Abstract
Recurrent copy number variations (CNVs) of human 16p11.2 have been associated with a variety of developmental/neurocognitive syndromes. In particular, deletion of 16p11.2 is found in patients with autism, developmental delay, and obesity. Patients with deletions or duplications have a wide range of clinical features, and siblings carrying the same deletion often have diverse symptoms. To study the consequence of 16p11.2 CNVs in a systematic manner, we used chromosome engineering to generate mice harboring deletion of the chromosomal region corresponding to 16p11.2, as well as mice harboring the reciprocal duplication. These 16p11.2 CNV models have dosage-dependent changes in gene expression, viability, brain architecture, and behavior. For each phenotype, the consequence of the deletion is more severe than that of the duplication. Of particular note is that half of the 16p11.2 deletion mice die postnatally; those that survive to adulthood are healthy and fertile, but have alterations in the hypothalamus and exhibit a “behavior trap” phenotype—a specific behavior characteristic of rodents with lateral hypothalamic and nigrostriatal lesions. These findings indicate that 16p11.2 CNVs cause brain and behavioral anomalies, providing insight into human neurodevelopmental disorders.
Keywords: Home-cage, stereotypic behavior, structural variation, brain MRI
Accumulating evidence suggests the importance of copy number variations (CNVs) in the etiology of neuropsychiatric disorders, including autism (1), schizophrenia (2–4), developmental delay (5), and other complex traits (6). The 16p11.2 region is particularly intriguing. Whereas deletion of 16p11.2 has been associated with autism (7–9), duplication of 16p11.2 has been associated with autism (9, 10) as well as schizophrenia (11). 16p11.2 CNVs have also been reported in patients with developmental delay, mental retardation, repetitive behaviors (12–16), and a highly penetrant form of obesity (17). A reciprocal effect of 16p11.2 dosage on head size has been noted, as deletions are associated with large head size or macrocephaly, whereas duplications are associated with microcephaly (16). These studies reveal the variability of symptoms in patients carrying the same 16p11.2 CNV, an extreme example being a family with three affected members with symptoms so heterogeneous that they were barely overlapping (18).
Mouse models allow direct assessment of CNVs while reducing variability caused by genetic and environmental factors. We and others have previously used chromosome engineering (19) to model genetic alterations found in complex human diseases including cancer (20) and genomic disorders (21–24), allowing identification of the causative gene and elucidation of the mechanism involved (20, 25–27). Here we used a similar approach to generate mouse models with deletion and duplication corresponding to those found in patients with 16p11.2 CNVs. Because of the evidence for clinical heterogeneity, we screened these models for multiple changes in brain anatomy and behavior by using a combination of high-resolution MRI (28) and a monitoring system that assesses multiple behaviors (29). We found that the deletion and the duplication affect behavior and brain anatomy in opposing ways, with deletion mice exhibiting behaviors that resemble sensorimotor deficits in rats with lateral hypothalamic and nigrostriatal lesions (30, 31). These findings provide evidence that brain anatomy and behavior depend on dosage of the region corresponding to 16p11.2.
Results
Generation of Mouse Models for Human 16p11.2 CNVs.
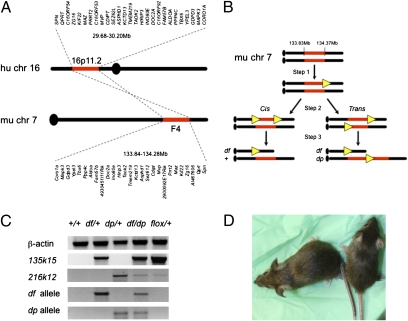
We asked whether altered dosage of the region corresponding to 16p11.2 causes abnormalities in mice. Genes mapping to the 0.52-Mb 16p11.2 CNV in humans cluster within a 0.44-Mb region of mouse chromosome 7 (Fig. 1A). Using chromosome engineering (19) as we have previously (20, 27, 32), we generated mice with one copy [heterozygous for a deletion or deficiency (df) allele], as well as mice with three copies [heterozygous for a duplication (dp) allele] of the region corresponding to 16p11.2 (Fig. 1B and Fig. S1). Endpoints for the rearrangement were selected based on human data (1), with each gene in the interval being conserved in mouse (Dataset S1). Gene targeting constructs were generated using MICER (33), and sequential targeting in mouse ES cells resulted in integration of loxP sites and selection cassettes at each endpoint (Fig. 1B and Fig. S1). Cre-mediated recombination and drug selection within eight independent doubly targeted clones revealed that three clones had been targeted in cis and five clones had been targeted in trans, which generated df/+ and df/dp ES cells, respectively (Fig. 1B and Figs. S1 and S2). Five independent df/dp clones were used for blastocyst injection, producing 40 different male chimeras that were crossed to +/+ females. Ten of these chimeras (representing two independent ES cell clones) produced df/+ and dp/+ mice that were identified by PCR (Fig. 1C). This approach provides mouse models for directly assessing the consequences of both the 16p11.2 CNV losses (i.e., deletion) and gains (i.e., duplication) found in humans.
Fig. 1.
Generation of 16p11.2 models. (A) Genes mapping to human 16p11.2 CNVs are conserved in mouse. (B) Schematic of the chromosome engineering strategy used to generate mouse models of 16p11.2 CNVs. Step 1 is gene targeting at the 135k15 locus; step 2 is gene targeting at the 216k12 locus in the 135k15-targeted ES cells; and step 3 is Cre-mediated recombination. Cis and trans indicate that loxP sites (yellow triangles) had integrated on the same or different chromosome homologues, respectively. (C) Molecular validation. PCR products using primers specific for the positive control (β-actin), targeting at the first and second endpoints (135k15 and 216k12 loci, respectively), the df allele, and the dp allele are shown. (D) Before weaning, df/+ mice (Right) tend to be smaller than their +/+ siblings (Left; 8.8 and 15.4 g, respectively, for the females shown). Note the light-colored tail and ears of the df/+, which is a result of the presence of the Agouti transgene. In A and B, chromosome positions are shown in megabases. Further information is provided in Figs. S1 and S2, Dataset S1, and SI Experimental Procedures.
We established both df/+ and dp/+ mice, but at weaning we noticed that df/+ mice were underrepresented and litter sizes were smaller than expected (Table S1). Before weaning, df/+ mice were sometimes small (Fig. 1D), but as adults, they were essentially the same size as their siblings and appeared healthy (SI Experimental Procedures). To determine whether df/+ mice were dying during embryogenesis, we crossed df/+ males to +/+ females, and harvested embryos at day 13.5 of development [i.e., embryonic day (E) 13.5] as well as just before birth (E17.5–E18.5); progeny from similar crosses using the same studs as well as their male siblings were also genotyped at weaning (Table S1). Whereas litter sizes during embryogenesis averaged 9.4 embryos and the ratio of df/+ embryos was Mendelian, litter sizes at weaning averaged only 5.0 mice and the ratio of df/+ mice was half that expected. In addition, litter sizes were normal and df/+ mice were present in expected ratios immediately after birth, whereas dead pups lacking a milk pouch were sometimes found later on. Therefore, some df/+ mice die after birth, indicating that 16p11.2 loss can compromise survival.
Gene Expression in Multiple Brain Regions Corresponds to 16p11.2 Dosage.
To validate the models, we analyzed gene expression profiles in the brain and determined whether expression corresponded with dosage. We measured mRNA intensities in 37 microarray hybridizations representing four brain regions (olfactory bulbs, cortex, cerebellum, and brainstem; five samples were hybridized twice for estimation of technical errors) in two df/+, three +/+, and three dp/+ male mice. All mice were F1 C57BL/6N:129Sv hybrids; therefore, other than the engineered CNV, their genomes were identical. A scatter plot of the gene expression intensity difference between dp/+ and +/+ vs. the difference between df/+ and +/+ indicated that genes within 16p11.2 displayed a large difference between df/+ and +/+ brain, and a much smaller difference between dp/+ and +/+ brain (Fig. S3A). Two-way ANOVA with brain region and dosage as main factors indicated that, of 33 genes in the engineered region, expression of 26 was affected directly by dosage (Dataset S2). Gdpd3, mapping within the engineered region, showed extreme up- and down-regulation; this reflected differences in Gdpd3 expression in C57BL/6N vs. 129Sv strains. Further analysis indicated that expression of genes in the region was significantly altered in each of the four brain regions analyzed, and that expression was affected more by deletion than by duplication (Fig. S3B). These findings indicate that copy number dictates gene expression levels in multiple brain regions, and that loss has the largest effect.
df/+ and dp/+ Mice Have Distinctive Behavioral Phenotypes.
General survey of behavior.
The clinical evidence that patients with 16p11.2 CNVs have highly heterogeneous symptoms suggested that if the corresponding genomic alteration did in fact cause behavioral alterations in mice, the phenotypes might also be highly variable. Therefore, we believed it imperative to monitor the 16p11.2 CNV models for multiple behaviors by using as quantifiable and unbiased approaches as possible. We used HomeCageScan, a system previously used to assess behavioral alterations caused by neurodegenerative disease, neurotoxic agents, and pain (29, 34–36). We investigated behavior of a cohort of 50 male and female mice. The mice were progeny from df/+ × dp/+ crosses, and therefore included +/+ and df/dp diploid controls (n = 15 and n = 9, respectively), df/+ (n = 13), and dp/+ (n = 13). The parents in these crosses came from two chimeras. Thirty-nine of these were later used for MRI (as detailed later). Recording was done in cages that were significantly larger, with a ceiling that was much higher, than a standard mouse cage. The reason for using large cages is that, by minimizing the physical constraints on the animals, a rich spectrum of behaviors evolves (37) and the dynamics of the change in behavior varies between genotypes (38). In this experimental paradigm, the recording cages posed a new environment to the mice being analyzed. In particular, mice had to adapt their climbing abilities to this new environment. In each session, the behavior of four individual mice was recorded simultaneously. Multiple sessions were performed so that the behavior of each of the 50 mice was analyzed. Mice were transferred into the recording cages before the last 2 h of the light period. The recording started immediately and continued for 60 h after the onset of the first dark period (i.e., over three 12-h dark periods and two 12-h light periods). We also tested social behavior and grip strength (Fig. S4). These analyses did not show significant differences and therefore we do not discuss those data.
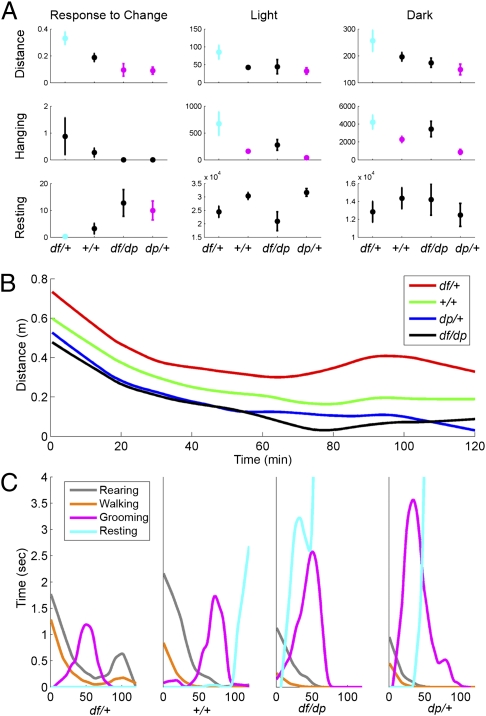
However, six of eight distinct behavioral measures revealed significant genotype differences. The changes were evident immediately after the mice were introduced into the new cages, as well as throughout the entire period of the test (Fig. 2A and Dataset S3). Five of these six differences were particularly interesting, as these behaviors were affected in opposite directions in df/+ and dp/+ mice relative to diploid (+/+ and df/dp) controls. As was the case for gene expression profiles, the effect of the deletion on behavior was more severe than that of the duplication. As 16p11.2 CNV-associated syndromes sometimes have a gender bias (16), we asked whether the changes in behavior were sex-specific. Two-way ANOVA did not reveal significant interactions between 16p11.2 dosage and sex for any of the behavioral measures (Dataset S3). Later we describe in detail the behaviors affected by 16p11.2 dosage.
Fig. 2.
Multiple behaviors depend on 16p11.2 dosage. (A) The relationship between 16p11.2 dosage and distance traveled (Top), hanging (Middle), and resting (Bottom) is shown during the 2-h period immediately after cage transfer (Left), the light periods (Middle), and the dark periods (Right). Averages and standard error of the mean (SEM) are displayed. Statistically significant pair-wise differences relative to the df/+ group (determined by ANOVA followed by Tukey's confidence intervals) are depicted by the following: df/+ is cyan when it differs significantly from at least one other cohort, but is otherwise black; cohorts that differ significantly from df/+ are magenta, but are otherwise black. Dataset S3 shows pair-wise comparisons between genotypes. (B) Median distance traveled vs. time during the first 2 h after cage transfer. (C) Detailed behaviors during the first 2 h after cage transfer. Medians of the cumulative time of four distinct behaviors (rearing, walking, grooming, resting) vs. time elapsed from the beginning of the trial are shown for each genotype. During this period, df/+ mice do not rest; instead, they have a second peak of activity (B and C). Further information is provided in Dataset S3 and SI Experimental Procedures.
Response to change in environment.
The 16p11.2 CNV models responded uniquely to environmental change: within the 2-h period after being transferred to the test cage, the distance traveled, as well as the time spent walking, lingering, and resting, depended on genotype (Pdistance = 0.0016, Pwalking = 0.003, Plingering = 0.021, Presting = 0.025; Dataset S3). Tukey's confidence intervals were calculated to determine pair-wise differences between genotypes while controlling for false discovery rate (FDR) (39). Distance traveled and time spent walking were significantly higher in df/+ relative to other groups, and the time spent lingering was significantly higher in df/+ relative to dp/+ mice. Resting was lower in df/+ and higher in dp/+ mice relative to controls. We further analyzed the time course of these changes during the first 2 h of the test (Fig. 2 B and C). Mice of all four genotypes were most active immediately after being transferred to the test cage, with activity gradually declining during the first hour as the mice habituated to their new environment (Fig. 2B). However, the df/+ cohort had a second burst of activity that did not occur in other groups. The response to being transferred to the test cage occurred in three sequential stages defined by the actions of the mice (Fig. 2C). For each group, the first stage was characterized by elevated walking and rearing, and the second stage was characterized by elevated grooming. The third stage was characterized by resting, which was significantly decreased and increased in df/+ and dp/+, respectively, relative to +/+; indeed, df/+ mice had a burst of walking and rearing, whereas resting was absent during this stage. Thus, the rate of certain behaviors is affected reciprocally by loss and gain of 16p11.2 dosage in response to environmental challenge. In addition, the sequence of these behaviors is disrupted in df/+ mice. Thus, 16p11.2 CNVs affect both the rate and the timing of specific behaviors.
Diurnal deficits.
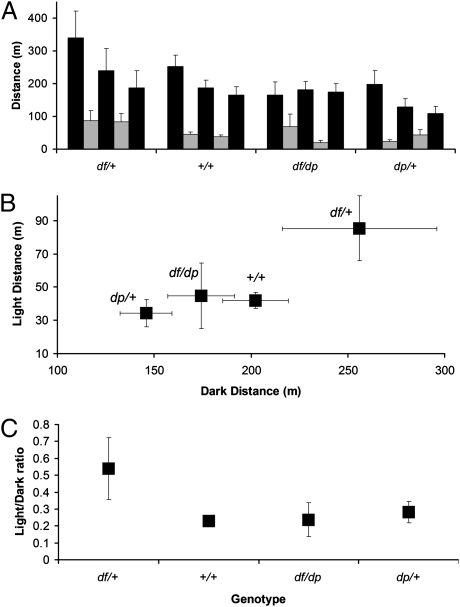
Sleeping disorders are frequently reported in many psychiatric disorders, including autism (40). Because we recorded behavior over sequential dark/light intervals, we could assess the effect of 16p11.2 CNVs on light and dark cycling by using previously established methods (29). Mice are nocturnal, and indeed, each genotype was most active during the dark periods (Fig. 3A). The +/+, df/+, and dp/+ mice were most active during the initial dark period, with activity decreasing in successive dark periods. Although activity of df/+ mice was highest in the first dark period, during subsequent dark cycles, the mice adapted and had activity levels similar to controls. The activity of df/dp and +/+ mice was indistinguishable in light and dark periods; dp/+ mice were notably less active in the dark (but not light) period (Fig. 3B). In striking contrast, df/+ mice were significantly more active than mice of other genotypes in both light and dark periods. Furthermore, df/+ mice were unique, as they had a higher ratio of light to dark activity compared with the other genotypes (Fig. 3C). These findings indicate that 16p11.2 CNVs affect diurnal behaviors.
Fig. 3.
Behavior/diurnal rhythms are affected in 16p11.2 CNV mice. Comparison of the distance traveled in cohorts of distinct genotypes (detailed in the text) during 60 h of alternating 12-h dark and 12-h light cycles (i.e., spanning three dark periods plus two light periods). (A) The distance traveled over successive dark (black bars) and light (gray bars) periods is shown in bins of 12 h. Five successive dark or light periods are shown. (B) The average distance traveled during light vs. the average distance traveled during dark. (C) The ratio of light-to-dark activity indicates that df/+ mice are unusual in that their activity is not as restricted to the dark periods as the controls are. Average and SEM are presented in all panels.
Climbing deficits.
The most significant genotype effect reported by HomeCageScan was that df/+ mice remained on the ceiling of the cage for extended periods (Phang = 0.000021; Fig. 2A). Therefore, we further investigated the climbing patterns of the mice. The ceiling-climbing behavior of controls was dynamic and changed over the course of the session. Shortly after being introduced into the test cage, diploid controls climbed up to the lower part of the V-shaped ceiling, remained there briefly, and then returned to the floor. During this early phase of testing, control mice returned to the floor with the rear part of their bodies leading, i.e., they hung on the ceiling with their forelimbs, touched the floor with their hindlimbs, and then left the ceiling (Movie S1). In subsequent climbing episodes, control mice traveled to higher and more distant locations on the ceiling, gradually progressing to the highest point of the cage. The climbing behavior of controls developed in two dimensions: first, they left the ceiling from different locations and returned to different places on the floor; second, they could climb down from the ceiling with their head and forepaws leading, i.e., they hung on the ceiling by their hindlimbs and then touched down on the floor with their forelimbs.
In contrast to the adaptability of controls, the ceiling-climbing behavior of df/+ mice was extremely stereotypic throughout the test period. Like control mice during the early phase of being introduced into the test cage, df/+ mice returned to the floor with their hindlimbs leading. However, in contrast to control mice, df/+ mice did not progress to the stage at which they were able to climb down from the ceiling with their head and forelimbs leading. In further contrast to controls, df/+ mice did not climb off the ceiling from different spots; they continued to go up to and down from the ceiling at the same location (Table 1). Some df/+ mice became “trapped” on the ceiling for extensive periods, apparently lacking the ability to return to the floor of the cage (Movie S2). Other df/+ mice developed stereotypic ways of coming down from the ceiling (Movie S3) that they repeated hundreds of times during the course of the session. This repetitive behavior continued throughout the recording period, even after the mice had performed hundreds of climbing episodes. This analysis revealed that 16p11.2 deletion mice show nonprogressive, stereotypic motor behavior that is similar to stereotypic behavior caused by lateral hypothalamic and nigrostriatal lesions (30, 31).
Table 1.
Number of mice from each cohort that performed different climbing behaviors
| Group | Travel on ceiling | Climb down from high point | Climb down with head leading | Stereotypic climb down |
| df/+ | 13/13 | 2/13 | 1/13 | 4/13 |
| +/+ | 15/15 | 15/15 | 15/15 | 0/15 |
| df/dp | 6/9 | 6/9 | 6/9 | 1/9 |
| dp/+ | 1/13 | 1/13 | 1/13 | 0/13 |
16p11.2 CNV Models Have Distinct Changes in Brain Architecture.
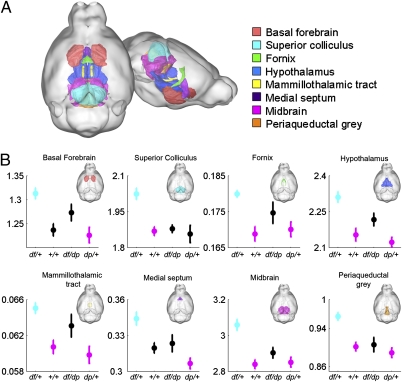
To identify brain regions altered in 16p11.2 CNV mice, we used MRI to analyze the brains of 39 mice from the cohort that had already been analyzed for behavioral phenotypes (Fig. 4) (28). We included both male and female mice in the cohort, which consisted of +/+ and df/dp diploid controls (n = 9 and n = 8, respectively), df/+ (n = 11), and dp/+ (n = 11). Anesthetized mice were perfused and euthanized, and the brain (which remained within the skull) was subjected to MRI. Sixty-two different brain regions (41) were examined, and their volumes were assessed as the percentage of total brain volume averaged for each of the four models (Dataset S4).
Fig. 4.
MRI identifies structural changes in brains of 16p11.2 CNV mice. The relative volume (percentage of total brain volume) of eight brain regions is increased in df/+ mice. (A) Three-dimensional representation of the mouse brain highlights eight regions (colored as in legend) affected by 16.p11.2 dosage. (B) Relative volumes (shown as percentage of total brain volume) are dependent on dosage. Mean and SEM are shown. Statistically significant pairwise differences to the df/+ group (determined by t test followed by Bonferroni–Holm procedure) are depicted as follows: cyan indicates that df/+ differs from at least one other cohort, magenta indicates cohorts that differ significantly from df/+, and black indicates groups that do not differ significantly from df/+. Full pair-wise comparisons are shown in Dataset S4.
Significant changes between brains of df/+ and +/+, as well as between brains of df/+ and dp/+ mice, were noted (Fig. 4 and Fig. S5). Although brains of +/+ and dp/+ mice were not significantly different, a clear trend was found for some regions. Brain structures significantly affected after stringent correction for multiplicity (with the Holm procedure) included the basal forebrain, superior colliculus, fornix, hypothalamus, mammillothalamic tract, medial septum, midbrain, and periaqueductal gray (Fig. 4 A and B). For each structure, the volumetric changes were more extensive between df/+ and dp/+ than between df/+ and +/+, indicating that loss and gain of 16p11.2 dosage affects these regions in opposite ways (Fig. 4 and Fig. S5).
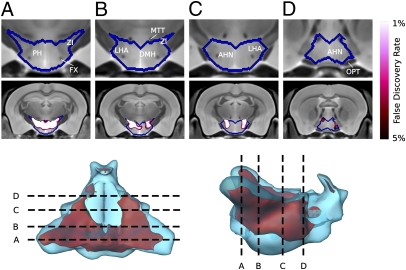
Because the “behavior trap” resembles a phenotype described in rats with lesions in the lateral hypothalamus, we performed detailed MRI analysis of the hypothalamus. Most changes between df/+ and dp/+ were located in the posterior region of the hypothalamus, with pronounced changes in the lateral zone (Fig. 5). These findings support the hypothesis that the lateral hypothalamus is affected in df/+ mice. In addition, we found that Mapk3—which maps within the region corresponding to human 16p11.2—is expressed robustly in specific brain regions including the lateral hypothalamus and the nigrostriatal tract (Fig. S6). These findings demonstrate that altered dosage of 16p11.2 causes changes in the size of several brain structures, and that deletion and duplication have opposing effects.
Fig. 5.
Details of alterations in the hypothalamus detected by MRI. Three-dimensional models of the surface of the hypothalamus (Bottom), coronal images showing the regions affected (Middle), and magnification focusing on the hypothalamus (Top). Red indicates voxels that differ significantly between df/+ and dp/+ cohorts with an FDR of 0.05. The sections performed along four locations marked A–D (A, most posterior; D, most anterior). Colors indicate voxels that differ significantly between df/+ and dp/+ cohorts, with brightness indicating the significance of the difference, as specified by the FDR. AHN, anterior hypothalamic nucleus; DMH, dorsomedial hypothalamus; FX, columns of the fornix; LHA, lateral hypothalamic area; MTT, mammillothalamic tract; OPT, optic tract; PH, posterior hypothalamic nucleus; ZI, zona incerta.
Discussion
16p11.2 CNV Models Provide Insight into Human Syndromes.
CNVs affecting 16p11.2 have been associated with autism and other neurodevelopmental/neuropsychiatric syndromes (1, 7, 9, 12–16), yet several issues remain unresolved. Are these conditions unique to humans? Do loss and gain cause the same syndrome? Does dosage of 16p11.2 affect brain architecture? Why are symptoms of patients with the same CNV so diverse? To begin to address these issues, we engineered mice heterozygous for deletion and duplication of the interval corresponding to 16p11.2 CNVs found in humans. The striking changes we discovered in gene expression profiles, viability, brain architecture, and most importantly behavior, provide functional evidence that 16p11.2 CNVs cause phenotypes in mice, that loss and gain have opposing effects, and that multiple brain regions and behaviors are affected. Our finding that brain volume size is affected reciprocally in deletion vs. duplication mice is concordant with the macrocephaly and microcephaly observed in human subjects with 16p11.2 deletion and duplication, respectively, indicating that our animal models recapitulate the human genomic disorders.
The finding that mice with the same CNVs present in humans have neuroanatomical and behavioral phenotypes indicates that 16p11.2 genes are important for brain function in mammals other than humans. For some human CNV-associated syndromes such as 7q11.23 deletion (i.e., Williams–Beuren syndrome) and the reciprocal duplication (42), loss and gain are associated with opposing clinical features. Indeed, this is the case for head size alterations associated with 16p11.2 CNVs (16), but certainly not for behavioral symptoms of these patients (10, 13, 16). In mice, we see that loss and gain of 16p11.2 cause distinct and opposing behavioral phenotypes. Similarly, mouse chromosome engineered models of human 17p11.2 deletion/duplication-associated syndromes had opposing phenotypes for some, but not all, clinical phenotypes studied (22).
The side-by-side comparison of mice with deletion and duplication of the region corresponding to human 16p11.2 reveals that expression of most genes within the engineered interval correlates directly with dosage, and that a number of neuroanatomical and behavioral phenotypes are affected in opposing directions by loss and gain. Deletion has a more severe effect than duplication on each phenotype—viability, gene expression, brain structure, and behavior—in keeping with the severity of deletion vs. duplication of 16p11.2 in humans. For examples, duplications are occasionally seen in asymptomatic carriers, but carriers of the deletion are extremely rare; in addition, patients with 16p11.2 deletions tend to be diagnosed earlier than those with duplications (16).
16p11.2 CNVs Affect Many Brain Regions.
Changes in head circumference and abnormal brain structure have been reported in patients with 16p11.2 CNVs (14, 16). By using MRI, we find significant volumetric changes in eight different brain regions. Brains of df/+ (but not dp/+) mice, have significant volumetric changes relative to controls, but the most extensive difference is between df/+ and dp/+ mice, emphasizing the opposing effects that 16p11.2 dosage has on brain architecture. Importantly, brains of df/dp diploid controls are not significantly different from +/+ controls, providing genetic evidence that the structural changes in df/+ and dp/+ models are dosage-dependent.
16p11.2 CNVs Affect Multiple Behaviors.
Several human studies compared the behavioral symptoms of patients with 16p11.2 deletions and duplications (10, 13, 16); however, to our knowledge, there is no evidence that loss and gain of 16p11.2 affect behavior in opposing ways. Even with patients harboring the same 16p11.2 lesion, there is a broad spectrum of clinical symptoms, some patients being severely affected and others highly functional. By simultaneously analyzing multiple behaviors in the context of a new environment, we identify a number of behaviors that are altered in 16p11.2 CNV mice, revealing that deletion and duplication have opposite consequences. These highly significant changes survive strict statistical analyses (37, 43). Each genotype responds to the new cage with heightened activity, but only df/+ mice have a second burst of activity at a time when controls are already resting. When control mice have become accustomed to their new environment, they have a gradual increase in freedom of movement on the ceiling over the course of the trial, i.e., a mobility gradient that recapitulates the ontogeny of movement (44). In contrast, df/+ mice do not show the mobility gradient: their ceiling-climbing behavior is restricted to specific locations and their movements are stereotypic. Interestingly, this ceiling-climbing behavior is similar to the behavior trap described in rats with lateral hypothalamic lesions and 6-hydroxydopamine–induced lesions (30, 31), a well characterized model of Parkinson disease. Other phenotypes of these rats are feeding problems (45, 46), sensory neglect, and abnormal gait (30, 31, 47–49). Indeed, abnormal gait and motor delay (13, 16, 18, 50), attention deficits (13), and feeding defects (16) are common in patients with 16p11.2 deletion. Moreover, motor development problems are common in autism spectrum disorders and may serve as an indicator for early intervention, as these features appear before the core symptoms that define autism (51).
Deletion of 16p11.2 Causes Lethality in Neonates.
A major finding of this work is that approximately half of df/+ neonates die after birth, a finding that may have relevance to autism incidence. The precise cause of death in df/+ mice could be related to feeding deficits, but this remains to be investigated. Based on our findings, we suggest that efforts be made to determine whether 16p11.2 deletion is associated with unexplained cases of infant death. If these findings generalize to other genotypes associated with autism, they may explain puzzling aspects of the human condition. The recent increase in autism incidence (52) might be partially attributable to factors that improve pre- and/or postnatal survival. Human studies are consistent with this idea, as it is much more common for inherited rare copy number polymorphisms that affect coding regions to be duplications than deletions (53).
Closing.
This work demonstrates the value of using mice to model CNVs found in human disorders. This approach provides functional evidence that 16p11.2 CNVs affect brain anatomy and behavior in mice, with loss and gain having opposing effects. Multiple brain regions are affected, with deletion of 16p11.2 causing profound behavioral changes such as hyperactivity, difficulty adapting to change, sleeping abnormalities, and repetitive or restricted behaviors. In addition, our findings suggest a potential link between 16p11.2 copy number alterations and infant mortality. Finally, we note a similarity in phenotype between 16p11.2 deletions and rats with lateral hypothalamic lesions. These 16p11.2 CNV models should prove valuable for elucidating the physiological basis of neurodevelopmental syndromes and for evaluating their treatments.
Experimental Procedures
Mice carrying rearrangements corresponding to the human CNVs (Dataset S1) were established by using chromosome engineering as described previously (19, 20, 27, 32). HomeCageScan system (CleverSys) was used to analyze behavior in a cohort of 50 adult df/+, +/+, df/dp, and dp/+ mice. Thirty-nine of these mice were also analyzed by MRI. Hypothesis testing was followed by correction for multiplicity (SI Experimental Procedures provides additional details).
Supplementary Material
Acknowledgments
We thank members of the A.A.M. laboratory, D. Yekutieli, and I. Golani for input; L. Bianco, G. Munoz, and N. Alston for animal husbandry; and C. Johns for microarray hybridizations. This project was funded by Simons Foundation Autism Research Initiative.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE32012).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114042108/-/DCSupplemental.
References
- 1.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, et al. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002;99:3717–3722. doi: 10.1073/pnas.042700699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefansson H, et al. GROUP Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stone JL, et al. International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moeschler JB. Medical genetics diagnostic evaluation of the child with global developmental delay or intellectual disability. Curr Opin Neurol. 2008;21:117–122. doi: 10.1097/WCO.0b013e3282f82c2d. [DOI] [PubMed] [Google Scholar]
- 6.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 7.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 8.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss LA, et al. Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez BA, et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2010;47:195–203. doi: 10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy SE, et al. Wellcome Trust Case Control Consortium Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bijlsma EK, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: Deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld JA, et al. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J Neurodev Disord. 2010;2:26–38. doi: 10.1007/s11689-009-9037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaaf CP, et al. Expanding the clinical spectrum of the 16p11.2 chromosomal rearrangements: three patients with syringomyelia. Eur J Hum Genet. 2011;19:152–156. doi: 10.1038/ejhg.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimojima K, Inoue T, Fujii Y, Ohno K, Yamamoto T. A familial 593-kb microdeletion of 16p11.2 associated with mental retardation and hemivertebrae. Eur J Med Genet. 2009;52:433–435. doi: 10.1016/j.ejmg.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Shinawi M, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y, et al. Intra-family phenotypic heterogeneity of 16p11.2 deletion carriers in a three-generation Chinese family. Am J Med Genet B Neuropsychiatr Genet. 2011;156:225–232. doi: 10.1002/ajmg.b.31147. [DOI] [PubMed] [Google Scholar]
- 19.Mills AA, Bradley A. From mouse to man: Generating megabase chromosome rearrangements. Trends Genet. 2001;17:331–339. doi: 10.1016/s0168-9525(01)02321-6. [DOI] [PubMed] [Google Scholar]
- 20.Bagchi A, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay EA, et al. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 22.Ricard G, et al. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walz K, et al. Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: Phenotypic consequences of gene dosage imbalance. Mol Cell Biol. 2003;23:3646–3655. doi: 10.1128/MCB.23.10.3646-3655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li HH, et al. Induced chromosome deletions cause hypersociability and other features of Williams-Beuren syndrome in mice. EMBO Mol Med. 2009;1:50–65. doi: 10.1002/emmm.200900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatani J, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anney R, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 28.Bock NA, et al. High-resolution longitudinal screening with magnetic resonance imaging in a murine brain cancer model. Neoplasia. 2003;5:546–554. doi: 10.1016/s1476-5586(03)80038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steele AD, Jackson WS, King OD, Lindquist S. The power of automated high-resolution behavior analysis revealed by its application to mouse models of Huntington's and prion diseases. Proc Natl Acad Sci USA. 2007;104:1983–1988. doi: 10.1073/pnas.0610779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schallert T, Whishaw IQ, Ramirez VD, Teitelbaum P. Compulsive, abnormal walking caused by anticholinergics in akinetic, 6-hydroxydopamine-treated rats. Science. 1978;199:1461–1463. doi: 10.1126/science.564552. [DOI] [PubMed] [Google Scholar]
- 31.Golani I, Wolgin DL, Teitelbaum P. A proposed natural geometry of recovery from akinesia in the lateral hypothalamic rat. Brain Res. 1979;164:237–267. doi: 10.1016/0006-8993(79)90019-2. [DOI] [PubMed] [Google Scholar]
- 32.Guo X, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams DJ, et al. Mutagenic insertion and chromosome engineering resource (MICER) Nat Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- 34.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 35.Roughan JV, Wright-Williams SL, Flecknell PA. Automated analysis of postoperative behaviour: Assessment of HomeCageScan as a novel method to rapidly identify pain and analgesic effects in mice. Lab Anim. 2009;43:17–26. doi: 10.1258/la.2008.007156. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Zhang X, Chen-Roetling J, Regan RF. Increased striatal injury and behavioral deficits after intracerebral hemorrhage in hemopexin knockout mice. J Neurosurg. 2011;114:1159–1167. doi: 10.3171/2010.10.JNS10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, et al. Ten ways to improve the quality of descriptions of whole-animal movement. Neurosci Biobehav Rev. 2010;34:1351–1365. doi: 10.1016/j.neubiorev.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Horev G, Benjamini Y, Sakov A, Golani I. Estimating wall guidance and attraction in mouse free locomotor behavior. Genes Brain Behav. 2007;6:30–41. doi: 10.1111/j.1601-183X.2006.00216.x. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507. [Google Scholar]
- 40.Bourgeron T. The possible interplay of synaptic and clock genes in autism spectrum disorders. Cold Spring Harb Symp Quant Biol. 2007;72:645–654. doi: 10.1101/sqb.2007.72.020. [DOI] [PubMed] [Google Scholar]
- 41.Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 42.Merla G, Brunetti-Pierri N, Micale L, Fusco C. Copy number variants at Williams-Beuren syndrome 7q11.23 region. Hum Genet. 2010;128:3–26. doi: 10.1007/s00439-010-0827-2. [DOI] [PubMed] [Google Scholar]
- 43.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 44.Golani I. A mobility gradient in the organization of vertebrate movement: The perception of movement through symbolic language. Behav Brain Sci. 1992;15:249–266. [Google Scholar]
- 45.Teitelbaum P, Epstein AN. The lateral hypothalamic syndrome: Recovery of feeding and drinking after lateral hypothalamic lesions. Psychol Rev. 1962;69:74–90. doi: 10.1037/h0039285. [DOI] [PubMed] [Google Scholar]
- 46.Ungerstedt U. Stereotaxic mapping of monoamine pathways in rat brain. Acta Physiol Scand Suppl. 1971;367:1–48. doi: 10.1111/j.1365-201x.1971.tb10998.x. [DOI] [PubMed] [Google Scholar]
- 47.Marshall JF, Teitelbaum P. Further analysis of sensory inattention following lateral hypothalamic damage in rats. J Comp Physiol Psychol. 1974;86:375–395. doi: 10.1037/h0035941. [DOI] [PubMed] [Google Scholar]
- 48.Marshall JF, Richardson JS, Teitelbaum P. Nigrostriatal bundle damage and the lateral hypothalamic syndrome. J Comp Physiol Psychol. 1974;87:808–830. doi: 10.1037/h0037223. [DOI] [PubMed] [Google Scholar]
- 49.Marshall JF, Turner BH, Teitelbaum P. Sensory neglect produced by lateral hypothalamic damage. Science. 1971;174:523–525. doi: 10.1126/science.174.4008.523. [DOI] [PubMed] [Google Scholar]
- 50.Hanson E, et al. 16p11.2 Study Group Clinicians Cognitive and behavioral characterization of 16p11.2 deletion syndrome. J Dev Behav Pediatr. 2010;31:649–657. doi: 10.1097/DBP.0b013e3181ea50ed. [DOI] [PubMed] [Google Scholar]
- 51.Lloyd M, Macdonald M, Lord C. Motor skills of toddlers with autism spectrum disorders. Autism. 2011 doi: 10.1177/1362361311402230. 10.1177/1362361311402230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newschaffer CJ, et al. The epidemiology of autism spectrum disorders. Annu Rev Public Health. 2007;28:235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- 53.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.