutation in the tumor, we used exome sequence data from 1,865 individuals sequenced at the University of Washington.
To identify genes with the highest rate of very rare germline variants (i.e., singletons), we tabulated the genes that were affected by rare variants (nov-nsSNVs, defined as protein-altering mutations seen uniquely in this individual relative to all other exomes in the set of 1,865) for each individual. We estimated the likelihood of seeing a rare protein-altering mutation in an individual by dividing the number of individuals with nov-nsSNVs in a given gene by the total number of individuals sampled.
We also used exome data on these 1,865 individuals to estimate the likelihood of observing recurrence in a gene as a result of germline polymorphism. Sixteen individuals were randomly selected in each iteration, and for each of these 16 exomes, we identified genes that were affected by nov-nsSNVs. We then looked for genes that recurrently contained nov-nsSNVs within the set of 16 individuals and repeated this process 20,000 times to generate an estimate for the probability that a given gene would be observed to contain recurrent nov-nsSNVs attributable to previously unobserved germline polymorphisms.
Assessments of Filtering Approaches.
To test the effectiveness of our method of filtering germline variants, we sequenced normal and tumor tissue corresponding to each of three xenografts. Sequence data were processed through the same mapping pipeline [mapping to the mouse and human (hg18) reference, variant calling using SAMtools] as was used for xenograft exome data. Positions called as a high-quality variant (position has 24-fold coverage and a Phred-like consensus probability of at least 50) in the xenograft line were queried within both nonxenografted metastasis and normal tissues. To increase accuracy, only those positions that also had at least 24-fold coverage in both the metastasis and normal tissue were considered for this analysis. A position was considered to be a “true” somatic mutation (i.e., arising before xenografting) if it was called as a variant within the xenograft tumor and metastasis but not within the normal tissue. To account for the possibility of low coverage resulting in a miscall within normal tumor tissue, we used less stringent criteria to determine if a position was variant within normal tissue (at least 10% or 10 reads covering this position support this call). A position was considered to be a somatic mutation that arose after xenografting if it was called as variant in the xenograft and invariant within metastasis and normal tissue. If a position was variant within the xenograft as well as within its corresponding metastasis and normal tissue, it was considered to be a germline polymorphism. This process was repeated only considering those positions previously determined to be nov-SNVs to estimate the sensitivity of the germline filtering approach.
Estimation of the Background Rate for Calling Genes as Recurrently Mutated.
To estimate the rate of calling genes as recurrently mutated as a result of rare germline variation, we used exome sequence data from 16 normal individuals that had been both captured and sequenced at the University of Washington in a similar manner to tumors in this study, although they had been sequenced to a modestly lower depth. Sequence data were processed through the same variant calling and filtering pipeline [mapping to the mouse and human (hg18) reference variant calling using SAMtools and manual validation using the IGV] as was used for xenograft exome data.
Genome Copy Number Analysis.
CNV analysis was carried out using Illumina Infinium 660W-Quad Beadchips following manufacturer's standard protocols. Genotyping calls were generated for six samples (3 hypermutated and 3 randomly chosen other xenografts) using the Illumina BeadStudio software with Illumina Human660W-Quad_v1_A.egt HapMap genotype cluster definitions. Data analysis was performed with Biodiscovery Nexus Copy Number 6.0 software. The SNP-FASST2 segmentation algorithm and default Illumina settings for significance, number of probes per segment, and gain and loss thresholds were used to identify regions of CNV for each sample. Statistical analysis was done using a two-sided t test assuming unequal variance.
Identification of CR-Specific Mutations.
To identify genes potentially involved in the development of CR, we compared the sequences of CR lines with their corresponding CS lines (SI Appendix, Fig. S4). Variants were called as mentioned in “Read mapping and base calling,” except positions were only considered if both CR and CS sequences had an eightfold coverage and base quality of 30 as determined by SAMtools. Resulting genotypes were annotated using the SeattleSeq server, and only nonsynonymous variants (missense/nonsense/splice-site mutations) were considered. This subset of genes was then validated manually using the IGV to ensure that variant alleles were not present in CS xenografts. We entered these genes into the MSigDB Web site (http://www.broadinstitute.org/gsea/msigdb/) using the “Investigate gene sets” option. We looked for overlap with “KEGG gene sets” and report the q-value from the Web site (18).
Supplementary Material
Acknowledgments
We deeply appreciate the participation of patients and their families in these studies. We thank and recognize the following ongoing studies that produced and provided exome variant calls for comparison: National Heart, Lung, and Blood Institute Lung Cohort Sequencing Project (Grant HL 1029230); National Heart, Lung, and Blood Institute Women's Health Initiative Sequencing Project (Grant HL 102924); National Institute on Environmental Health Sciences SNPs (Grant HHSN273200800010C); National Heart, Lung, and Blood Institute/National Human Genome Research Institute SeattleSeq (Grant HL 094976); and Northwest Genomics Center (Grant HL 102926). We also thank J. Hiatt, C. Igartua, J. Kitzman, N. Krumm, M. Mynsberge, B. O'Roak, J. Schwartz, I. Stanaway, and E. Turner for thoughtful discussions. This work was supported by the US Department of Defense (Grant W81XWH-10-1-0589), National Institutes of Health (Grant PO1 CA085859), Richard M. Lucas Foundation, Prostate Cancer Foundation, and Pacific Northwest Prostate Cancer Specialized Programs of Research Excellence (Grant P50 CA097186). A.K. is supported by an Achievement Rewards for College Scientists Fellowship. J.S. is the Lowell Milken Prostate Cancer Foundation Young Investigator.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRA037395). Additional accession numbers are provided in SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108745108/-/DCSupplemental.
References
- 1.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holcomb IN, et al. Genomic alterations indicate tumor origin and varied metastatic potential of disseminated cells from prostate cancer patients. Cancer Res. 2008;68:5599–5608. doi: 10.1158/0008-5472.CAN-08-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robbins CM, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helgeson BE, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 8.Holcomb IN, et al. Comparative analyses of chromosome alterations in soft-tissue metastases within and across patients with castration-resistant prostate cancer. Cancer Res. 2009;69:7793–7802. doi: 10.1158/0008-5472.CAN-08-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Weerden WM, Bangma C, de Wit R. Human xenograft models as useful tools to assess the potential of novel therapeutics in prostate cancer. Br J Cancer. 2009;100:13–18. doi: 10.1038/sj.bjc.6604822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark MJ, et al. U87MG decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin RM, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corey E, et al. LuCaP 35: A new model of prostate cancer progression to androgen independence. Prostate. 2003;55:239–246. doi: 10.1002/pros.10198. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 16.Lohmueller KE, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper GM, et al. NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speiser P, et al. A constitutional de novo mutation in exon 8 of the p53 gene in a patient with multiple primary malignancies. Br J Cancer. 1996;74:269–273. doi: 10.1038/bjc.1996.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez-Solana B, et al. The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulates each other's activities. Biochim Biophys Acta. 2011;1813:1153–1164. doi: 10.1016/j.bbamcr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Scherer PE, et al. Cab45, a novel (Ca2+)-binding protein localized to the Golgi lumen. J Cell Biol. 1996;133:257–268. doi: 10.1083/jcb.133.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang H, Escudero-Esparza A, Douglas-Jones A, Mansel RE, Jiang WG. Transcript analyses of stromal cell derived factors (SDFs): SDF-2, SDF-4 and SDF-5 reveal a different pattern of expression and prognostic association in human breast cancer. Int J Oncol. 2009;35:205–211. doi: 10.3892/ijo_00000330. [DOI] [PubMed] [Google Scholar]
- 23.Filmus J. Glypicans in growth control and cancer. Glycobiology. 2001;11:19R–23R. doi: 10.1093/glycob/11.3.19r. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto K, et al. Common variation in GPC5 is associated with acquired nephrotic syndrome. Nat Genet. 2011;43:459–463. doi: 10.1038/ng.792. [DOI] [PubMed] [Google Scholar]
- 25.Williamson D, et al. Role for amplification and expression of glypican-5 in rhabdomyosarcoma. Cancer Res. 2007;67:57–65. doi: 10.1158/0008-5472.CAN-06-1650. [DOI] [PubMed] [Google Scholar]
- 26.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 27.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timmermann B, et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS ONE. 2010;5:e15661. doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun C, et al. Androgen receptor mutation (T877A) promotes prostate cancer cell growth and cell survival. Oncogene. 2006;25:3905–3913. doi: 10.1038/sj.onc.1209424. [DOI] [PubMed] [Google Scholar]
- 33.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: Clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. discussion 3557. [DOI] [PubMed] [Google Scholar]
- 34.Loeb LA. Human cancers express mutator phenotypes: Origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan SY, et al. Three novel missense germline mutations in different exons of MSH6 gene in Chinese hereditary non-polyposis colorectal cancer families. World J Gastroenterol. 2007;13:5021–5024. doi: 10.3748/wjg.v13.i37.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corey E, Quinn JE, Vessella RL. A novel method of generating prostate cancer metastases from orthotopic implants. Prostate. 2003;56:110–114. doi: 10.1002/pros.10235. [DOI] [PubMed] [Google Scholar]
- 37.van Weerden WM, Bangma C, de Wit R. Human xenograft models as useful tools to assess the potential of novel therapeutics in prostate cancer. Br J Cancer. 2009;100:13–18. doi: 10.1038/sj.bjc.6604822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corey EV, Vessella RL. Prostate cancer: biology, genetics, and the new therapeutics. In: Chung LWK, Isaacs WB, Simons JW, editors. Contemporary Cancer Research. 2nd Ed. Totowa, NJ: Humana; 2007. pp. 3–32. [Google Scholar]
- 39.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.