Abstract
The deleted in liver cancer 1 (DLC1) tumor suppressor gene, which is frequently inactivated in cancer, encodes a Rho-GAP (GTPase activating protein) focal adhesion protein whose negative regulation of Rho-GTPases is necessary but not sufficient for its full tumor suppressor activity. Here, we report that DLC1 forms a complex with two prooncogenic focal adhesion proteins, talin and the focal adhesion kinase (FAK). We identified an 8-aa sequence (residues 469LDDILYHV476) in DLC1 and designated it an LD-like motif, because it shares homology with the LD motifs of paxillin. This motif was necessary for DLC1 binding to talin and FAK, because a DLC1 mutant, from which six of the residues have been deleted, and another mutant carrying amino acid substitutions in three of the residues are deficient for binding both proteins and localization of DLC1 to focal adhesions. FAK binding was independent of talin and vice versa. In bioassays, both DLC1 mutants were less active than wild-type (WT) DLC1, although the ability of the mutants to negatively regulate overall Rho-GTP was not impaired. We conclude that the LD-like motif, which binds talin and FAK, is required for the full tumor suppressor activity of DLC1 and contributes to the association of DLC1 with focal adhesions.
Cancer develops as a multifactorial process that may include the activation of oncogenes and antiapoptotic genes as well as the inactivation of tumor suppressor genes and proapoptotic genes (1). Deleted in liver cancer 1 (DCL1) is a tumor suppressor gene that is inactivated by deletion, mutation, or methylation in a variety of tumors, including lung, breast, and prostate cancer (2, 3). It is the prototypic member of a multigene family, and the two other genes, DLC2 and DLC3, encode closely related proteins. DLC2 and DLC3 have been studied less intensively than DLC1, but the available data suggest that they are frequently down-regulated in tumors (2).
The functional basis for the frequent inactivation of DLC1 in cancer has not been explored in detail. The DLC1 protein localizes to focal adhesions, which can regulate normal and neoplastic cell movement and signaling through mechanisms that are incompletely understood (4). DLC1 contains several motifs, including a Rho-GAP catalytic domain that negatively regulates Rho-GTPases by accelerating their intrinsic GTPase activity. Rho-A and Rho-C may be up-regulated in human tumors (5), and studies of cultured cells have shown that the Rho-GAP activity of DLC1 participates in its inhibition of cell migration and anchorage-independent growth (6). However, although the Rho-GAP function of the DLC proteins seems to be necessary for their full biological activity, it is not sufficient. For example, tumor-associated loss of function mutants have been described that seem to have WT Rho-GAP activity (7), and other Rho-GAPs, such as p190 Rho-GAP, do not seem to be frequently inactivated in cancer.
These observations suggest that DLC1–3 may possess additional activities that contribute to their frequent inactivation in cancer. Consistent with this possibility, DLC1–3 have been shown to bind directly to proteins of the tensin family (8–10), which colocalize to focal adhesions. A DLC1 point mutant deficient for tensin binding displays impaired biological activity in assays of colony formation, cell migration, and anchorage-independent growth. In addition, the Rho-GAP catalytic domain of DLC1 has been found to interact with the SH3 domain of p120Ras-GAP, and this binding can interfere with the Rho-GAP activity of DLC1 (11).
In this communication, we identify the adhesion protein talin (12) and the nonreceptor tyrosine kinase FAK as binding partners for DLC1 and DLC3. We also determine that their binding to DLC1 requires a sequence in DLC1 that has homology to LD motifs in paxillin and other proteins (13, 14) and that mutation of this motif in DLC1 impairs its tumor suppressor activity.
Results
Endogenous DLC1 and Talin Form a Complex in Mammalian Cells.
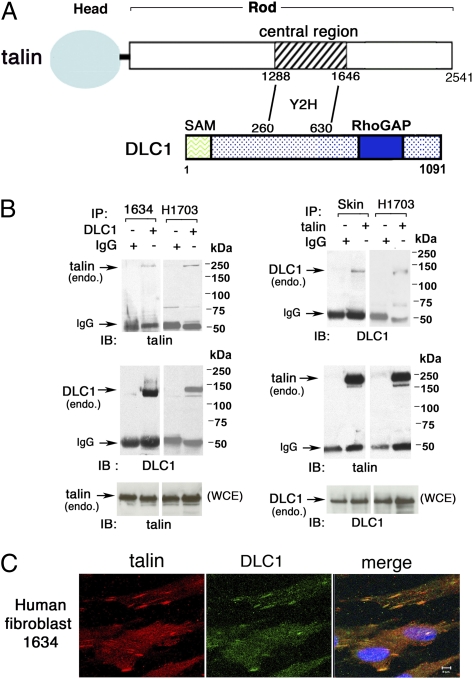
A yeast two-hybrid screen was carried out with a human DLC1 bait fragment encoding amino acids 260–630, which includes many of the amino acids required for the localization of DLC1 to focal adhesions (7), and a human cDNA library as prey. This screen identified an interaction of DLC1 with amino acids 1,288–1,646 of talin in the central part of the talin rod domain (Fig. 1A). To establish that endogenous DLC1 and talin interact in mammalian cells, we first determined that the 1634 human fibroblast cell line and the H1703 nonsmall cell lung cancer (NSCLC) cell line express endogenous DLC1 as well as talin, and then, showed by coimmunoprecipitation that the two proteins form a complex in these cells (Fig. 1B). Immunofluorescent analysis of the 1634 line confirmed colocalization between talin and DLC1 in punctate cytoplasmic structures consistent with focal adhesions (Fig. 1C).
Fig. 1.
Immune complex formation between endogenous DLC1 and talin in human cell lines. (A) Schematic representation of talin and DLC1 proteins and identified yeast two-hybrid (Y2H) interaction between a central region (residues 1,288–1,646) in the talin rod and DLC1 (residues 260–630). (B) Immune complex formation between endogenous talin and DLC1 in human cells was analyzed by reciprocal immunoprecipitation in 1634 fibroblasts and H1703 NSCLC cells (B Left) and human skin fibroblast line and H1703 (B Right). (C) Colocalization of endogenous DLC1 with talin; 1634 cells were stained with anti-talin (red) and anti-DLC1 (green) antibodies. The confocal images are representative of the majority of cells. (Scale bar: 5 μm.)
LD-Like Motif in DLC1 Is Required for Efficient Interaction with Talin.
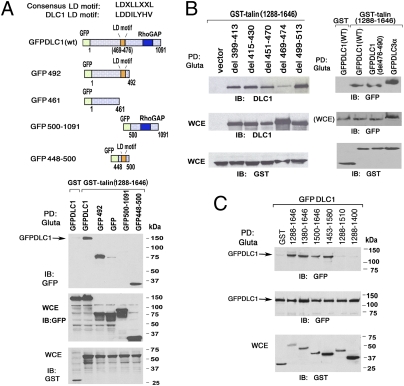
We also verified that the polypeptide from the talin rod domain identified in the yeast two-hybrid system can interact with DLC1 in mammalian cells by determining, in an in vivo pull-down assay, that a GST fusion protein of the talin polypeptide [GST-talin (1,288–1,646)] forms a complex with full-length GFP-DLC1 (GFPDLC1) and DLC3 (GFPDLC3α) in HEK 293T cells (Fig. 2 A and B). To further define the DLC1 sequences required for the interaction, several GFP-DLC1 fragments were coexpressed with GST-talin (1,288–1,646). DLC1 fragments composed of amino acids 1–492 (GFP-492) or amino acids 448–500 (GFP 448–500) formed complexes with the GST-talin polypeptide, whereas the DLC1 N-terminal amino acid 461 (GFP-461) or amino acids 500–1,091 (GFP 500–1,091) were negative (Fig. 2A).
Fig. 2.
The LD-like motif in DLC1 is responsible for the interaction with the central region of talin. (A) Mapping the region in DLC1 required for binding the talin central region. (A Upper) Schematic representation of full-length GFP-DLC1 and its derivatives; the LD-like motif is shown to highlight its location in DLC1 and its presence or absence in the derivatives. (A Lower) After transient transfection of 293T cells, the GFP-DLC1 constructs were pulled down by GST or GST-talin (1,288–1,646) fusion protein followed by immunoblotting (IB) with anti-GFP. The whole-cell extracts (WCE) containing transfected proteins are shown as loading controls. (B) Pull-down assays using DLC1 internal deletion mutants in transfected 293T cells. The WT DLC1 and DLC1 with small internal deletions within or outside the LD motif as well as GFPDLC3α were pulled down by GST or GST-talin (1,288–1,646). (C) Mapping the minimal talin fragment in the central region (1,288–1,646) required for DLC1 binding in pull-down assays. Transfected GST-talin fragments with N- or C-terminal truncations of 1,288–1,646 were used to pull down GFPDLC1. Expression of the transfected proteins is shown as loading controls.
These results indicate that DLC1 amino acids 448–500 are sufficient for talin binding and imply that some residues between 462 and 491 are necessary for binding. Inspection of this latter segment indicated that it contains sequences with homology to LD domains (an LD-like motif), sites of potential protein–protein interaction that have been studied particularly in paxillin (14). The consensus LD motif is LDXLLXXL, where X can be any amino acid; the homologous DLC1 sequence is 469LDDILYHV476 (Fig. 2A Upper). To determine if this DLC1 sequence is involved in the interaction, a mutant was constructed in which the first 6 of 8 aa were deleted in full-length DLC1 (del 469–474; also designated delLD) (Fig. 2B Left). Several other deletions near the LD-like motif were also constructed (del 399–413, del 415–430, del 451–470, del 476–490, and del 499–513), in which the respective delimited amino acids have been deleted. When these mutants were coexpressed with GST-talin (1,288–1,646), the del 469–474 mutant lacking the LD-like motif exhibited markedly reduced interaction with the GST-talin fusion protein, whereas the GST-talin interaction with the other DLC1 mutants was similar to WT DLC1 (Fig. 2B). Therefore, sequences in the LD-like motif are required for efficient binding of DLC1 to talin amino acids 1,288–1,646.
Several GST-talin fusion constructs smaller than 1,288–1,646 were also studied to narrow the amino acids in the talin rod required for interaction with DLC1 (Fig. 2C). When coexpressed with GFPDLC1 in 293T cells, GST-talin (1,380–1,646), GST-talin (1,500–1,646), and GST-talin (1,453–1,580) formed a complex with efficiency similar to the efficiency of GST-talin (1,288–1,646). However, GST-talin (1,288–1,510) and GST-talin (1,288–1,400) were negative, indicating that some talin amino acids between residues 1,510 and 1,580 are required for complex formation.
DLC1 Mutants Deficient in Talin and/or Tensin Binding Retain Rho-GAP Activity but Display Impaired Colocalization with Focal Adhesion Proteins.
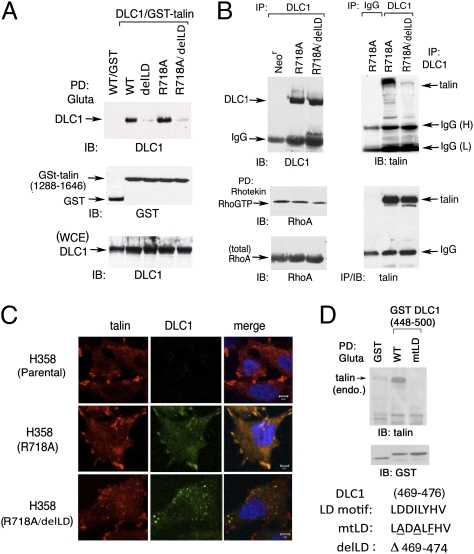
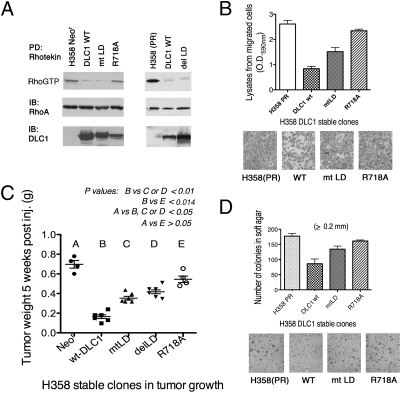
We examined whether the DLC1–talin interaction and their colocalization in focal adhesions were dependent on the Rho-GAP activity of DLC1. For this purpose, we compared a GAP dead DLC1 mutant (R718A) with a compound mutant that also carried the delLD deletion (R718A/delLD). We first determined in 293T cells that, as found previously with WT DLC1 and the delLD mutant, the DLC1 R718A mutant bound GST-talin (1,288–1,646) efficiently, whereas the DLC1 R718A/delLD mutant was deficient for this binding (Fig. 3A). We then confirmed that analogous coimmunoprecipitation results were also seen with endogenous talin when R718A and R718A/delLD were stably expressed at similar levels in the H358 NSCLC line, which is deficient for endogenous DLC1 (Fig. 3B) (10). Consistent with the binding results, the DLC1 R718A/delLD compound mutant colocalized poorly with endogenous talin in H358, which contrasts with the colocalization between talin and the DLC1 R718A mutant (Fig. 3C).
Fig. 3.
The LD mutation but not the GAP dead mutation in full-length DLC1 reduces complex formation and colocalization with endogenous talin. (A) Pull-down assay in transfected 293T cells. The WT DLC1 and the GAP dead DLC1 R718A with or without deletion of LD motif (delLD) were pulled down by GST-talin (1,288–1,646) followed by immunoblotting. The expression of transfected proteins from WCE is shown as loading controls. (B) Complex formation of DLC1 with endogenous talin in stably transfected H358 cells, which do not express detectable endogenous DLC1. Characterization of H358 stable clones expressing R718A with or without delLD: expression, Rho-GTP, and coimmunoprecipitation with endogenous talin. (C) Colocalization of DLC1 with endogenous talin in H358 stable cell lines. Coimmunostaining of endogenous talin (red) and DLC1 (green) in H358 parental cells and stable clones expressing DLC1-R718A with or without delLD mutation. The confocal images are representative of the majority of cells observed. (Scale bar: 10 μm.) (D Upper) Pull down of endogenous talin in 293T cells cotransfected with GST or GST-fusion and DLC1 fragment (448–500) containing WT or mutant LD motif (mtLD) followed by immunoblotting. (D Lower) The conserved WT LD-like motif in DLC1, the three substitution mutations (underlined) in mtLD, and the deleted LD amino acids in delLD are shown.
Additional experiments in 293T, NIH 3T3, and A549 NSCLC cells suggested that the interaction between DLC1 and talin is not required for the DLC1-dependent regulation of Rho-GTP in cells. Both the DLC1 delLD mutant and WT DLC1 reduced Rho-GTP and the Rho-dependent activities of stress fiber formation and myosin light-chain phosphorylation to the same degree, in contrast to the R718A GAP dead mutant (Fig. S1 A–C). A phenotype similar to the delLD mutant was also seen with a DLC1 substitution mutant, where 3 aa in the LD-like motif were replaced with nonhomologous amino acids (mtLD) (Fig. 3D and Fig. S1 A–C).
We previously determined that a point mutant (Y442F) with reduced binding of DLC1 and tensin also contributes to the association of DLC1 with focal adhesions (10). When a compound mutant containing the Y442F and mtLD mutations was tested (Y442F/mtLD), it colocalized with paxillin in focal adhesions less efficiently than either single mutant (Fig. S2). Thus, the interactions of DLC1 with talin and tensin make separate contributions to the association of DLC1 with focal adhesions.
LD-Like Motif Interacts with FAK.
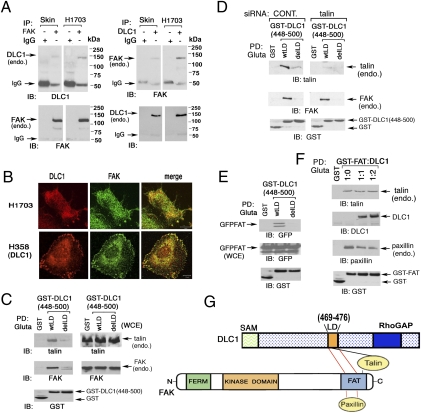
LD motifs in paxillin can interact with several proteins. Here, we examined whether the LD-like motif in DLC1 can form a complex with vinculin or FAK, two ligands that bind some LD motifs in paxillin (14, 15). Although the DLC1 448–500 fragment, which contains the LD-like motif, did not form a detectable complex with endogenous vinculin under the conditions tested (Fig. S3A), it did with FAK (Fig. 4C); the interaction depended on the LD-like motif, because the delLD mutant in the 448–500 fragment or the full-length DLC1 was deficient for complex formation (Fig. 4C and Fig. S3B). In addition, endogenous DLC1 and FAK formed a complex with each other in a human skin fibroblast line and the H1703 line, and they were colocalized in H1703 and H358 cells (Fig. 4 A and B) (H358 was stably transfected with DLC1).
Fig. 4.
DLC1 binds FAK and interferes with paxillin binding to the FAT domain of FAK: dependence on the LD-like motif. (A) Endogenous DLC1 and FAK form a complex in human cell lines. A skin fibroblast line (Skin) and NSCLC line H1703 were analyzed by reciprocal coimmunoprecipitation. (B) Colocalization of DLC1 with FAK in NSCLC lines. Endogenous DLC1 in H1703 cells or transfected DLC1 in H358 cells were stained with anti-DLC1 antibody (red) and endogenous FAK with anti-FAK antibodies (green). The confocal images are representative of the majority of cells observed. (Scale bar: 10 μm.) (C) Association of DLC1 with talin or FAK depends on the LD-like motif. Pull down of endogenous talin and FAK by GST-fusion DLC1 fragment (448–500) with WT LD-like motif or delLD in 293T cells. (D) Interaction between DLC1 and FAK is talin-independent. Pull-down of 293T cells, with or without talin siRNA treatment, of endogenous FAK and talin by DLC1 GST (448–500) WT or delLD. (E) DLC1-FAK interaction is through the FAT-domain of FAK and requires the LD-like motif of DLC1. Pull down of GFP-FAT, derived from FAK, by GST-tagged DLC1 fragment (448–500) with WT LD-like motif or delLD in 293T cells. (F) DLC1 can compete with paxillin but not with talin for binding the FAT-domain. GST-FAT was cotransfected with increasing amounts of DLC1 at the indicated ratios in 293T cells. The pull down of DLC1, endogenous paxillin, and endogenous talin are shown, and the transfected proteins are also shown. (G) Schematic representation of the similar region in the FAT domain of FAK binds paxillin and the LD-like motif, whereas talin binds a distinct region of the FAT domain as well as the LD-like domain.
FAK is known to bind talin (16, 17). To determine whether FAK binding to DLC1 was primarily through an interaction with talin or though its putative direct binding to DLC1, cells were treated with a talin siRNA to reduce endogenous talin. This reduction had only marginal effects on endogenous FAK binding to endogenous DLC1 in H1703 cells, the transfected full-length DLC1, or the 448–500 fragment in 293T cells, although it reduced the level of talin sufficiently to reduce the amount of talin bound to DLC1 (Fig. 4D and Fig. S3 C and D), which strongly implies that most of the FAK–DLC1 interaction is direct.
Paxillin and talin are both reported to bind the so-called focal adhesion targeting (FAT) domain of FAK, although to distinct residues within the FAT domain (16). Given that the LD motifs of paxillin mediate its binding to the FAT domain of FAK and that the LD-like motif of DLC1 binds FAK, we speculated that FAK binding to DLC1 was through the FAT domain of FAK and that at least some of the same FAT residues would be involved in binding paxillin and DLC1. If this speculation were correct, DLC1 would bind to the FAT domain of FAK through the LD-like motif of DLC1, and DLC1 would also compete with paxillin for binding the FAT domain; however, talin would not. It was first verified by pull-down assay with a GST-DLC1 448–500 fragment that it bound the FAT domain of FAK and that the binding depended on the LD-like motif (Fig. 4E). We then determined experimentally that the hypothesized competition between DLC1 and paxillin, but not with talin, was seen when different levels of input DLC1 were cotransfected with a set input of GST-FAT (Fig. 4F). The competition depended on the LD-like motif, because the DLC1 448–500 fragment with the delLD mutation did not compete with paxillin binding to the FAT domain, which contrasts with the WT version of the 448–500 fragment (Fig. S4A). Therefore, DLC1 binds to the FAT domain of FAK through at least some of the FAT sequences that bind paxillin, although it remains possible that other regions of FAK may also interact with DLC1 (Fig. 4G). We also determined that DLC3 binds the FAT domain (Fig. S4B).
LD-Like Motif Contributes to the Biological Activity of DLC1.
The influence of the LD-like motif on the biological activity of DLC1 was studied in the H358 NSCLC line stably expressing mtLD DLC1. Although the Rho-GAP activity in cells expressing mtLD was as high as the activity in the DLC1 WT expressers, the cells expressing mtLD were less active in suppressing cell migration in vitro, xenograft tumor formation in vivo, and anchorage-independent growth in vitro (Fig. 5 A–D). However, they had greater biological activity than the R718A GAP dead mutant in each assay. When a series of DLC1 mutants were tested in NIH 3T3 cells that had been transformed by an activated c-Src mutant (c-SrcY527F), the anchorage-independent growth of mtLD and delLD was less inhibitory than WT DLC1, whereas the anchorage-independent growth of Y442F/mtLD was even less inhibitory than either respective single mutant (Fig. S5). Therefore, the LD-like motif contributes to the biological activity of DLC1 in a human NSCLC line and an oncogene-transformed mouse fibroblast line.
Fig. 5.
Mutant DLC1 is less active than WT in inhibiting the bioactivity of H358 cells in vitro and in vivo. (A) Rho-GAP activity in H358 cells. H358 parental cells and derived DLC1 stable clones were analyzed by Rhotekin pull-down assay. The Rho-GTP level, total Rho-A protein, and expression level of DLC1 in each stable clone are shown. (B and D) H358 parental cells and derived DLC1-expressing cells were assayed by transwell migration (B) and soft agar colony growth (D). The migrated cells and the number of colonies are shown, respectively. (C) Xenographic tumors of H358 cells expressing WT or mutant DLC1. H358 stable clones were injected s.c. into NOD/SCID mice. Tumors were weighted 5 wk later. The results are expressed as means ± SD (tests shown in A and E had four animals, and tests shown in B–D had six animals). Statistical significance was analyzed by the nonparametric Mann–Whitney t test.
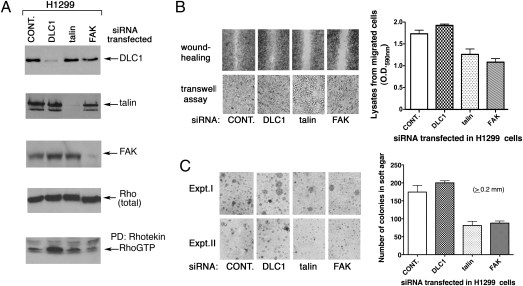
Both FAK and talin are reported to be up-regulated in tumors and prooncogenic proteins (18, 19), and we confirmed, by specific siRNA knockdown of their endogenous levels, that each makes a positive contribution to the cell migration and anchorage-independent growth properties of the H1299 NSCLC line, which contrasts with endogenous DLC1 (Fig. 6 A–C).
Fig. 6.
The DLC1 targets talin and FAK contribute to cell migration and anchorage-independent growth. (A) Verification of siRNA knockdown in NSCLC line H1299. The specific siRNA knockdown of DLC1, talin, and FAK was confirmed by immunoprecipitation/IB of anti-DLC1, anti-talin, and anti-FAK blots. The Rho-GTP level was also analyzed, and the totalt Rho-immunoblo is shown as loading control. (B) Effect of indicated siRNA on cell migration. H1299 cells were analyzed by wound healing and transwell assays 48 h after siRNA transfection. The quantitation of migrated cells in the transwell assay is shown. (C) Effect of indicated siRNA on anchorage-independent cell growth. Equal numbers of siRNA-transfected H1299 cells were seeded in soft agar for growth and quantitation.
Discussion
Recognition that the Rho-GAP activity of DLC1 does not account for the full tumor suppressor function of DLC1 led us to identify a physiologic interaction between DLC1 and two major focal adhesion proteins, talin and FAK, that here join another focal adhesion protein, tensin, as binding partners for DLC1. The region of DLC1 found to be necessary for binding talin and FAK is distinct from the region required for tensin binding, because the LD-like motif is located just downstream from sequences previously identified as being required for the interaction between DLC1 and tensin (9, 10, 20). Our DLC1 mutant analysis indicates that each of these two regions makes an independent contribution to the tumor suppressor activity of DLC1 and its association with focal adhesions, because a compound mutant involving these two regions is more deficient for both of these parameters than mutants of either region alone.
We initially identified the interaction between DLC1 and talin in a yeast two-hybrid assay and confirmed it in mammalian cells. Additional analysis identified a previously unrecognized 8-aa motif, 469LDDILYHV476, in DLC1, with homology to the LD motifs in paxillin and related proteins. This LD-like motif is required for efficient complex formation between DLC1 and talin, because a DLC1 mutant, from which 6 of 8 aa that form the motif were deleted, and a mutant carrying substitution in 3 aa were deficient for complex formation with talin. The ability of FAK to bind some paxillin LD motifs led us to determine that DLC1 also binds FAK and that the DLC1 mutants deficient for talin binding are also deficient for FAK binding. The LD-like motif that is required for these interactions is biologically relevant, because the DLC1 mutants with amino acid deletion or substitution within the LD-like motif suppress cell migration, anchorage-independent cell growth, and xenograft tumor formation less efficiently than WT DLC1; 7 of 8 aa in the DLC1 LD-like motif are conserved in DLC2 and DLC3 (Fig. S6A), and we found that DLC3 also forms a complex with talin and the FAT domain of FAK. Although we have not tested DLC2, the identity between its LD-like motif and the motif of DLC3 implies that talin and FAK will also bind DLC2.
There are several interactions between paxillin, FAK, talin, and DLC1 that we have characterized here. Because each of these proteins is found in focal adhesions, they are likely to form complexes that include at least the four of them (4). Paxillin contains five LD motifs, two of which (LD2 and LD4) bind directly to the FAT domain of FAK through two cooperative paxillin binding subdomains (PBS1 and PBS2) located in the FAT domain (15, 21). Interestingly, although talin also binds the FAT domain of FAK, it binds to sequences other than PBS1 and PBS2 (16). Our data with GST pull-down assays of the FAT domain of FAK strongly suggest that DLC1 binds at least one PBS in FAT, because DLC1 competes with paxillin but not with talin for binding the FAT domain of FAK. Additional confirmation of the direct interaction between DLC1 and FAK comes from the siRNA knockdown of endogenous talin, which was sufficient to reduce the amount of talin bound to DLC1 but only marginally affected FAK binding to DLC1. A direct interaction between talin and DLC1 is strongly supported by the yeast two-hybrid results. In addition, talin amino acids 1,559–1,574, which fall in the region between amino acids 1,500 and 1,580 experimentally identified as being required for DLC1 binding, have some homology to the consensus sequence for binding paxillin LD motifs (Fig. S6B).
The precise mechanism by which the LD-like motif, through its ability to bind talin and FAK, may contribute to the tumor suppressor activity of DLC1 remains to be determined. It seems likely to be related to the stronger localization of DLC1 to focal adhesions or other putative effects resulting from the DLC1 binding of talin, FAK, and possibly other unknown ligands. FAK and talin are reported to be up-regulated in tumors (18, 19), and it is plausible to speculate that binding to DLC1 may attenuate their prooncogenic activity, analogous to what has been shown for the DLC1 binding of tensin (22). The mechanism does not seem to be attributable to overall changes in Rho-GTP, because mutation of the LD-like motif was not associated with an increase in Rho-GTP or Rho-dependent downstream activities. However, it might be attributable to a localized effect on Rho-GTP.
In conclusion, we have identified two prooncogenic focal adhesion proteins, talin and FAK, as binding partners for the tumor suppressor protein DLC1, shown that the binding of both proteins depends on a short motif in DLC1, and determined that this motif contributes to the localization of DLC1 to focal adhesions and its tumor suppressor activity.
Materials and Methods
DNA Constructs, Antibodies, Yeast Two-Hybrid Assay, Cell Culture, and Transfection.
Full-length GFP-tagged mouse talin, a gift from Kenneth Yamada (National Institute of Dental and Craniofacial Research, Bethesda, MD) (23), was used as a PCR template to construct various talin fragments. They were then subcloned into a eukaryotic expression vector, PEBG (24), using BamHI and NotI sites to generate the GST-talin (1,288–1,646) fusion and its N- and C-terminal derivatives shown in Fig. 2C. GFP-tagged DLC1 1–492, 1–461, 400–1,092, 500–1,092, and 448–500 were constructed by PCR and subcloned into a modified pEGFP–C1 vector (Clontech) through Kpn1–NotI sites. The pcDNA3-DLC1 WT, GAP dead mutant R718A, compound talin and tensin binding mutant Y442F/mtLD, and pEGFP-DLC1expression vectors were engineered as previously described (10). Two full-length DLC1 mutants of the 469-LDDILYHV-476 LD-like motif were generated. In one mutant, a substitution mutant was constructed (469-LADALFHV-476; designated mtLD) by site-directed alanine mutagenesis of conserved residues 470, 472, and 474 using overlapping PCR followed by subcloning. In the other mutant (designated delLD), residues 469–474 were deleted by PCR and subcloning. Other DLC1 mutants with small internal deletions (pcDNA3-DLC1 with deletion of 399–413, 415–430, 451–470, 476–490, and 499–513) were made by designing PCR fragments and subcloning them back into the DLC1 expression vector. GST-FAT (amino acids 930–1,052) and GFP-FAT (amino acids 930–1,052) were generated by PCR cloning using a human FAK clone (clone ID: 4812915; Open Biosystems) as template and subcloned into PEBG and pEGFP expression vector, respectively. All these constructs were verified by DNA sequencing. To make the anti–DLC1-specific antibody (428), the cDNA encoding a DLC1 polypeptide (amino acids 82–251) was subcloned into the bacterial expression vector PGEX-4T-1 (Pharmacia) using EcoRI and XhoI, and its encoded GST fusion protein was induced by isopropyl β-d-1-thiogalactopyranoside (IPTG) in bacteria, purified by a Glutathione Sepharose 4B column, and used to immunize rabbits.
In a yeast two-hybrid screen between DLC bait fragments and a human lung cDNA prey library, an interaction was identified between a DLC1 fragment (amino acids 260–630) and a talin fragment (1,288–1,646) (Myriad Genetics). NIH 3T3, HEK 293T (293T), which expresses SV40 mT (25), human fibroblast 1634, and human skin fibroblastic cells were maintained in DMEM containing 10% FBS. NSCLC cell lines (gift from Curt Harris, National Cancer Institute, National Institutes of Health, Bethesda, MD) with endogenous DLC1 expression (H1299 and H1703) or undetectable DLC1 expression (A549 and H358) were cultured in RPMI-1640 supplemented with 10% FBS. Cells were cultured at 37 °C in a humidified 5% CO2 atmosphere. Transient transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Stable clones, derived from H358 and expressing WT or mutant DLC1, were generated by transfection with lipofectamine followed by selection with G418 (0.9 mg/mL).
siRNA Transfection.
Control siRNA and validated siRNAs for human DLC1 (siRNA-5 and -11), human talin (siRNA-5), and human FAK (siRNA-10) were all from Qiagen. The target sequences are 5′-GCCGATGTCGTAATTCCTATA-3′ and 5′-CTGGAGTGTAGGAATTGACTA-3′ for DLC1, 5′-AACAGACCCCTGAAGATCC-3′ for talin, and 5′-CCGGTCGAATGATAAGGTGTA-3′ for FAK. Cells were transfected with siRNAs as indicated for 18–24 h using lipofectamine 2000 followed by a change of media. In some cases, DNA transfection was also carried out after the cells had recovered from siRNA transfection; 2–3 d after siRNA transfection, cells were counted to set up various bioassays. Reduced expression by each siRNA was monitored by immunoblotting with the appropriate antibody, except for DLC1, which was analyzed by immunoprecipitation followed by immunoblotting because of the low endogenous expression level in the H1299 human NSCLC line.
Wound Healing, Transwell Cell Migration, and Soft Agar Assays.
For wound healing, 0.6 × 105 cells/well were evenly plated in duplicate 12-well dishes and subsequently scratched in the center of the well with standard pipette tips followed by a change of media. The migrated cells within the scratched area were observed and photographed at fixed times. Transwell cell migration assays were performed with 6.5-mm-diameter Falcon cell culture inserts (8-μm pore size; Becton Dickinson) precoated with 0.01% gelatin in 24-well cell culture plates. Cells were trypsinized, resuspended in serum-free media, and transferred to the upper chamber (2.0–2.3 × 105 cells in 350 μL depending on cell type), and 900 μL media containing 10% FBS were added to the lower chamber. After incubation for 24 h, cells remaining on the upper surface of the filter were removed with a cotton swab; cells that had migrated to the lower surface were fixed, stained with 1% crystal violet for 10 min, destained, visualized microscopically, and photographed. The migrated cells were then solublized overnight with 1% Triton X-100 (Triton). The collected lysates were quantified colorometrically in a spectrophotomer using OD590. For soft agar colony assays, 1 × 105 cells were mixed with complete medium containing 0.4% agar (Difco) and placed over 0.6% basal agar in 60-mm dishes. Cells were grown for 3 wk, and colonies were photographed microscopically and quantified with a colony counter.
Mouse Tumorigenesis Studies.
The mouse studies were approved by the National Cancer Institute Animal Care and Use Committee and conducted in compliance with the approved protocols. For tumor xenographs, H358 stable clones expressing WT and mutant DLC1 were washed with cold PBS, diluted to 108/mL with serum-free medium/Matrigel basement membrane matrix (Becton Dickinson) at a ratio of 3:1, and injected s.c. into NOD/SCID mice (107 cells/injection). The animals were monitored for tumor growth, and tumor mass was weighed (in grams) 5 wk postinjection.
In Vivo Pull-Down Assay, Coimmunoprecipitation, and Immunoblotting.
The pull-down assay has been described previously (22). Briefly, cells were transiently transfected with plasmids expressing GST, various GST-talin fragments, GST-DLC1 (448–500), GFP-DLC1, or GFP-DLC3 as indicated in the relevant figures; 2 d after transfection, cells were lysed with Golden Lysis Buffer (20 mM Tris, pH 7.9, 137 mM NaCl, 10% glycerol, 1% Triton, 5 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 10 mM NaF, 1 mM sodium pyrophosphate, 0.5 mM β-glyceroposphate, protease inhibitor mixture tablet; Roche). The cleared supernatants were collected, and protein was estimated with a BCA kit (Pierce). For pull-down assays, equal amounts of protein from cell extracts were used, to which 30 μL glutathione Sepharose-4B slurry (GE Healthcare) were added, with rotation for 3 h at 4 °C. The pellets were washed one time with Golden Lysis Buffer, one time with high-salt HNTG (20 mM Hepes, 500 mM NaCl, 0.1% Triton-X-100, 10% Glycerol), and two times with low-salt HNTG (20 mM Hepes, 150 mM NaCl, 0.1% Triton-X-100, 10% glycerol), and they were incubated with Laemmli sample buffer; 85% of each sample was used for detecting the pull-down proteins, including DLC1 and endogenous talin or FAK. The remaining 15% of the pull-down samples was used as a loading control to document the input of GST-fusion protein. For coimmunoprecipitation experiments, equal amounts of protein lysates were precleared with Protein A/G slurry (Pierce) and then incubated with specific IgG antibodies or control IgG matched for subtype. Twenty-five microliters protein A/G slurry were added to each immune reaction and rotated overnight at 4 °C. The immunopellets were washed four times as above. Separation of protein samples by SDS/PAGE was followed by immunoblotting using specific antibodies as indicated. For each blot, HRP-conjugated anti-rabbit or anti-mouse IgG (GE Heathcare) was used for the second reaction at 1:10,000 dilution. Immunocomplexes were visualized by ECL using an ECL kit (GE Healthcare).
Rhotekin-Rho Binding Domain (RBD) Pull-Down Assays.
WT DLC1 and its mutants were transiently transfected in 293T cells. Cell extracts were collected using Magnesium lysis buffer supplemented with 1 mM Na3VO4 and protease inhibitor mixture tablet (Roche Diagnostics). Equal amounts of protein lysates were used for the pull-down assay by Rhotekin RBD agarose (Upstate Biotechnology). The pellets were washed three times with lysis buffer, resuspended in Laemmli sample buffer, and then, separated by 15% SDS/PAGE. Anti–Rho-A antibody (Cytoskeleton) was used to detect Rho-AGTP. The same assays were used for DLC1 stable clones derived from human NSCLC line H358 and the siRNA-transfected H1299 cell line.
Immunofluorescent Staining and Microscopy.
Cells, some of which had been transfected with GFP-tagged human DLC1, were seeded on glass coverslips, incubated for 24 h, and fixed in 2% formaldehyde at room temperature. After rinsing with PBS, cells were incubated with 1:50 anti-DLC1 rabbit polyclonal antibody 428 (as described), anti-paxillin (1:100; BD Biosciences), anti-talin (clone 8d4, 1:25; Sigma), anti-FAK (1:100; BD Biosciences), or anti-phosphomyosin light chain 2 (Thr18/Ser19) antibody (1:100; Cell Signaling) for 2 h followed by incubtion rhodamine (or FITC) -conjugated secondary antibodies (1:200; Jackson ImmunoResearch) for 1 h. Rhodamine-conjugated phalloidin was used at 1:800 dilution. Images were visualized with an LSM510 confocal microscope. For scoring quantitation of colocalization, a minimum of 50 cells of each type was analyzed for colocalization of paxillin and DLC1 by two blinded people, with excellent agreement. If the proteins were predominantly colocalized, the cell was scored as positive. If there were not colocalized, the cell was scored as negative.
Supplementary Material
Acknowledgments
We thank Zhizhong Fei and Lyra Olson for technical assistance, Nicolae Popescu for providing yeast two-hybrid assay information, Curt Harris for NSCLC lines, and Kenneth Yamada for helpful discussions and reagents. This research was supported by the Intramural Research Program, National Institutes of Health, National Cancer Institute, and Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112122108/-/DCSupplemental.
References
- 1.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33(Suppl):238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 2.Durkin ME, et al. DLC-1: A Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim TY, Vigil D, Der CJ, Juliano RL. Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev. 2009;28:77–83. doi: 10.1007/s10555-008-9167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubash AD, et al. Chapter 1. Focal adhesions: New angles on an old structure. Int Rev Cell Mol Biol. 2009;277:1–65. doi: 10.1016/S1937-6448(09)77001-7. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Wong CM, et al. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- 7.Liao YC, Shih YP, Lo SH. Mutations in the focal adhesion targeting region of deleted in liver cancer-1 attenuate their expression and function. Cancer Res. 2008;68:7718–7722. doi: 10.1158/0008-5472.CAN-08-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yam JW, Ko FC, Chan CY, Jin DY, Ng IO. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 2006;66:8367–8372. doi: 10.1158/0008-5472.CAN-05-2850. [DOI] [PubMed] [Google Scholar]
- 9.Liao YC, Si L, deVere White RW, Lo SH. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J Cell Biol. 2007;176:43–49. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian X, et al. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc Natl Acad Sci USA. 2007;104:9012–9017. doi: 10.1073/pnas.0703033104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XY, et al. p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth-suppressing activities. Oncogene. 2009;28:1401–1409. doi: 10.1038/onc.2008.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 13.Brown MC, Curtis MS, Turner CE. Paxillin LD motifs may define a new family of protein recognition domains. Nat Struct Biol. 1998;5:677–678. doi: 10.1038/1370. [DOI] [PubMed] [Google Scholar]
- 14.Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tachibana K, Sato T, D'Avirro N, Morimoto C. Direct association of pp125FAK with paxillin, the focal adhesion-targeting mechanism of pp125FAK. J Exp Med. 1995;182:1089–1099. doi: 10.1084/jem.182.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox BD, Natarajan M, Stettner MR, Gladson CL. New concepts regarding focal adhesion kinase promotion of cell migration and proliferation. J Cell Biochem. 2006;99:35–52. doi: 10.1002/jcb.20956. [DOI] [PubMed] [Google Scholar]
- 17.Hall JE, Fu W, Schaller MD. Focal adhesion kinase: Exploring Fak structure to gain insight into function. Int Rev Cell Mol Biol. 2011;288:185–225. doi: 10.1016/B978-0-12-386041-5.00005-4. [DOI] [PubMed] [Google Scholar]
- 18.Sakamoto S, McCann RO, Dhir R, Kyprianou N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 2010;70:1885–1895. doi: 10.1158/0008-5472.CAN-09-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwock J, Dhani N, Hedley DW. Targeting focal adhesion kinase signaling in tumor growth and metastasis. Expert Opin Ther Targets. 2010;14:77–94. doi: 10.1517/14728220903460340. [DOI] [PubMed] [Google Scholar]
- 20.Chan LK, Ko FC, Ng IO, Yam JW. Deleted in liver cancer 1 (DLC1) utilizes a novel binding site for Tensin2 PTB domain interaction and is required for tumor-suppressive function. PLoS One. 2009;4:e5572. doi: 10.1371/journal.pone.0005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheswohl DM, et al. Multiple paxillin binding sites regulate FAK function. J Mol Signal. 2008;3:1–11. doi: 10.1186/1750-2187-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian X, et al. The Tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell. 2009;16:246–258. doi: 10.1016/j.ccr.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green JA, Berrier AL, Pankov R, Yamada KM. Beta1 integrin cytoplasmic domain residues selectively modulate fibronectin matrix assembly and cell spreading through talin and Akt-1. J Biol Chem. 2009;284:8148–8159. doi: 10.1074/jbc.M805934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anborgh PH, et al. Ras-specific exchange factor GRF: Oligomerization through its Dbl homology domain and calcium-dependent activation of Raf. Mol Cell Biol. 1999;19:4611–4622. doi: 10.1128/mcb.19.7.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient intracellular assembly of papillomaviral vectors. J Virol. 2004;78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.