Abstract
During each molting cycle of insect development, synthesis of new cuticle occurs concurrently with the partial degradation of the overlying old exoskeleton. Protection of the newly synthesized cuticle from molting fluid enzymes has long been attributed to the presence of an impermeable envelope layer that was thought to serve as a physical barrier, preventing molting fluid enzymes from accessing the new cuticle and thereby ensuring selective degradation of only the old one. In this study, using the red flour beetle, Tribolium castaneum, as a model insect species, we show that an entirely different and unexpected mechanism accounts for the selective action of chitinases and possibly other molting enzymes. The molting fluid enzyme chitinase, which degrades the matrix polysaccharide chitin, is not excluded from the newly synthesized cuticle as previously assumed. Instead, the new cuticle is protected from chitinase action by the T. castaneum Knickkopf (TcKnk) protein. TcKnk colocalizes with chitin in the new cuticle and organizes it into laminae. Down-regulation of TcKnk results in chitinase-dependent loss of chitin, severe molting defects, and lethality at all developmental stages. The conservation of Knickkopf across insect, crustacean, and nematode taxa suggests that its critical roles in the laminar ordering and protection of exoskeletal chitin may be common to all chitinous invertebrates.
Keywords: RNAi, nikkomycin, phylogenetic tree, transmission electron microscopy, chitin synthase
During development, insects must undergo periodic molting to accommodate growth and to overcome the rigid constraints imposed by portions of their chitinous exoskeletons (1, 2). This process entails the complete replacement of the entire outer shell of the insect, including digestion; resorption and recycling of the inner, more pliable layers; and shedding of the outer, more highly sclerotized and waterproofed layers, which are either discarded or, in some cases, ingested for further recycling (1–4). The molting process is hormonally initiated by 20-hydroxyecdysone and begins with the epidermis secreting what will become the outer layers of the new cuticle that separate the epidermal layer from the overlying old cuticle. An “apolytic space” then forms, separating new (inner) from old (outer) cuticles (5). With the delicate epidermis now protected by the first layers of new cuticle, the molting fluid in the apolytic space can digest the inner layers of the outer (old) cuticle. According to long-held dogma, protection of the new cuticle from degradation by molting fluid enzymes is conferred by a thin, nonchitinous envelope (previously termed the “cuticulin” layer or the “outer epicuticle”) deposited by the epidermal cells just before the secretion of new cuticular chitin underneath (3). This envelope was believed to form a protective barrier against proteolytic and chitinolytic enzymes of the molting fluid, thereby confining their actions to the proximal layers of the old (outer) exoskeleton while preventing digestion of the newly deposited layers of the replacement cuticle (2, 3).
However, the role of the envelope in preventing inward diffusion of molting enzymes has never been convincingly demonstrated, and molecular determinants that ensure selective degradation of old cuticle while protecting the new cuticle are still a matter of speculation. A finding that chitinolytic or proteolytic enzymes in molting fluid are excluded from the nascent procuticle would lend some support to the view that the envelope forms a barrier for protection of newly secreted layers of chitin or protein. Zheng et al. (6) immunolocalized a chitinase in spruce budworm [orthologous to chitinase 5 of Tribolium castaneum (TcCht-5) (7, 8)] in the molting fluid and the integument but did not determine whether this chitinase was colocalized with chitin in the newly synthesized cuticle. In this study, we refute the venerable notion that access of molting enzymes to nascent cuticle is restricted by the presence of a protective envelope and provide evidence for an entirely unexpected alternative mechanism. We show that the insensitivity of new cuticle to molting enzymes, specifically chitinase, is conferred by the protective effects of the protein Knickkopf (Knk) and that Knk has an additional essential role in the proper organization and layering of the exoskeletal laminae.
Results and Discussion
Colocalization of Chitinase with Chitin in the Newly Synthesized Cuticle.
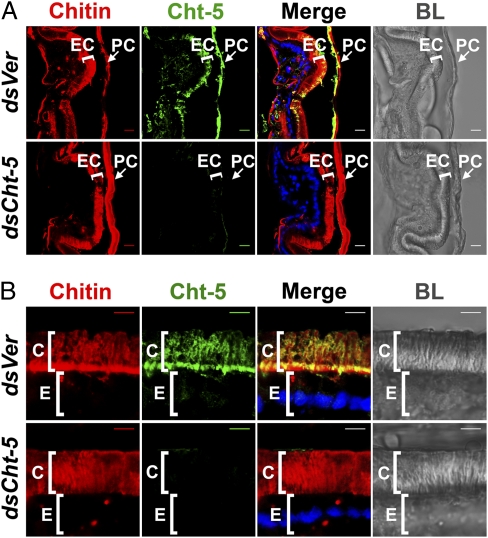
To establish the spatial relation between chitin in the newly synthesized cuticle and molting fluid chitinases, we simultaneously localized both of them within several cuticles of the red flour beetle, T. castaneum, before the adult molt. The results revealed that chitinase is associated not only with the old (pupal) cuticular chitin but also with chitin in the newly formed pharate adult elytral cuticle (Fig. 1A). A closer examination of the newly formed abdominal body wall cuticle (Fig. 1B) showed that chitinase actually permeates the site of new chitin accumulation just outside the epidermal cell layer and throughout the entire procuticle. This unexpected discovery challenges the long-held view of an impermeable envelope that surrounds the new cuticle, limiting accessibility and protecting it from molting fluid enzymes.
Fig. 1.
Chitinase colocalizes with chitin in the newly synthesized cuticle. Cuticular chitin (red) in elytra (A) and lateral (B) abdominal body walls of pharate adults subjected to RNAi for TcVer (dsVer) and TcCht-5 (dsCht-5) were detected with a fluorescent rhodamine-conjugated chitin-binding protein probe. TcCht-5 (Cht-5, green) was immunostained with polyclonal antibodies raised against M. sexta group I chitinase. Bright-field images (BL) were used as a guide to delineate cellular and cuticular regions. EC, new adult elytral cuticle; PC, old pupal cuticle; C, cuticle; E, epithelial cells. (Scale bars: A, 10 μm; B, 5 μm.)
Effect of the T. castaneum Knk (TcKnk) on Molting and Chitin Content.
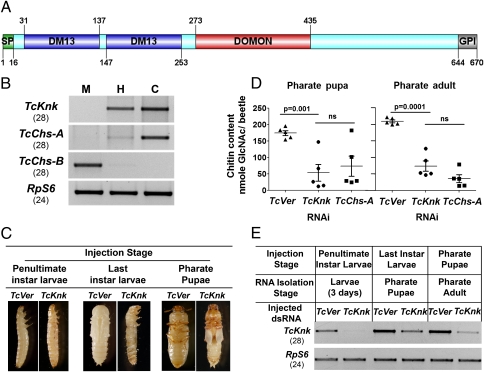
Deletion mutants of the gene knickkopf (knk) exhibit cuticular defects and embryonic lethality in Drosophila melanogaster (9). Knk has subsequently been found to be essential for the normal laminar organization of endocuticle at embryonic stages of growth (10). However, the possible contributions of Knk to postembryonic cuticle morphogenesis were unknown. To investigate these possible aspects of Knk function, we identified its ortholog in T. castaneum and named the gene TcKnk (Fig. S1A). The encoded protein has a domain organization similar to that of D. melanogaster Knk (10, 11) (Fig. 2A and Fig. S2), and its mRNA is expressed throughout all postembryonic stages of development up to the young adult stage, when cuticle deposition is essentially complete (Fig. S1B). Transcripts of TcKnk were observed only in cells of epidermal origin, namely the hindgut and carcass (whole body without gut), but were notably absent in the endodermally derived midgut, which is suggestive of a specialized role for TcKnk in cuticle formation (Fig. 2B). As demonstrated previously with D. melanogaster Knk, recombinant full-length TcKnk protein expressed in an insect cell line can be released into the culture medium by treatment with a phosphatidylinositol-specific phospholipase C (PI-PLC), indicating that it is a GPI-anchored membrane protein (Fig. S3).
Fig. 2.
Domain organization, expression, and role of TcKnk in maintenance of cuticular chitin. (A) TcKnk is composed of an N-terminal signal peptide (SP), two DM13 domains, a dopamine monooxygenase N-terminal like (DOMON) domain, and a C-terminal GPI anchor. Domains were annotated with the SMART database and graphed with the protein-domain illustrator program DOG 1.0.3. (B) Tissue-specific expression pattern of TcKnk in the last-instar larval midgut (M), hindgut (H), and carcass (C) of T. castaneum. TcChs-A and TcChs-B transcripts were used as carcass-specific and midgut-specific markers, respectively, to assess the purity of the tissue preparations, and RpS6 was an internal loading control used to normalize for the differences in the concentrations of the cDNA templates. Numbers in parentheses indicate the cycle number for each of transcripts shown. (C) RNAi was carried out by injecting ∼200 ng of TcKnk dsRNA into penultimate instar larvae, last-instar larvae, or pharate pupae. Representative images of terminal phenotypes are displayed along with controls injected with dsRNA for TcVer (n = 20 each). (D) Quantitative analysis of chitin content from whole animals at pharate pupal and pharate adult stages was carried out by using a modified Morgan–Elson assay. Data are reported as mean ± SE (n = 5). Statistical significance was computed with Student's t test. ns, not significant. (E) At 4 d after injections, the effect of TcKnk dsRNA on transcript levels was determined by collecting dsRNA-treated insects (n = 4) from each treatment at the indicated stage of development. Depletion of transcripts was detected by extracting total RNA from each treatment followed by cDNA preparation and RT-PCR. RpS6 transcript was used as an internal loading control. Numbers in parentheses indicate the cycle number for each of transcripts shown.
Consistent with a putative function in postembryonic cuticle morphogenesis, dsRNA-mediated knockdown of TcKnk transcripts led to lethal molting defects at all stages of Tribolium development (larval–larval, larval–pupal, and pupal–adult molts) (Fig. 2 C and E). Molting defects in TcKnk dsRNA-treated beetles closely resembled those previously observed after dsRNA-mediated knockdown of T. castaneum chitin synthase A (TcChs-A), which is essential for cuticular chitin synthesis (12). Quantitative chemical analysis revealed that insects subjected to RNAi for TcKnk were severely deficient in total chitin, comparable to insects in which chitin synthesis was prevented by RNAi for TcChs-A (Fig. 2D). Examination of cryosections after RNAi for TcKnk confirmed a near-complete loss of cuticular chitin in the body wall similar to that observed after TcChs-A knockdown (Fig. 3 A and B, column 1). These data reveal a crucial role of TcKnk in the deposition or maintenance of chitin in the newly formed cuticle.
Fig. 3.
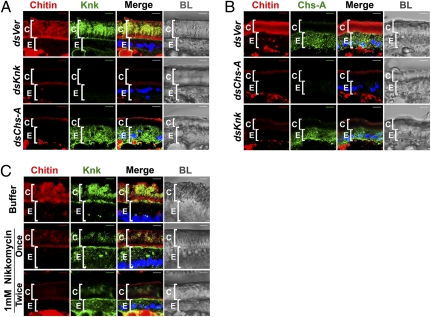
TcKnk is mislocalized after disruption of chitin synthesis. (A) Cryosections of T. castaneum pharate adult lateral body walls (20 μm thick) of control TcVer (dsVer), TcKnk (dsKnk), and TcChs-A (dsChs-A) dsRNA-treated insects were immunostained with D. melanogaster Knk antiserum (green). (B) Distribution of TcChs-A was determined by using anti–TcChs-A antibody (green) in various dsRNA-treated insects. Specificities of the D. melanogaster Knk and TcChs-A antibodies as well as the chitin probe were ascertained by using insects subjected to RNAi for these two genes as controls. The punctate distribution of both TcChs-A and TcKnk within cells suggests vesicular localization. (C) Nikkomycin (1 mM, 0.2 μL per insect) was injected either once at the pharate pupal stage or twice (on days 1 and 3 of the pupal stage) before cryosectioning and immunostaining for TcKnk (green) and staining for chitin (red). Chitin, red; proteins, green; DAPI, blue; C, cuticle; E, epithelial cells. (Scale bars: 5 μm.)
Procuticular Chitin Associates with TcKnk and Dictates Its Cellular Distribution.
Immunostaining of the lateral body wall of pharate adults with polyclonal anti-Knk antibodies revealed a layer of TcKnk at the junction between the inner layer of the endocuticle and the apical surface of the epidermal cells, as expected for a membrane-anchored protein. However, immunostaining was most intense throughout the newly synthesized chitinous procuticle, suggesting that TcKnk becomes incorporated into the cuticle after detachment from the plasma membrane by enzymatic cleavage of its GPI anchor (Fig. 3A, top row). RNAi for TcChs-A prevented synthesis of cuticular chitin as expected but unexpectedly also resulted in the mislocalization of the TcKnk protein, preventing its secretion and incorporation into the newly forming cuticle. Instead, the vast majority of TcKnk was retained within the epidermal cells (Fig. 3A, bottom row). Conversely, down-regulation of TcKnk transcripts did not alter the normal cellular distribution of TcChs-A (Fig. 3B, column 2). Based on these observations, we hypothesized that TcChs-A may facilitate the proper targeting of TcKnk to the new cuticular chitin, either through direct protein–protein interaction(s) or via its product, chitin. To distinguish between these two possibilities, we inhibited chitin synthesis at the pharate adult stage by administering lethal doses of nikkomycin, a structural analog of the UDP-GlcNAc substrate of chitin synthases, and monitoring its effect on TcKnk trafficking. We reasoned that nikkomycin would specifically inhibit the production of chitin by TcChs-A without perturbing any possible protein–protein interactions between TcChs-A and TcKnk. As expected, nikkomycin-treated insects had progressively diminished levels of chitin in the procuticle, whereas the level and cellular distribution of TcChs-A were unaffected (Fig. S4). Importantly, TcKnk was mislocalized (retained) within the epidermal cells (Fig. 3C, column 2), just as we had observed after TcChs-A RNAi. These results suggest that TcChs-A has no direct involvement in the normal secretion of TcKnk or its incorporation into the extracellular cuticular matrix but, rather, that the presence of extracellular chitin itself is a precondition for the normal cuticular localization of TcKnk.
TcKnk Protects Cuticular Chitin from Degradation by Chitinases and Organizes Chitin into Laminae.
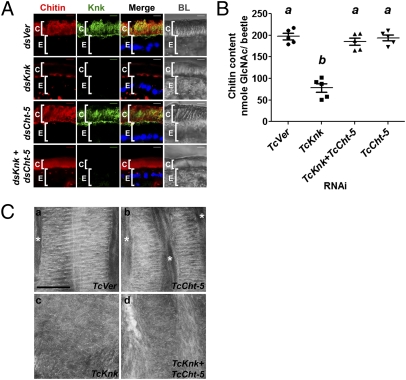
Because the reduction in chitin content brought about by RNAi for TcKnk could not be attributed to altered levels or distribution of TcChs-A (Fig. 3B), we suspected that TcKnk might have a previously unrecognized function in protection of nascent cuticle from molting fluid, a role previously attributed exclusively to the envelope. We performed TcKnk RNAi and assessed whether the resulting chitin depletion could be rescued by simultaneous down-regulation of transcripts for either or both of the two chitinase-encoding genes TcCht-5 and TcCht-10 because these two genes have been shown to be critical for molting and turnover of chitin in the old cuticle (8). Remarkably, in agreement with the above hypothesis, confocal microscopy and quantitative chitin content analysis revealed that chitin levels could be restored to normal when both TcCht-5 and TcCht-10 were down-regulated along with TcKnk (Fig. 4 A and B and Fig. S5 A and B). RNAi of TcCht-5 alone was more effective than down-regulation of TcCht-10 in rescuing dsRNA-TcKnk–mediated chitin depletion, indicating a more critical role for TcCht-5 in chitin turnover (Fig. 4 A, row 4, and B). Transmission electron microscopy of the adult elytral cuticle additionally indicated a role for TcKnk in the organization of cuticular chitin because down-regulation of TcCht-5 and TcCht-10 along with TcKnk failed to rescue the normal laminar architecture of the elytral procuticle even though chitin content was restored to normal levels (Fig. 4C and Fig. S5C). These results suggest that TcKnk is essential for the laminar organization of chitin in the elytral procuticle. Similar results were also observed for the ventral body wall cuticle, which has fewer nascent laminae (Fig. S6).
Fig. 4.
TcKnk aids laminar organization of the procuticular chitin and prevents its degradation by molting fluid chitinases. (A) Chitin depletion resulting from TcKnk RNAi was rescued after double RNAi for TcKnk and TcCht-5 (dsKnk + dsCht-5). Chitin, red; TcKnk, green; DAPI, blue; C, cuticle; E, epithelial cells. (Scale bars: 5 μm.) (B) Analysis of total chitin content by a modified Morgan–Elson assay. Data are reported as mean ± SE (n = 5 each). Statistical significance was computed with Student's t test. Means identified by different letters (a and b) are significantly different at P < 0.05. (C) Ultrastructure of pharate adult elytral cuticle. (a) Elytra from Vermilion dsRNA-treated animals (TcVer) consist of horizontal brick-wall–like chitin laminae that are occasionally separated by vertical channels (*) that contain a fibrous material, probably chitin. (b) Cuticle after TcCht-5 dsRNA treatment is normal. (c) Elytral cuticle of pharate adults treated with dsRNA for TcKnk, by contrast, is disorganized. (d) Concomitant depletion of TcCht-5 along with TcKnk transcripts does not restore the laminar organization of chitin. (Scale bar: 500 nm.)
It is conceivable that TcKnk directly interacts with chitin to enable its laminar organization. To test this hypothesis, recombinant TcKnk (with or without the GPI anchor) was extracted, either directly or by PI-PLC treatment, from the membrane fraction of Hi-5 cells expressing TcKnk and was assayed for binding to colloidal chitin as previously described for Manduca sexta chitinase (13). Both forms of TcKnk bound to colloidal chitin more tightly than did M. sexta chitinase 5, which has a peritrophin A-type chitin-binding domain. Chitin-bound TcKnk could not be eluted with Calcofluor but required treatment with hot SDS for dissociation from colloidal chitin (Fig. S7). The high affinity of TcKnk for chitin, its colocalization with chitin in the cuticle, and its ability to prevent cuticle degradation by chitinases strongly suggest a critical role for this protein in cuticular chitin maintenance during the molting process.
Susceptibility of Old Cuticle to Digestion During Molting Correlates with Reduced TcKnk Immunostaining.
Because the old endocuticular chitin must be partially digested by chitinases to promote shedding of the old cuticle as the exuvia during a molt, one might expect a depletion of TcKnk in the old cuticle at the time of apolysis. Indeed, sections of pharate adults that contain both old pupal cuticle and new elytral cuticle clearly reveal localization of TcKnk predominantly in the new cuticle (Fig. S8). The substantial reduction of TcKnk in the old cuticle indicated by the immunostaining may account for the susceptibility of the old cuticle to molting fluid chitinolytic enzymes. It is unknown what factor or factors account for the disappearance of detectable TcKnk in mature cuticle. Its depletion might be attributable to digestion by molting fluid proteases. In that regard, it is interesting to point out that RNAi of transcripts for a chymotrypsin-like protein that is present in cast cuticles results in molting defects (14). Immunolocalization of the proteolytic enzyme(s) involved in cuticular protein turnover will lend further support to our conclusion that the envelope alone cannot account for the stability of the newly synthesized cuticle in the presence of molting enzymes. An alternative explanation, that cuticle sclerotization reduces the permeability of the cuticle to Knk antibodies, seems unlikely because chitinase 5 can be detected in old pupal cuticle by immunostaining (Fig. 1).
Knk Is Widely Distributed in Invertebrates with Chitinous Extracellular Matrices.
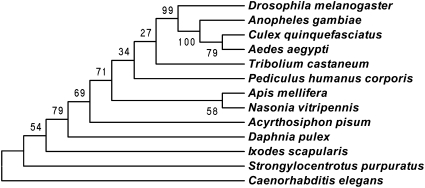
TcKnk orthologs are present in all animal groups examined that secrete chitinous extracellular matrices, including representative species of insects, ticks, crustaceans, echinoderms, and nematodes (Fig. 5). Fungi are the only eukaryotes with chitinous extracellular matrices that lack TcKnk orthologs. All of these putative Knk proteins have the same domain organization, which is suggestive of an essential conserved function in those taxa.
Fig. 5.
Phylogenetic analysis of Knk-like proteins from different taxa. Phylogenetic analysis of Knk homologs from arthropods, including several orders of insect species, sea urchin, and C. elegans as an outlier was done with MEGA 4.0 (16). Bootstrap analysis of 5,000 replications was carried out on trees inferred from the neighbor-joining method. Bootstrap values are shown in the cladogram at the branching points. Drosophila melanogaster, fruit fly (NP_649981.1); Anopheles gambiae, African malaria mosquito (XP_313797.4); Aedes aegypti, yellow fever mosquito (XP_001657332.1); Culex quinquefasciatus, Southern house mosquito (XP_001863519.1); Pediculus humanus corporis, head louse (XP_002430062.1); Tribolium castaneum, red flour beetle (AEM60136.1); Apis mellifera, honey bee (XP_394084.4); Nasonia vitripennis, parasitic wasp (XP_001606352.1); Acyrthosiphon pisum, pea aphid (XP_001950825.2); Daphnia pulex, water flea (EFX88131.1); Ixodes scapularis, deer tick (XP_002407022.1); Strongylocentrotus purpuratus, sea urchin (XP_001187728.1); Caenorhabditis elegans, roundworm (NP_508959.1).
Concluding Remarks
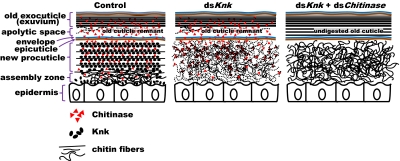
Our work has revealed essential and unexpected roles for TcKnk during the exoskeletal molting process. This protein colocalizes with nascent chitin and organizes it into laminae to confer selective protection of the new cuticle from degradation by chitinases while simultaneously allowing digestion of chitin in the inner layers of the old procuticle that enshrouds it. This discovery casts strong doubt on the long-held hypothesis that an impermeable envelope is sufficient to ensure this selective susceptibility of old versus new cuticle to molting enzymes. A model representing the dynamic changes in chitin degradation and deposition in the old and newly forming insect cuticle, respectively, during molting, including the role of Knk and chitinases, is presented in Fig. 6. The mechanism described herein is probably conserved in all chitinous invertebrates.
Fig. 6.
Model of chitin deposition and degradation in the composite insect cuticle during molting. Note that chitinases are present in both the old exocuticle and the newly forming cuticle. The envelope and epicuticle layers do not exclude chitinases from the chitinous procuticle. Knk protects chitin from chitinases by acting as a chitin-binding protein and masking the chitin. Knk also facilitates formation of laminae by an unknown mechanism. Note the absence of Knk protein in the old exocuticle, presumably as a result of turnover by molting-associated proteases.
Materials and Methods
Tribolium Strains.
The T. castaneum GA-1 strain was used for all experiments. Insects were reared in whole wheat flour containing 5% brewer's yeast at 30 °C under standard conditions as described previously (15).
Identification, Cloning, and Phylogenetic Analysis of TcKnk.
The D. melanogaster Knk protein sequence was used as the query to detect putative homologs in the T. castaneum genome database with the tBLASTn program, resulting in the identification of a presumptive orthologous gene, which we have named TcKnk (GenBank accession no. JN314843.1). TcKnk-specific primers (forward: 5′-GCGGTTCTCGTGTAACTATGTG-3′; reverse: 5′-TCTCAGATGTTTATGGCCTCTCTG-3′) were used to amplify the 2,013-bp-long complete coding sequence of TcKnk using pupal-stage cDNA as template. The amplification product was subsequently cloned into pGEM-T vector (Promega), and its identity was confirmed by DNA sequencing. Sequence alignment of the predicted Knk proteins from different insects as well as other arthropod and nematode species was carried out with ClustalW software before phylogenetic analysis. MEGA 4.0 (16) was used to construct the consensus phylogenetic tree by using the neighbor-joining method.
Determination of Expression Profile of TcKnk.
With the RNeasy Mini Kit (Qiagen), total RNA was prepared from pools of embryos, young larvae, last-instar larvae, pharate pupae, pupae, young adults (0 d old), and mature adults (10 d old). For determining tissue specificity of expression, RNA was extracted from midgut, hindgut, and carcass (whole body without gut) of the last-instar feeding-stage larvae. cDNAs were synthesized from total RNA by using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) as described previously (17). RT-PCR was done to check the expression profile using the gene-specific primer pair (forward: 5′-AAAGATTTCCAAGTGTCGTGG-3′; reverse: 5′-CCAAATAGGCTTTAAAAGTCCG-3′). cDNA amplification product for T. castaneum ribosomal protein S6 (RpS6) was used as an internal loading control for RT-PCR (18).
dsRNA Synthesis.
dsRNAs were prepared as described previously (17). Two unique regions within TcKnk were chosen as templates for the synthesis of gene-specific dsRNAs (Table S1). The dsRNAs were synthesized from linearized templates by using the AmpliScribe T7-Flash Transcription Kit (Epicentre Technologies).
Injection of dsRNA into T. castaneum.
dsRNAs were injected into young larvae, last-instar larvae, and pharate pupae by using a microinjection system and a dissection stereomicroscope as described previously (19).
Measurement of Transcripts for TcKnk After dsRNA Treatment.
Total RNA was prepared from pools of three insects after injection of dsRNA with the RNeasy Mini Kit (Qiagen). cDNA was synthesized from total RNA by using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). RT-PCR with gene-specific primers was carried out to determine the effect of dsRNA injections on level of transcripts. A pair of T. castaneum RpS6 gene primers was used as an internal control to monitor equal loading of cDNA for analysis of transcript levels.
Expression of TcKnk.
A full-length cDNA clone of TcKnk was used as a template to amplify a sequence corresponding to the complete coding region of the TcKnk protein. The primer pair containing appropriate restriction enzyme sites (forward: 5′-GAGACCCGGGATGTTACTACTCTTCGTT-3′; reverse: 5′-GAGATCTAGATTACACGAATTGCTTAAC-3′) was used to facilitate directional cloning of the TcKnk ORF in the pVL1393 expression vector (BD Pharmingen). PCR-amplified TcKnk ORF DNA and pVL1393 vector DNA were digested with the same pair of restriction enzymes and ligated as described previously (7). Recombinant baculovirus for expression of TcKnk protein was constructed with BaculoGold DNA and 1 μg of the recombinant baculovirus transfer vector pVL1393 DNA containing the ORF for the TcKnk protein and amplified to obtain a high-titer virus. Hi-5 cells (Trichoplusia ni cell line) were used to express the TcKnk protein as described previously (7).
PI-PLC Treatment and Western Blot Analysis.
Hi-5 cells were infected with a recombinant baculovirus containing the TcKnk ORF and incubated at 30 °C for 3 d for expression of TcKnk protein. At 3 d postinfection, medium (1 mL) and Hi-5 cells were harvested from each well as a control to check the levels of TcKnk protein at the beginning of the experiment. Subsequently, 1 mL of fresh medium containing 100 μL of PI-PLC from Bacillus cereus (7.89 units/mg; Sigma Aldrich) or medium alone (mock) was added. PI-PLC– or mock-treated cells were incubated at 30 °C for 4 h followed by collection of the medium and cells. Cells were then resuspended in 1 mL of PBS and lysed in sample buffer, and aliquots of the supernatant were analyzed by SDS/PAGE on 4–12% gradient SDS gel (Invitrogen). TcKnk released as a result of PI-PLC treatment was detected by Western blotting using the anti-Knk antibody.
Immunohistochemistry and Confocal Analysis.
dsRNA-treated pupae were collected at the pharate adult stage (day 5) and fixed in 4% paraformaldehyde at 4 °C overnight followed by treatments with a series of solutions with increasing sucrose concentrations (12%, 15%, 18%, and 20% sucrose). Cryosections (20 μm thick) of the fixed pharate adult elytral and body wall samples were stained for TcKnk and TcChs-A proteins by using D. melanogaster Knk rabbit antiserum (1:100) and T. castaneum Chs-A rabbit antiserum (1:50) as primary antibodies, respectively. Alexa Fluor 488 goat anti-rabbit IgG (1:1,000 dilution) was used as secondary antibody for fluorescence detection of the respective proteins. Rhodamine-conjugated chitin-binding probe (1:100 dilution; New England Biolabs) and DAPI (1:15) were used for the chitin and nuclei staining, respectively. Confocal microscopy was performed on a Zeiss LSM 510 META laser scanning confocal microscope equipped with lasers capable of 405 nm, 488 nm, and 543 nm excitation. An oil objective (40×, 1.3 N.A.) with 8× zoom was used for the present study.
Chitin Content Analysis.
dsRNA-treated pharate pupal and pharate adult insects were collected for chitin content analysis. Chitin content was measured with a modified Morgan–Elson method as described previously (12). Insects treated with dsRNA for T. castaneum Vermilion (TcVer) and TcChs-A were used as negative and positive experimental controls, respectively (n = 5).
Colloidal Chitin-Binding Assay.
For colloidal chitin-binding assay, TcKnk expressed in Hi-5 cells was extracted with the Mem-PER Kit per the manufacturer's instructions (Pierce) and referred to as TcKnk with GPI anchor (GPI+). TcKnk without GPI anchor was extracted with PI-PLC (Sigma) from Hi-5 cells expressing TcKnk. Colloidal chitin prepared from crab shell chitin was used for the binding assay at a final concentration of 1 μg/μL (7). The colloidal chitin-binding assay was carried out as described previously (7) with the following modifications. Colloidal chitin (50 μL) was mixed with membrane protein extracts containing ∼100 ng of protein. This mixture was incubated at room temperature for 1 h with end-to-end rotation and then centrifuged for 10 min at 4 °C. The pellet was then washed with 10 mM sodium phosphate buffer (pH 8.0) followed by centrifugation for 4 min. The supernatant was saved as a flow-through fraction for loading on the gel. The relative affinity of TcKnk for the chitin pellet was further tested by two additional treatments of 10 mM sodium phosphate buffer containing 1 M NaCl (pH 8.0), followed by elution with a solution containing 1% Calcofluor, which is known to elute some chitin-binding proteins. Finally, the pellet was resuspended in 50 μL of SDS gel loading buffer and boiled for 10 min. The presence of TcKnk in each fraction was detected by Western blot analysis using the anti-Knk polyclonal antibody and the horseradish peroxidase-conjugated secondary antibody detection system (Bio-Rad).
Transmission Electron Microscopy.
dsRNA-treated insects were collected at the pharate adult stage of development and fixed in 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2–7.4) for 24 h at room temperature with constant rotation. Samples were then washed three times for 5 min each in 0.1 M sodium cacodylate buffer at room temperature with constant rotation followed by postfixation with 1–2% osmium tetroxide in 0.1 M sodium cacodylate buffer at room temperature with constant rotation for 1–2 h. Samples were then washed with 0.1 M sodium cacodylate buffer at room temperature three times for 5 min each with constant rotation and stained with 2% uranyl acetate for 1 h. Samples were rinsed with 0.1 M sodium cacodylate buffer, dehydrated with an ascending acetone gradient (50%, 60%, 70%, 80%, 90%, 95%, and 100%), infiltrated with EMBED 812/Araldite resin (1:1 propylene oxide:resin mixture for 10 min followed by a 1:2 propylene oxide:resin mixture for 20 min and 100% resin for 16 h), vacuum-infiltrated for 1 h, and embedded in flat molds. The resin was cured in a drying oven at 60 °C for 24–48 h. Samples were then trimmed, thin-sectioned (silver to gold section), absorbed onto 200 mesh copper grids, and imaged on a CM-100 transmission electron microscope (FEI).
Supplementary Material
Acknowledgments
We thank Drs. Judith Willis, Hans Merzendorfer, and Vinai Chittezham Thomas for critical reading and editing of this manuscript. We thank Joel Sanneman and Dr. Philine Wangemann for the training and use of the Center of Biomedical Research Excellence (COBRE) Confocal Microfluorometry and Microscopy Core Facility that is supported by Kansas State University and National Institutes of Health Grant P20-RR017686. This work was supported by National Science Foundation Grant IOS-1022227 and Grant DFG MO1714/2-1 (to B.M.). This is contribution 11-366-J from the Kansas Agricultural Experiment Station.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. JN314843.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112288108/-/DCSupplemental.
References
- 1.Chang ES. Comparative endocrinology of molting and reproduction: Insects and crustaceans. Annu Rev Entomol. 1993;38:161–180. doi: 10.1146/annurev.en.38.010193.001113. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds SE, Samuels RI. Physiology and biochemistry of insect moulting fluid. In: Evans PD, editor. Advances in Insect Physiology. Vol 26. London: Academic; 1996. pp. 157–232. [Google Scholar]
- 3.Locke M. The Wigglesworth Lecture: Insects for studying fundamental problems in biology. J Insect Physiol. 2001;47:495–507. doi: 10.1016/s0022-1910(00)00123-2. [DOI] [PubMed] [Google Scholar]
- 4.Merzendorfer H, Zimoch L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003;206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 5.Locke M, Huie P. Apolysis and the turnover of plasma membrane plaques during cuticle formation in an insect. Tissue Cell. 1979;11:277–291. doi: 10.1016/0040-8166(79)90042-9. [DOI] [PubMed] [Google Scholar]
- 6.Zheng YP, et al. Temporal, spatial and induced expression of chitinase in the spruce budworm, Choristoneura fumiferana. J Insect Physiol. 2003;49:241–247. doi: 10.1016/s0022-1910(02)00271-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Q, Arakane Y, Beeman RW, Kramer KJ, Muthukrishnan S. Characterization of recombinant chitinase-like proteins of Drosophila melanogaster and Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:467–477. doi: 10.1016/j.ibmb.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Q, Arakane Y, Beeman RW, Kramer KJ, Muthukrishnan S. Functional specialization among insect chitinase family genes revealed by RNA interference. Proc Natl Acad Sci USA. 2008;105:6650–6655. doi: 10.1073/pnas.0800739105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostrowski S, Dierick HA, Bejsovec A. Genetic control of cuticle formation during embryonic development of Drosophila melanogaster. Genetics. 2002;161:171–182. doi: 10.1093/genetics/161.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moussian B, et al. Drosophila Knickkopf and Retroactive are needed for epithelial tube growth and cuticle differentiation through their specific requirement for chitin filament organization. Development. 2006;133:163–171. doi: 10.1242/dev.02177. [DOI] [PubMed] [Google Scholar]
- 11.Moussian B. Recent advances in understanding mechanisms of insect cuticle differentiation. Insect Biochem Mol Biol. 2010;40:363–375. doi: 10.1016/j.ibmb.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Arakane Y, et al. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol Biol. 2005;14:453–463. doi: 10.1111/j.1365-2583.2005.00576.x. [DOI] [PubMed] [Google Scholar]
- 13.Arakane Y, Zhu Q, Matsumiya M, Muthukrishnan S, Kramer KJ. Properties of catalytic, linker and chitin-binding domains of insect chitinase. Insect Biochem Mol Biol. 2003;33:631–648. doi: 10.1016/s0965-1748(03)00049-3. [DOI] [PubMed] [Google Scholar]
- 14.Broehan G, et al. Chymotrypsin-like peptidases from Tribolium castaneum: A role in molting revealed by RNA interference. Insect Biochem Mol Biol. 2010;40:274–283. doi: 10.1016/j.ibmb.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Beeman RW, Stuart JJ. A gene for lindane + cyclodiene resistance in the red flour beetle (Coleoptera, Tenebrionidae) J Econ Entomol. 1990;83:1745–1751. [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 17.Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc Natl Acad Sci USA. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arakane Y, et al. Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochem Mol Biol. 2011;41:42–50. doi: 10.1016/j.ibmb.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575–578. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.