Abstract
We report a chip-scale lensless wide-field-of-view microscopy imaging technique, subpixel perspective sweeping microscopy, which can render microscopy images of growing or confluent cell cultures autonomously. We demonstrate that this technology can be used to build smart Petri dish platforms, termed ePetri, for cell culture experiments. This technique leverages the recent broad and cheap availability of high performance image sensor chips to provide a low-cost and automated microscopy solution. Unlike the two major classes of lensless microscopy methods, optofluidic microscopy and digital in-line holography microscopy, this new approach is fully capable of working with cell cultures or any samples in which cells may be contiguously connected. With our prototype, we demonstrate the ability to image samples of area 6 mm × 4 mm at 660-nm resolution. As a further demonstration, we showed that the method can be applied to image color stained cell culture sample and to image and track cell culture growth directly within an incubator. Finally, we showed that this method can track embryonic stem cell differentiations over the entire sensor surface. Smart Petri dish based on this technology can significantly streamline and improve cell culture experiments by cutting down on human labor and contamination risks.
Keywords: lensless imaging, time-lapse microscopy, on-chip cellular imaging, stem cell differentiation tracking, superresolution algorithm
Recent rapid advances and commercialization efforts in CMOS imaging sensor has led to broad availability of cheap and high pixel density sensor chips. In the past few years, these sensor chips enabled the development of new microscopy implementations that are significantly more compact and cheaper than traditional microscopy designs. The optofluidic microscope (1–3) and the digital in-line holographic microscope (4–9) are two examples of these new developments. Both of these technologies are designed to operate without lenses and, therefore, circumvent their optical limitations, such as aberrations and chromaticity. Both technologies are suitable for imaging dispersible samples, such as blood, fluid cell cultures, and other suspensions of cells or organisms. However, neither can work well with confluent cell cultures or any sample in which cells are contiguously connected over a sizable length scale.
In the case of the optofluidic microscope, imaging requires the microfluidic flow of the specimens across a scanning area. Adherent cells are simply incompatible with this imaging mode. In digital in-line holographic microscopy, the interference intensity distribution of a target under controlled light illumination is measured and then an image reconstruction algorithm is applied to render microscopy images of the target. There have been two major types of algorithms that have been reported (10–12). In both cases, the image quality depends critically on the extent of the target, the scattering property and the signal-to-noise ratio (SNR) of the measurement processes (5, 7, 8, 13–15). The method works well for well-isolated targets, such as diluted blood smear slides. However, to our knowledge, such approaches have not been applied to targets that occupy more than 0.1 mm2 in total contiguous area coverage with submicron resolution (4–7, 16). The reason of this limitation is well-known: the loss of phase information during the intensity recording process. In order to recover the phase information, object support has to be used in the iterative phase recovery algorithm, which involves the light field propagation back and forth between the imaging domain (where the intensity data are applied) and object domain (where a priori object constrains are applied) (11). When the test object is real or nonnegative, it is easy to apply the powerful nonnegativity support constraint to extract the phase information from the recorded diffraction intensity (11). However, for digital in-line holography, light field in the object domain is complex valued and, therefore, the phase recovery is possible only if the support of the object is sufficiently isolated (i.e., sparsity constrains) (14, 15, 17, 18) or the edges are sharply defined (true boundary) (14, 15, 18). Furthermore, the interference nature of the technique implies that coherence-based noise sources, such as speckles and cross-interference, would be present and would need to be addressed (7, 8, 19). While methods for mitigating these have been reported (13, 14, 20), the generated images are, nevertheless, identifiably different from images acquired with conventional microscopes due to coherence based noise sources.
The need for a high-quality, autonomous, and cost-effective microscopy solution for imaging confluent cell culture samples, especially for longitudinal studies, is a strong one (21). To name a few specific examples, the determination of daughter fates before the division of neural progenitor cells (22), the existence of haemogenic endothelium (23), neural and hematopoietic stem and progenitor divisional patterns and lineage choice (24, 25), the in vitro tissue culture studies using the neutral red dye (26), the studies of dynamics of collective cell migration (27), detection of toxic compound (28), and drug screening (29, 30). In these cases, the labor-intensive nature of these experiments and the challenge of efficiently imaging large assays have typically plagued this type of experiment format.
A chip-scale microscopy method that can automatically image growing or confluent cell cultures can significantly improve Petri dish-based cell culture experiments. In fact, with this approach providing a compact, low-cost, and disposable microscopy imaging solution, we can start to transit Petri dish-based experiments from the traditionally labor-intensive process to an automated and streamlined process. This technological shift from an inert Petri dish to a self-imaging Petri dish, which we term ePetri, is appropriately timely as well, because, the cost of high performance CMOS imaging sensors (which are widely used in cellphone cameras and webcams) have recently reached a price point where they can be used as recyclable or disposable components. We believe that such a self-imaging Petri dish can significantly affect cell culture-based procedures in both medicine and science.
In this paper, we report on such a chip-scale microscopy method and demonstrate a proof-of-concept self-imaging Petri dish solution (ePetri). This system has the ability to automatically image confluent cell sample with subcellular resolution over a large field of view. As such, it is well suited to long-term cell culture imaging and tracking applications. This paper is structured as follows. We will first present our prototype setup and the principle of subpixel perspective sweeping microscopy (SPSM). Then we will report on our large-field-of-view imaging experiment on Giemsa-stained confluent HeLa cell samples. We will then report on our experimental demonstration of long-term cell imaging and tracking of HeLa cell and embryonic stem cell culture growth with the ePetri platform. Next, we will discuss the resolution and limitations of the subpixel perspective sweeping microscopy (SPSM) method. Finally, we will discuss the application advantages of the ePetri platform.
Results
Principle of SPSM.
Conceptually, this method of microscopy imaging, SPSM, is simple to understand. Geometrically, we simply culture cells or place cells of interest directly on the surface of a CMOS image sensor. To start, consider an idealized image sensor that has a high density grid of infinitesimally small pixels. In such a case, as long as the cells are right on the sensor, this idealized sensor would be able to collect a high-resolution shadow image of the cells with excellent acuity. Unfortunately, currently available sensor chips have rather large pixels (2.2 μm in our particular experiment). This implies that the direct shadow images we collect with our sensor chips are intrinsically coarse (31, 32). Specifically, the raw resolution of the shadow image would be no better than two times of the pixel size (as dictated by Nyquist criterion considerations). To address this, we take the following approach to improve resolution or, more specifically in our case, to generate a denser grid of smaller “virtual” pixels.
First, we take note of the fact that there is a thin transparent passivation layer that separates the cells from the actual light sensitive region of the sensor chip. With this recognition in mind, we sequentially tilt/shift an incoherent illumination source above the sample and acquire a sequence of raw images. With the incremental tilt/shift of the illumination, the target cells’ shadow will incrementally shift across the sensor pixels (Fig. 1 A–C). The amount of shadow shift is proportional to the passivation layer thickness and the tilt/shift extent of the light source. As long as the shadow shift between each raw image frame is much smaller than the physical pixel size, we can then combine the information from multiple sub-pixel-shifted low-resolution (LR) shadow images to create a single high-resolution (HR) image with a pixel superresolution algorithm (33–36). The algorithm we used in this experiment is a simple, fast and noniterative method (35) that preserves the estimation optimality in the maximum-likelihood sense (Supplementary Note 1). The computation complexity of this approach is O(n ∗ log(n)), where n is the number of pixels.
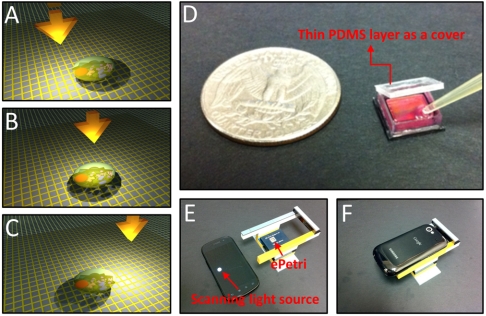
Fig. 1.
Principle of SPSM and the ePetri prototype. (A–C) With the incremental tilt/shift of the illumination, the target cells’ shadow will incrementally shift across the sensor pixels. These subpixel shifted low-resolution images will be used to reconstruct the high-resolution image by using pixel superresolution algorithm. (D) The ePetri prototype. A thin PDMS layer is used as a cover to prevent the evaporation of the culture media while allowing for CO2 exchange between the well and exterior. (E–F) The ePetri imaging platform. We used the LED screen of a smartphone as the scanning light source. The holder is built with Lego blocks.
Our ePetri prototype based on SPSM imaging is shown in Fig. 1D. This prototype was built on a commercial available CMOS image sensor with a 6 mm × 4 mm imaging area filled with 2.2-μm pixels (Aptina MT9P031). The microlens layer and color filter on the sensor surface were removed to provide us with direct access to the sensor pixels. In a separate experiment, we determined that the sensor top passivation layer was about 0.9 μm thick. We glued a homemade square plastic well to the image sensor with poly dimethylsiloxane (PDMS) (see Methods for details). We then used a thin PDMS layer (approximately 100 μm) as a cover for this ePetri prototype. The thin PDMS layer served to prevent the evaporation of the culture media while allowing for CO2 exchange between the well and exterior. For illumination, we used the LED screen of a smartphone (Google Nexus S) as the scanning illumination light source, as shown in Fig. 1 E–F. A holder was built with Lego building blocks to house the image sensor socket board and the smartphone. The screen of smartphone was set at about 2.0 cm away from the image sensor. In this method, the alignment between the smartphone and the image sensor is not a critical consideration. During imaging, we arranged the perspective illumination angle from −60 ° to +60 ° with respect to the surface of the image sensor. The entire platform can be placed in an incubator for automatic long-term cell imaging and tracking, as we shall report in a later section.
We used a smartphone screen as illumination to highlight the point that the light intensity requirement of this imaging scheme is low. The scheme can flexibly work with an LED display panel, a television screen, or an LED matrix. In our experiments, the average light intensity incident on a sensor pixel was 0.015 W/m2. As a point of reference, a halogen-lamp based conventional microscope typically delivers intensity of 20 W/m2 on a sample.
Large-Field-of-View Color Imaging of the Confluent Cell Sample.
To demonstrate the ability of this ePetri prototype to image confluent cell samples, we cultured HeLa cells on the ePetri platform (Fig. 1F) for about 48 h. The sample was then stained with Giemsa (detailed procedures of fixation and staining are explained in Methods). The entire image area was 6 mm × 4 mm. We used 15 × 15 scanning steps (Fig. S1) for each color illumination (the scanning video for the smart phone is provided in Movie S1). The image capture rate was set at 10 frames per second with the pixel clock of the image sensor running at 70 MHz. The entire data acquisition process took about 20 s.
Fig. 2A shows the reconstructed color image of the confluent HeLa cell sample. The image enhancement factor used in the algorithm to generate the image was set at 13. In other words, each pixel at the low-resolution raw image level (2.2 μm) was enhanced into a 13 × 13 pixel block in the reconstructed image. The entire image of Fig. 2A contains about 8.45 × 108 pixels. The prototype took about 22 s to capture each raw image set for each color (a video showing the captured raw image sequence and the reconstructed image is provided in Movie S2). Given the sheer amount of data generated, the data transfer rate of approximately 100 MB/s between the image sensor and the computer via ethernet connection imposed a throughput limit. After transferring the raw data into the computer, it took us 2–3 min to reconstruct the entire high-resolution image using a personal computer with an Intel i7 CPU. We note that, the solution for the reconstructed image was noniterative, deterministic, and was optimized in the maximum-likelihood sense. The relative long time for image reconstruction was simply attributable to the fact that we were dealing with a large amount of data. However, with the use of a GPU unit, we expect the image processing time can be cut down to less than 1 s for the entire image. As we believe the primary use of ePetri would be for tracking cell culture growth directly from within an incubator, we do not believe that the current data transfer limitation or the current processing speed of the prototype will be the bottleneck for the proposed platform.
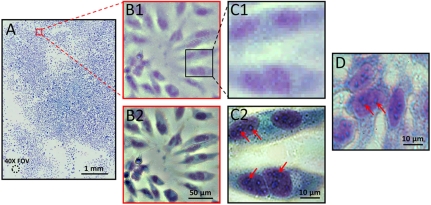
Fig. 2.
(A) Large-field-of-view color imaging of the confluent cell sample. The field of view of a 40× objective lens is also shown in left bottom. (B1 and C1) Raw images of a small region of A. (B2 and C2) The reconstructed high-resolution images corresponding to B1 and C1. (D) The conventional microscopy image, with 40× objective lens (0.66 N.A.), acquired from similar cells cultured on a Petri dish. The slight color difference between D and C2may be due to the reflective surface of the image sensor of the ePetri platform.
The amount of details in the reconstructed color image is too large to fully display on a computer screen or print on a printer; we have provided vignette views of selected regions for comparison in Fig. 2. Fig. 2B1 and C1 shows the raw images from a small region of Fig. 2A. Fig. 2 B2 and C2 shows the corresponding reconstructed high-resolution image of B1 and C1. From the reconstructed high-resolution image in Fig. 2 B2 and C2, we can readily discern organelles within the HeLa cell, such as multiple nuclear granules (indicated by red arrows) and the nucleus. The images also closely corresponded to conventional microscopy images acquired from similar cells cultured on a Petri dish (see Fig. 2D; image acquired with an Olympus BX 51 microscope with a 40×, NA = 0.66 objective). The conventional microscopy image of Fig. 2C2 is provided in Fig. S2). This strongly indicates that the ePetri can directly replace and improve (by providing a wide field of view) upon the conventional microscope for cell culture analysis.
Longitudinal Cell Imaging and Study Using the ePetri Platform.
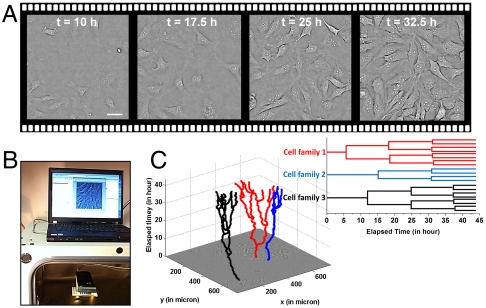
Here, we report on our demonstration of using our ePetri prototype to perform longitudinal cell imaging and study from within an incubator. In the first experiment, we seeded HeLa cells onto the ePetri and the entire imaging platform (as shown in Fig. 3B) was placed into the incubator. An ethernet cable connected our prototype to a personal computer outside the incubator for data transfer. In this experiment, we took a complete image set at 15 min interval for the entire growth duration of 48 h. The number of cells grew from 40+ to hundreds in this period. Fig. 3A shows the reconstructed images of the cells from a specific sublocation acquired at t = 10 hr, t = 17.5 hr, t = 25 hr, and t = 32.5 hr. Based on the time-lapse cell imaging data, we can detect and track each individual cell’s movements in space and time and generate corresponding lineage trees (i.e., mother-daughter relationship). For example, Fig. 3C shows tracking trajectories of three cell families annotated by a biologist (Movie S3). The lineage trees for these cell families are also shown in Fig. 3C.
Fig. 3.
(A) Time-lapse imaging of HeLa cell culture on the ePetri platform. Scale bar, 20 μm. (B) The experimental setup. The ePetri platform was placed into the incubator; the data was read out by an ethernet cable to a personal computer. A customized program was created to automatically reconstruct and display the image onto the screen for user monitoring. (C) The tracking trajectories of three cell families and the corresponding lineage trees for these cell families (see also Movie S3).
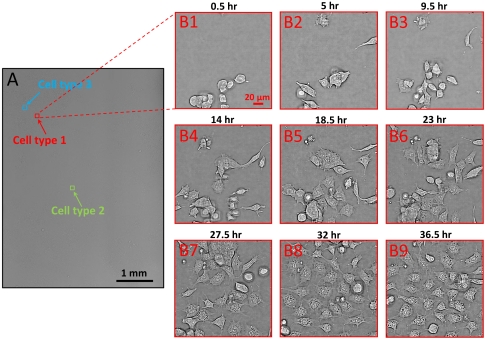
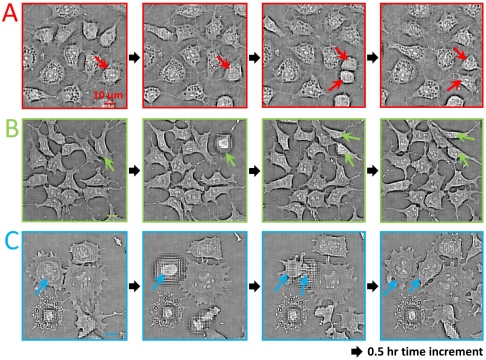
In order to further demonstrate the versatility of the ePetri as a general platform for culturing various types of cells, we perform a second experiment using embryonic stem (ES) cells. These cells are derived from the inner cell mass of developing embryos. ES cells offer tremendous biomedical potential on two interrelated levels: First, they provide a model system for uncovering the fundamental mechanisms governing cell fate decision-making and differentiation; second, they are the basis for a new generation of regenerative therapies for a wide range of neurodegenerative, autoimmune, and hematopoietic diseases, among others. In order to visualize the process of stem cell differentiation and the spatial heterogeneity in cell fate decisions, we cultured them on ePetri and followed changes in cell morphology during the differentiation process. We used the E14 mouse ES cell line as a specific example of ES cells. We cultured cells in vitro and imaged them both under stem cell maintaining conditions and under differentiation-inducing conditions. Initially, we imaged stem cells while maintaining their pluripotent state (see Methods for detail) (Fig. S3). Then, in the second stage of this experiment, we imaged the differentiation process and the dynamical morphological changes in stem cells. Media were being replaced every two days until cells differentiated and began to exhibit various morphologies (see Methods for details). Fig. 4A shows the reconstructed images of ES cells at the differentiation stage. Fig. 4 B1–B9 shows a specific sublocation (corresponded to cell type 1) acquired at different times. We were able to identify at least three cell variations in the reconstructed image (denoted by an arrow in Fig. 4A). From the morphologies, we estimate the likely identities of the cells in Fig. 5A were adipocytes, the cells in Fig. 5B were undifferentiated ES cells, and the cells in Fig. 5C were neural progenitor cells. Based on the time-lapse cell imaging data, we can track the cell division event for each type of cell, as shown in Fig. 5 A–C. The time increment between each image frame is about 0.5 h. This experiment clearly demonstrates that the ePetri can collect microscopy resolution images over the entire area of the sensor, which is orders of magnitude larger than the field of view of a conventional microscope with comparable resolution.
Fig. 4.
(A) Time-lapse imaging of embryonic stem cell culture on the ePetri platform. Based on the morphologies, at least three types of cells were found in the reconstructed image, denoted by the red, green and blue arrows. (B1–B9) A specific sublocation for cell type 1 (adipocytes) acquired at different time. The observable differentiation for this cell type occurred at about 20 h after the stem cell plating.
Fig. 5.
Tracking of cell division (denoted by the arrows) for cell type 1 (A), type 2 (B), and type 3 (C). The defocus effect in some of the images is due to the cell detaching from the sensor surface when cell division occurs. The time increment between each image frame is about 0.5 h. The location of these cell types are denoted at Fig. 4A.
Thus we were able to visualize dynamically one of the striking characteristics of ES cells—their intrinsic heterogeneity. In addition to morphological heterogeneity, ES cells are known to exhibit wide variations in gene expression. Furthermore, individual cells often differentiate at different times and locations and choose different fates even when exposed to the same media conditions in the same Petri dish. Consequently, the ability to continually monitor ES cells over time across a very large area could provide qualitatively new insights into the behavior of these cells across a variety of protocols and experiments.
Resolution.
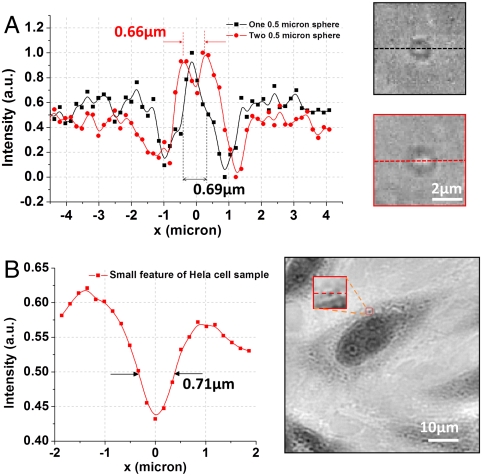
The optical resolution of the ePetri platform was investigated by imaging 500-nm microspheres (Polysciences) that were directly placed on the image sensor surface. The imaging process was identical to the one previously described for HeLa cell culture. For a single 500-nm microsphere, the bright center of the microsphere was clearly resolved (shown in Fig. 6A), with the full-width at half maximum (FWHM) of 690 nm. Because microscopy resolution is formally defined based on a given microscope’s ability to resolve two closely spaced feature points, we further analyzed the case of two closely spaced microspheres to better establish our prototype’s resolution. Fig. 6A also showed the reconstructed images of two closely packed 500-nm microspheres with center-to-center distance of 660 nm. The data trace clearly showed a valley between the two peaks and, thus, established that the resolution of our prototype was 660 nm or better. To further verify this point, Fig. 6B shows the magnified small feature of the stained HeLa cell sample (Movie S2), and the FWHM of this feature was estimated to be 710 nm, which was in good agreement with the estimated resolution limit.
Fig. 6.
Resolution of the proposed platform. (A) The line traces of images of one 500-nm microsphere (black line) and two 500-nm microspheres (red line). (B) The line traces of the small feature of reconstructed high-resolution HeLa cell image (left; see also Movie S2).
We do, hereby, note that the resolution of the SPSM will deteriorate if the target samples are placed at a substantial distance above the sensor surface. The exact resolution-to-height function is not trivially expressible and instead depends on the angular distribution function of the sample scattering, presence/absence and characteristics of pixel lens, characteristics of the pixel structure, sensor passivation layer thickness, and the physical dimensions of the light sensitive area on each sensor pixel. The last four parameters are proprietary information that are not publicly disclosed by the chip maker. We believe the method we just described provides an adequate resolution characterization recipe for readers interested in building their own ePetri, especially if a different sensor chip type is used.
Discussion
We have developed a lensless microscopy imaging method, SPSM, that is able to image confluent cell culture with high-resolution and incoherent light sources. The images are closely comparable with those obtained with a conventional microscope. Our prototype has a demonstrated resolution of 660 nm. This imaging method can be applied to implement a smart Petri dish, ePetri, which is capable of performing high-resolution and autonomous imaging of cells plated on or growing on a low-cost CMOS sensor chip. Our preliminary cell culture experiment indicates that the ePetri can be a useful tool for in vitro long-term cell observations. To demonstrate that this imaging platform can be easily assembled, our prototype was constructed out of Lego blocks, a smartphone, and an imaging sensor chip.
There are three aspects to the ePetri technology that are worth further investigation:
At present, the ePetri prototype is limited to nonfluorescence imaging. In principle, we can create a fluorescence-capable ePetri by simply coating the sensor chip with an appropriate filter material. However, the fluorescence emission (i.e., the image) cannot be subpixel shifted by sweeping the plane wave illumination. In the near future, we plan to examine several ePetri design permutations by using pattern illumination (37, 38), which is able to circumvent this problem.
Our ePetri prototype sustained cell culture growth by immersing the cells in a nutrient-filled fluid rather than providing them with a solid growth substrate. A thick solid growth substrate would have compromised the lateral resolution. For experiments that absolutely require thick solid growth substrate, we encourage the user to experimentally seek an appropriate compromise that can sustain cell growth without deteriorating resolution beyond the user’s requirements or use a pattern illumination (37, 38) instead of the simple plane wave illumination.
Our current ePetri can acquire a full set of data (approximately 20 seconds) to render a high-resolution image in a time period of 2–3 min. We did not optimize our system for speed because our focus here is on tracking cell culture growth—a relatively slow process. Interested users can certainly optimize the system for speed by improving on all aspects of the system.
There are several advantages associated with this technology that are worth noting:
Low cost. The ePetri uses a CMOS imaging sensor as the base substrate for cell culture growth. Post-experiment, the sensor can either be disposed or washed and reused. Given the low cost of these sensor chips, they are unlikely to represent a major cost component in most cell culture experiments.
Disposable. In certain biohazardous experiments, the ability to treat the sensor chips as disposable units would significantly reduce any associated risks.
Direct readout from the incubator. As our demonstration experiment shows, the ePetri is sufficiently compact to fit comfortably in a typical incubator. In fact, given its footprint, it would be possible to fit multiple ePetri units into the same incubator. Upon connecting the ePetri to an exterior processor via an appropriate data cable, a user can start to collect images of the growing cell culture without removing the unit from the incubator. This advantage saves labor and cuts down on the perturbations the cell culture is subjected to. It is also possible to design a compact and portable incubator ePetri combination that is suitable for point-of-care diagnostic and/or other uses.
Continuous from-the-incubator monitoring. On a related point, an ePetri user would be able to monitor cell growth continuously. In bioscience research, this represents a good means for performing longitudinal studies. In medical applications, this can significantly cut down on the diagnostic time for medical procedures that requires culture growth-based assessment. As an example, the ePetri can replace the standard Petri dish for tuberculosis, staph, and other bacteria infection diagnosis. Whereas standard medical practice would initiate a bacteria culture growth and then check the growth at relatively long time intervals (checking frequently would be too time consuming), a modified ePetri may potentially be able to continuously and autonomously monitor for growth changes and notify the user to examine the sample when significant changes have been detected.
Platform technology. Finally, we note that the ePetri is a platform technology. Because the top surface of the sensor chip is unmodified, a user is free to build upon it. It is very possible to simply use the ePetri as an imaging platform for a large number of sophisticated lab-on-a-chip designs, such as microorganism detection based on the use of closed dielectrophoretic cages (39), droplet-based platforms for cell encapsulation and screening (40), microfluidics-based phenotyping imaging and screening of multicellular organisms (41), and high throughput malaria infected erythrocyte separation and imaging (42). It is also possible to modify the ePetri to serve as a self-contained incubator and imaging unit.
Methods
ePetri Prototype.
The device is composed of three parts: a CMOS image sensor, a homemade square plastic wall, and a PDMS thin layer. We used MT9P031 (2.2 mm pixel, Aptina) for the image sensors. We removed the color filter and the microlens layer by treating the sensor under oxygen plasma for 10 min (80 W). The PDMS thin layer is prepared by mixing 1∶10 with base and curing agent, then spin coated on a 3 in. silicon wafer followed by baking at 80 °C for 1 h. The homemade square plastic wall was glued to the image sensor by using PDMS (1∶10 with base and curing agent), followed by baking at 80 °C for 1 h.
HeLa Cell Culture.
To promote the cell adhesion, the surfaces of ePetri chips were treated with Poly-L-lysine (0.01%, Sigma-Aldrich) for 15 min and washed three times with distilled water. HeLa cells were first cultured in Dulbecco’s modified eagle medium (DMEM, Invitrogen) supplemented with 1-glutamine (4 mM), penicillin (100 units/mL), streptomycin (100 μg/mL), and 10% (v/v) fetal calf serum in culture dishes and maintained in 5% CO2 humidified atmosphere at 37 °C. During the logarithmic growth period, cells were harvested by trypsin (0.05% trypsin with EDTA*4Na, Invitrogen), resuspended in DMEM, and then seeded onto the ePetri prototype.
HeLa cell staining.
We use the following steps to stain the HeLa cell culture:
Fix the air-dried sample in absolute methanol by dipping the ePetri briefly (two dips) in a Coplin jar containing absolute methanol.
Remove and let air dry.
Stain with diluted Giemsa stain (1∶20, vol/vol) for 20 min.
Wash by briefly dipping the ePetri in and out of a Coplin jar of distilled water (one or two dips).
Embryonic Stem Cell Culturing.
Initially, we imaged E14 mouse stem cells while maintaining their pluripotent state. For this stage, cells were resuspended using 0.25% trypsin plated and 104 cells were plated on the ePetri chip (2 × 104 cells per cm2). We precoated the ePetri with fibronectin (5 ug/mL) for 3 h prior to plating the cells, in order to allow cells to adhere efficiently to the sensor surface. Cells were then maintained in a standard stem cell medium (high glucose DMEM, supplemented with 15% FBS, L-glutamine/Pen/Strep, NEAA, sodium pyruvate, and 0.1 mM 2-mercaptoethanol) enriched with LIF (1,000 U/mL, Millipore) in order to sustain pluripotency. The media were replaced daily to resupply nutrients and maintain a proper pH level.
In the differentiation stage, cells were first plated at low density (approximately 5,000 cells) on a fibronectin-coated ePetri. Initially, cells were maintained in pluripotency-sustaining media for 24 h to allow adherence of the cells. After that point, the media was replaced with N2B27, a defined serum free media. In order to induce differentiation, pluripotency-sustaining signaling molecules were not included. Media were being replaced every 2 d until cells differentiated and began to exhibit various morphologies.
Supplementary Material
Acknowledgments.
We thank Dr. Benjamin Judkewitz for HeLa cell tracking and Mr. Samuel Yang for analyzing some of the data. We acknowledge funding support from the Coulter Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110681108/-/DCSupplemental.
References
- 1.Heng X, et al. Optofluidic microscopy—method for implementing a high resolution optical microscope on a chip. Lab Chip. 2006;6:1274–1276. doi: 10.1039/b604676b. [DOI] [PubMed] [Google Scholar]
- 2.Cui X, et al. Lensless high-resolution on-chip optofluidic microscopes for Caenorhabditis elegans and cell imaging. Proc Natl Acad Sci USA. 2008;105:10670–10675. doi: 10.1073/pnas.0804612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng G, Lee SA, Yang S, Yang C. Sub-pixel resolving optofluidic microscope for on-chip cell imaging. Lab Chip. 2010;10:3125–3129. doi: 10.1039/c0lc00213e. [DOI] [PubMed] [Google Scholar]
- 4.Repetto L, Piano E, Pontiggia C. Lensless digital holographic microscope with light-emitting diode illumination. Opt Lett. 2004;29:1132–1134. doi: 10.1364/ol.29.001132. [DOI] [PubMed] [Google Scholar]
- 5.Mudanyali O, et al. Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab Chip. 2010;10:1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu W, Jericho M, Meinertzhagen I, Kreuzer H. Digital in-line holography for biological applications. Proc Natl Acad Sci USA. 2001;98:11301–11305. doi: 10.1073/pnas.191361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Sucerquia J, et al. Digital in-line holographic microscopy. Appl Opt. 2006;45:836–850. doi: 10.1364/ao.45.000836. [DOI] [PubMed] [Google Scholar]
- 8.Malek M, Allano D, Coëtmellec S, Lebrun D. Digital in-line holography: Influence of the shadow density on particle field extraction. Opt Express. 2004;12:2270–2279. doi: 10.1364/opex.12.002270. [DOI] [PubMed] [Google Scholar]
- 9.Isikman SO, et al. Lens-free optical tomographic microscope with a large imaging volume on a chip. Proc Natl Acad Sci USA. 2011;108:7296–7301. doi: 10.1073/pnas.1015638108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G, Scott P. Phase retrieval and twin-image elimination for in-line Fresnel holograms. J Opt Soc Am A. 1987;4:159–165. [Google Scholar]
- 11.Fienup JR. Reconstruction of an object from the modulus of its Fourier transform. Opt Lett. 1978;3:27–29. doi: 10.1364/ol.3.000027. [DOI] [PubMed] [Google Scholar]
- 12.Koren G, Polack F, Joyeux D. Iterative algorithms for twin-image elimination in in-line holography using finite-support constraints. J Opt Soc Am A. 1993;10:423–433. [Google Scholar]
- 13.Lai S, King B, Neifeld MA. Wave front reconstruction by means of phase-shifting digital in-line holography. Opt Commun. 2000;173:155–160. [Google Scholar]
- 14.Rodenburg J, Hurst A, Cullis A. Transmission microscopy without lenses for objects of unlimited size. Ultramicroscopy. 2007;107:227–231. doi: 10.1016/j.ultramic.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Fienup JR. Reconstruction of a complex-valued object from the modulus of its Fourier transform using a support constraint. J Opt Soc Am A. 1987;4:118–123. [Google Scholar]
- 16.Biener G, et al. Combined reflection and transmission microscope for telemedicine applications in field settings. Lab Chip. 2011;11:2738–2743. doi: 10.1039/c1lc20169g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denis L, Lorenz D, Thiébaut E, Fournier C, Trede D. Inline hologram reconstruction with sparsity constraints. Opt Lett. 2009;34:3475–3477. doi: 10.1364/OL.34.003475. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F, Pedrini G, Osten W. Phase retrieval of arbitrary complex-valued fields through aperture-plane modulation. Phys Rev A. 2007;75:043805. [Google Scholar]
- 19.Xu L, Miao J, Asundi A. Properties of digital holography based on in-line configuration. Opt Eng. 2000;39:3214–3219. 10.1117/1.1327503. [Google Scholar]
- 20.Micó V, García J, Zalevsky Z, Javidi B. Phase-Shifting Gabor Holographic microscopy. J Disp Technol. 2010;6:484–489. [Google Scholar]
- 21.Schroeder T. Long-term single-cell imaging of mammalian stem cells. Nat Methods. 2011;8:S30–S35. doi: 10.1038/nmeth.1577. [DOI] [PubMed] [Google Scholar]
- 22.Cohen AR, Gomes FLAF, Roysam B, Cayouette M. Computational prediction of neural progenitor cell fates. Nat Methods. 2010;7:213–218. doi: 10.1038/nmeth.1424. [DOI] [PubMed] [Google Scholar]
- 23.Eilken HM, Nishikawa SI, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- 24.Costa MR, et al. Continuous live imaging of adult neural stem cell division and lineage progression in vitro. Development. 2011;138:1057–1068. doi: 10.1242/dev.061663. [DOI] [PubMed] [Google Scholar]
- 25.Dykstra B, et al. High-resolution video monitoring of hematopoietic stem cells cultured in single-cell arrays identifies new features of self-renewal. Proc Natl Acad Sci USA. 2006;103:8185–8190. doi: 10.1073/pnas.0602548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Repetto G, del Peso A, Zurita JL. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat Protoc. 2008;3:1125–1131. doi: 10.1038/nprot.2008.75. [DOI] [PubMed] [Google Scholar]
- 27.Angelini TE, et al. Glass-like dynamics of collective cell migration. Proc Natl Acad Sci USA. 2011;108:4714–4719. doi: 10.1073/pnas.1010059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borenfreund E, Puerner JA. Toxicity determined in vitro by morphological alterations and neutral red absorption. Toxicol Lett. 1985;24:119–124. doi: 10.1016/0378-4274(85)90046-3. [DOI] [PubMed] [Google Scholar]
- 29.Cavanaugh PF, et al. A semi-automated neutral red based chemosensitivity assay for drug screening. Invest New Drugs. 1990;8:347–354. doi: 10.1007/BF00198590. [DOI] [PubMed] [Google Scholar]
- 30.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 31.Lange D, Storment CW, Conley CA, Kovacs GTA. A microfluidic shadow imaging system for the study of the nematode Caenorhabditis elegans in space. Sens Actuators B Chem. 2005;107:904–914. [Google Scholar]
- 32.Wei L, Knoll T, Thielecke H. On-chip integrated lensless microscopy module for optical monitoring of adherent growing mammalian cells; 2010 Annual International Conference of the IEEE; Piscataway, NJ: Engineering in Medicine and Biology Society; 2010. pp. 1012–1015. [DOI] [PubMed] [Google Scholar]
- 33.Milanfar P. Super-Resolution Imaging. Boca Raton, FL: CRC Press; 2010. [Google Scholar]
- 34.Hardie R, Barnard K, Armstrong E. Joint MAP registration and high-resolution image estimation using asequence of undersampled images. IEEE Trans Image Process. 1997;6:1621–1633. doi: 10.1109/83.650116. [DOI] [PubMed] [Google Scholar]
- 35.Elad M, Hel-Or Y. A fast super-resolution reconstruction algorithm for puretranslational motion and common space-invariant blur. IEEE Trans Image Process. 2001;10:1187–1193. doi: 10.1109/83.935034. [DOI] [PubMed] [Google Scholar]
- 36.Farsiu S, Robinson M, Elad M, Milanfar P. Fast and robust multiframe super resolution. IEEE Trans Image Process. 2004;13:1327–1344. doi: 10.1109/tip.2004.834669. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, et al. Wide field-of-view microscope based on holographic focus grid illumination. Opt Lett. 2010;35:2188–2190. doi: 10.1364/OL.35.002188. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Zheng G, Li Z, Yang C. Focal plane tuning in wide-field-of-view microscope with Talbot pattern illumination. Opt Lett. 2011;36:2179–2181. doi: 10.1364/OL.36.002179. [DOI] [PubMed] [Google Scholar]
- 39.Medoro G, et al. A lab-on-a-chip for cell detection and manipulation. IEEE Sens J. 2003;3:317–325. [Google Scholar]
- 40.Clausell-Tormos J, et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Crane MM, Chung K, Stirman J, Lu H. Microfluidics-enabled phenotyping, imaging, and screening of multicellular organisms. Lab Chip. 2010;10:1509–1517. doi: 10.1039/b927258e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou HW, et al. Deformability based cell margination-A simple microfluidic design for malaria-infected erythrocyte separation. Lab Chip. 2010;10:2605–2613. doi: 10.1039/c003873c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.