Abstract
With the potential wide uses of nanoparticles such as carbon nanotubes in biomedical applications, and the growing concerns of nanotoxicity of these engineered nanoparticles, the importance of nanoparticle–protein interactions cannot be stressed enough. In this study, we use both experimental and theoretical approaches, including atomic force microscope images, fluorescence spectroscopy, CD, SDS-PAGE, and molecular dynamics simulations, to investigate the interactions of single-wall carbon nanotubes (SWCNTs) with human serum proteins, and find a competitive binding of these proteins with different adsorption capacity and packing modes. The π-π stacking interactions between SWCNTs and aromatic residues (Trp, Phe, Tyr) are found to play a critical role in determining their adsorption capacity. Additional cellular cytotoxicity assays, with human acute monocytic leukemia cell line and human umbilical vein endothelial cells, reveal that the competitive bindings of blood proteins on the SWCNT surface can greatly alter their cellular interaction pathways and result in much reduced cytotoxicity for these protein-coated SWCNTs, according to their respective adsorption capacity. These findings have shed light toward the design of safe carbon nanotube nanomaterials by comprehensive preconsideration of their interactions with human serum proteins.
Keywords: competitive adsorption, nanoparticle-protein corona, hydrophobic interactions, conformational flexibility
Carbon nanotubes (CNTs) have a broad range of potential applications, including nanoelectronics (1), biosensors (2), biomolecular recognition devices and molecular transporters (3, 4), and cancer therapy and diagnoses (5–7), because of their extraordinary mechanical, electronic, optical, geometric, and biological properties. Both the pharmacological (8, 9) and toxicological (10–15) profiles of CNTs have attracted much attention in recent years. Nanomedicine provides a great potential in fighting many diseases but a global effort is much needed to safely translate laboratory innovations into clinic trials. Seven grand challenges have been proposed for this endeavor (16). CNT-based multimodality imaging and therapy have been proposed for a variety of medical applications, such as carriers for chemotherapeutic drugs (17, 18), Raman detection and imaging (19, 20), photoacoustic imaging, and photothermal therapy of cancers (6, 21). Meanwhile, of equal importance there is the emerging field of nanotoxicology, which aims to address the biosafety of these engineered nanoparticles, in particular, how these nanoparticles interact with human proteins in vivo (22–25).
Many studies have been performed on the biological effects of carbon nanotubes, and the interactions between proteins and CNTs (nanoparticle–protein corona) are believed to play an important role in the biological effects of CNTs (26–28). For example, CNTs could bind with pulmonary surfactant proteins A and D and then lead to susceptibility of lung infection and emphysema in mice (29). When the functionalized CNTs were bound to the α-chymotrypsin, the complex could inhibit enzymatic activity (30). On the other hand, there could be potential positive outcomes as well. Promising uses of nanoparticles include increasing protein stability toward enzyme degradation and increasing the activity of enzymes via immobilization at protein surfaces (31). Interactions of CNTs with some proteins can also enhance their biocompatibility (25) or enable the protein-modified nanotubes to be nontoxic or less toxic than the pristine CNTs (32).
Protein binding to the surface of nanoparticles depends on their surface characteristics, composition, and method of preparation. However, no previous efforts have been made to unveil the underlying mechanism that governs the interaction processes and adsorption capacities of different blood proteins when they competitively bind onto CNT surfaces. In biomedical applications or environmental inhalation exposure of CNTs, our blood circulatory system will most likely be the first interaction organ exposed to these CNTs or CNT-based nanomaterials. Therefore, a better understanding of the interactions between CNTs and blood proteins may help to better clarify the potential risks of CNTs, as well as the related cellular trafficking and systemic translocation. In this study, we employed both experimental [fluorescence spectroscopy, CD, atomic force microscopy (AFM), and NMR spectroscopy] and theoretical approaches (molecular dynamics simulations; refs. 33, 34) to investigate the single-wall carbon nanotube (SWCNT)–protein interactions. We found a surprisingly competitive binding of different blood proteins onto the surface of SWCNTs, with different adsorption capacities and packing modes. The results indicate that these competitive binding behaviors of blood proteins on the SWCNT surfaces are governed by each protein’s unique structure and the amount of hydrophobic residues that each protein contains. These different protein-coated SWCNTs will then have different cytotoxicity by influencing the subsequent cellular responses.
Results and Discussion
AFM Images of Proteins After Incubation with SWCNTs.
High-purity SWCNTs were used and their characteristic details, including impurity, catalyst metal residues, and morphology, are listed in Fig. S1. AFM has been widely applied to monitor interactions between biological species and nanoparticles (35, 36). The AFM images of native proteins were provided in Fig. S2. We found that the adsorption models of SWCNT for different proteins were different and greatly depended on their protein structures (Table S1).
Bovine Fibrinogen (BFG).
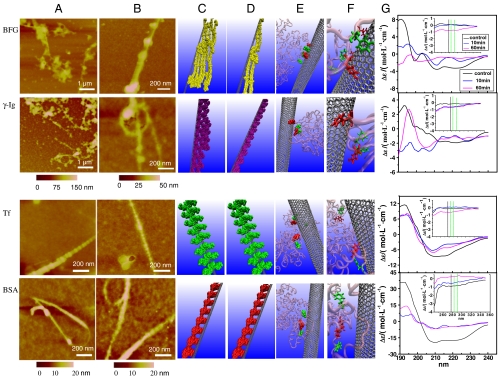
BFG molecules are found to bind to SWCNTs nonuniformly during the initial adsorption process (Fig. 1A, Top), and were prone to interwinding and forming aggregations onto the surfaces of SWCNTs, probably because of their large molecular length and cord-like structure. Accordingly, the CD spectrum of BFG had obvious changes when incubated for only 10 min (Fig. 1G, Top). For example, the α-helices and random coils are significantly reduced, whereas β-sheets are increased (see Table S2). With the increase of the incubation time, these changes in the protein secondary structures did not have much recovery. The nonuniform adsorption of BFG protein molecules might be related to the large contact surfaces of BFG available to SWCNTs (thus might not be in a final thermodynamically stable state yet). After 5 h, BFG protein molecules tended to form a well rdered and thermodynamically stable state (Fig. 1B, Top). Note that the BFG-SWCNT complex shows a node shape, which suggests that the chain of BFG protein tends to enwind along the surface of the SWCNT, and all the studied proteins (except BSA) on the SWCNT surfaces have similar node-like shapes. Very interestingly, the interval (distance) between two neighbor nodes changes regularly with the change of proteins, which may correlate with the number of hydrophobic amino acids on the protein surface (Table S1).
Fig. 1.
Interactions between BFG, Ig, Tf, BSA, and SWCNTs. AFM images of proteins after incubation with SWCNTs for 10 min (A) and 5 h (B). Molecular modeling illustrations for proteins (in beads representation) binding to SWCNTs after incubation for 10 min (C) and 5 h (D). (E) Locations of the most preferred binding sites on proteins for SWCNTs. Residues highlighted in van der Waals representation corresponding to tyrosine colored in red and phenylalanine colored in green. Other parts of protein are presented in transparent pink with new cartoon drawing method. (F) The detailed orientations of aromatic rings of tyrosine and phenylalanine residues interacted to six-member rings of SWCNTs colored in silver. The tyrosine residues are rendered as licorice representation and colored in red, and phenylalanine residues are in green. (G) The far-UV CD spectra of proteins after incubation with SWCNTs and the insets are near-UV CD spectra of proteins incubated with SWCNTs.
Gamma globulin (Ig).
The gamma Ig is also a relatively large globular protein (molecular mass is 150 kDa) and has two antigenic determinants. Similar to the behavior of BFG, Ig molecules bond themselves to the surface of the SWCNT in nonuniform form in the beginning (Fig. 1A, Second Row). Thermodynamically unstable nonuniform adsorption of SWCNT for protein aggregations tended to convert to thermodynamically stable close-packed aggregates on the surface (Fig. 1 A and B, Second Row). After 10 min, the CD spectra of Ig had significant changes (Fig. 1G), with significantly reduced α-helix and increased β-sheets (Table S2). With the increase of incubation time, these changes in protein secondary structure persisted. These results indicate that, in a relatively short period of time (< 10 min), the nonuniform adsorption of Ig and BFG on SWCNTs caused unrecoverable changes in their protein secondary structures.
Transferrin (Tf).
The Tf consists of two similar globular structures and has two binding sites of iron, which are located in the N and C terminus, respectively. During the interaction process, Tf molecules adsorbed onto the SWCNT surface to form a complex aggregate like nodes, which nearly uniformly bound to the surface of the SWCNTs (Fig. 1A, Third Row). With the increase of interaction time, the protein adsorption reached the thermodynamic equilibrium, and the Tf–SWCNT binding model remained unchanged during the succeeding 5 h (Fig. 1B, Third Row). These phenomena were confirmed by the Tf CD spectra, which had no meaningful changes in the succedent incubation time of 5 h (Fig. 1A, Third row), indicating few changes on the secondary structure of Tf (Table S2).
Bovine serum albumin.
The BSA molecule has a smaller and relatively spherical structure, and it is found to uniformly bind to the surfaces of SWCNTs. With more and more BSA molecules absorbing to the SWCNT surface, there was no space on the SWCNT surface to bind with more BSA. Therefore, the later-arriving BSA molecules could only stack onto the earlier ones, and accumulated on the SWCNT surface layer by layer quickly (Fig. 1A, Bottom). Similarly, with the increase of the interaction time, the protein adsorption reached the thermodynamic equilibrium, thus the BSA–SWCNT binding model remained unchanged during the 5 h (Fig. 1B, Bottom). After an incubation time of 10 min, the CD spectrum of BSA also had noticeable changes (Fig. 1G, Bottom) with reduced α-helix and increased β-sheets. However, different from BFG and Ig cases, with the increase of incubation time, these changes in the protein secondary structure had obvious recoveries (see Table S2). These findings indicate that the relatively uniform adsorption of Tf and BSA on SWCNTs caused less and/or recoverable changes in protein secondary structures, which are different from BFG and Ig.
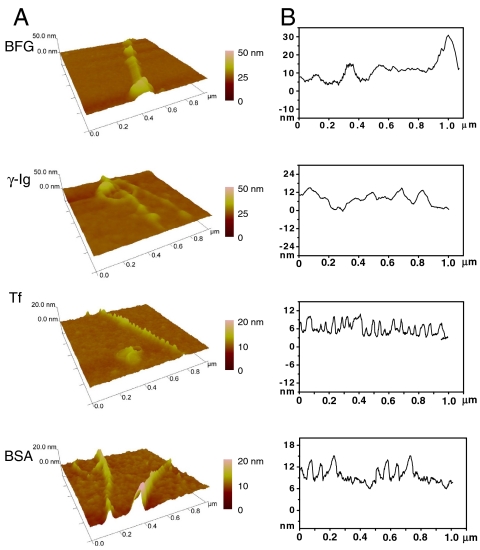
To understand the AFM morphology of the protein–SWCNTs complex for different blood proteins, we measured the number of molecules of each blood protein adsorbed on the surface of one SWCNT. The height profile of protein molecules on the SWCNTs after 5 h of interaction is displayed in Fig. 2. BFG and Ig that have many hydrophobic regions and larger amounts of Trp, Tyr, and Phe amino acids can be attached to the SWCNTs with higher quantities (more discussions below). In fact, they further form thicker layers and more regularly packed and ordered structures, with roughly 15–30 nm in height, on the SWCNT surfaces with the increase of interaction time (Fig. 1 A and B). On the other hand, the ordered structures of Tf and BSA can be formed quickly and be less than 20 nm in height (Figs. 1 and 2) after 5 h, respectively. A single protein molecule on an SWCNT surface was about 2.5 nm in height. The height analysis demonstrated that the numbers of each protein binding to SWCNT surfaces are about five, three, two, and two molecules on average for BFG, Ig, Tf, and BSA, respectively, which seems to show a linear correlation to the number of hydrophobic amino acid on the protein surface.
Fig. 2.
AFM images (A) and height profiles (B) of protein molecules on SWCNTs after incubation for 5 h. The height analysis shows that the numbers of protein molecules on top of SWCNTs are about 5, 3, 2, and 2, for BFG, Ig, Tf, and BSA, respectively.
Molecular Dynamics Modeling of Protein Adsorption by SWCNT.
To elucidate the mechanism of protein adsorption by the SWCNT, up to 150 ns molecular dynamics (MD) simulations, which are widely used in the modeling of both biomolecules (37–40) and nanoparticles (41–44), have been performed for each protein–SWCNT complex in explicit solvent. For illustration, we have also plotted some diagrams to indicate the different bindings of various proteins on the SWCNT at 10 min (Fig. 1C) and 5 h (Fig. 1D). For example, BFG and Ig proteins bound to the surface of SWCNT in a much looser aggregation state in a short incubation time of 10 min (Fig. 1C) than that in a longer incubation time of 5 h (Fig. 1D). This looser state might be thermodynamically unstable and tended to convert to a thermodynamically stable state—i.e., a well-ordered “rod-like” structure as shown in Fig. 1D. However, Tf and BSA protein molecules binding to SWCNT could form a stable well-ordered “node-like” structure even in a short incubation time of 10 min (Fig. 1C) and keep this structure intact for a long time (Fig. 1D).
The snapshots of MD simulation results describe the preferred binding sites on proteins for SWCNT in detail (Fig. 1E). We highlighted the hydrophobic residues Tyr, Phe, and Trp within 5-Å distance of the surface of SWCNT in red, green, and yellow, respectively, in van der Waals representation as the key adsorption hydrophobic residues. Only Tyr and Phe were observed in the adsorption region contact to the surface of SWCNT directly, and Trp residues made little contribution for adsorption in the present MD simulations, which are still short in timescale as compared to experiments (because of the enormous computational power needed to carry out these kind of simulations). Fig. 1F further illustrates the detailed orientations of aromatic rings of Tyr (red) and Phe (green) residues as licorice representation interacted with the six-member ring of the SWCNT in different manners for BFG, Ig, Tf, and BSA, respectively, during the strong adsorption between SWCNT and the surface of these blood proteins. There is a positive correlation between the total numbers of Trp, Tyr, and Phe residues and the adsorption content of the proteins, respectively (Fig. 1 C–F).
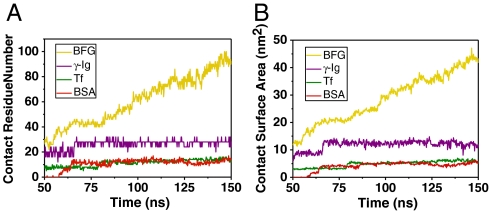
In order to illuminate the above positive correlation relationship more clearly, we calculated the contact residue number and contact surface area of various proteins, in adsorption of SWCNT versus time in Fig. 3. We used the average contact residue number (ACRN) of the protein to quantify its adsorption ability with SWCNT. The ACRN for BFG during 140–150 ns is 94. The ACRN for Ig after its saturation during 70–150 ns is 28. The ACRN for Tf after its saturation during 80–150 ns is 13. The ACRN for BSA after its saturation during 65–150 ns is 12. The relationship of ACRN of various protein complex systems shows an order of BFG > Ig > Tf > BSA, which is compatible with the above simple analysis using aromatic hydrophobic residues. Similarly, we computed the average contact surface area (ACSA) for these blood proteins, which is defined as the difference between the whole solvent-accessible surface area of the protein and the partial solvent-accessible surface area of its exposed portion in the corresponding protein–SWCNT complex. The ACSA for BFG, Ig, Tf, and BSA during the responding time ranges are 43.60, 12.40, 5.81, and 4.68 nm2, respectively, which is in good agreement with the experimental analysis results.† The importance of the hydrophobic residue–CNT interactions has also been found in a previous study on peptides’ confinement within a carbon nanotube using Langevin dynamics simulations (45). Brooks and coworkers had shown that the stability of different helix-forming sequences upon confinement to a carbon nanotube is determined by several factors, including sequence, solvent conditions, strength of nanotube–peptide interactions, and the nanotube diameter (45).
Fig. 3.
The contact residue number (A) and contact surface area (B) of various proteins in adsorption of SWCNT versus time. The ACRN and ACSA of each protein are used to quantify its adsorption ability with SWCNT.
The most abundant Tyr and Phe residues on the surface of the BFG molecule can be attributed to the highest protein quantities binding to SWCNT. Interestingly, the π–cation interactions are also found to play a role, though smaller, in the BFG–SWCNT binding. The average Arg and Lys residues in contact with SWCNT (ACRN-Arg, ACRN-Lys) are found to be about 1.3 and 3.5, respectively (out of the total approximate 94 ACRN above). The other three proteins Ig, Tf, and BSA show similar results, with the (ACRN-Arg, ACRN-Lys) having values of (0.0, 0.2), (1.4, 3.2), (0.1, 2.4), respectively. These findings are consistent with a previous report by Kagan et al., where they found that the interaction sites between human myeloperoxidase and the carboxylated nanotubes were both arginine (π–π-cation interaction) and tyrosine (π–π interaction) residues (46). It should be noted that, in our current classical force field simulations, there is no polarization captured in the simulations, therefore, the π–cation interactions might be underestimated. Nevertheless, both our experiments and simulations show that the π–π stacking interactions are the driving forces for the competitive binding of human serum proteins onto SWCNTs.
Main Factors Controlling the Interactions Between Blood Protein and SWCNTs.
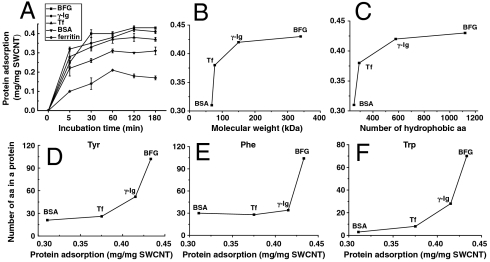
The kinetics curves of protein adsorption on SWCNTs are showed in Fig. 4A. In the first 5 min, the adsorption rate of each protein was on the order of Ig > Tf > BFG > BSA > ferritin, however, shortly after approximately 5 min, the adsorption rate of each protein was on the order of BFG > Ig > Tf > BSA > ferritin. Thus, a detailed investigation of the main factors controlling the interactions between blood proteins and SWCNTs (and resulting adsorption rates) is of great interest. First, a quick analysis of the number of hydrophobic surface residues shows that there is a similar order for these proteins in terms of their hydrophobic surface residues: 88(BFG) > 37(Ig) > 29(Tf) > 20(BSA). These simple analyses and detailed MD simulations both indicate that the serum protein–CNT interactions may be governed by hydrophobic residues within each protein. Note that, because the adsorption capacity and adsorption rate of ferritin to SWCNTs were too low, we did not study ferritin in other tests.
Fig. 4.
The adsorption kinetics and the main factors controlling the protein adsorption on SWCNTs. (A) The kinetics curves of protein adsorption on SWCNTs. The positive correlations between protein adsorption capacity and protein molecular weight (B) or the number of hydrophobic amino acids (C). The positive correlations between protein adsorption capacity and the number of Tyr (D), Trp (E), and Phe (F). The total numbers of (Trp, Tyr, Phe) residues are (70, 102, 104) in BFG; (28, 52, 34) in Ig; (8, 26, 28) in Tf; and (3, 21, 30) in BSA, respectively. Amino acid is abbreviated as “aa” in the figure.
Second, there is a significantly positive correlation between the adsorption amount of protein by SWCNTs and molecular weight or size of protein as shown in Fig. 4B. The overall molecular size is about 6.1 × 8.5 × 7 nm for BSA, 13.2 × 8.9 × 11.8 nm for Tf, 25.3 × 6.5 × 53.5 nm for BFG, and 13.7–27.4 × 16.6 × 6.3 nm for Ig. Thus, the protein volume is approximately on the order of BFG > Ig > Tf > BSA, which is on the same order of the adsorption capacity found above. Although fibrinogen consists of a linear array of three nodules, the carboxyl-terminal region of each of the three chains of fibrinogen is folded independently into a globular domain. The large adsorption capacity of SWCNTs for BFG is probably due to its long size and cord-like structure.
Third, there is a significantly positive correlation between the adsorption amount of protein by SWCNTs and the total number of hydrophobic residues in the protein (Fig. 4C), indicating that not just the surface hydrophobic residues are involved in these interactions, but the inner ones might be involved as well because of CNT-induced protein conformational changes. Among the 20 amino acids, there are nine hydrophobic amino acids, and their numbers in a protein molecule are listed in Table S1. Proteins containing more hydrophobic amino acids were more accessible to binding onto SWCNT surface via hydrophobic interactions, consistent with our MD simulations. Among various hydrophobic amino acids, Trp, Tyr, and Phe contain aromatic rings and can be adsorbed by SWCNTs through π–π stacking interactions. We observed a significant positive correlation between the adsorption content of protein onto SWCNTs and the numbers of Trp, Tyr, and Phe residues in a protein molecule (Fig. 4 D–F). Interestingly, there is also some correlation between the adsorption kinetic efficiency and the degree of aromatic residue burial. For example, protein BFG has the slowest kinetics among all the proteins examined (takes 5 h to form well-ordered rod-like structures), and we find it has the most buried aromatic residues (total 276, with only 88 on surface; see Table S1); meanwhile, BSA has much faster kinetics, and it has only 54 aromatic residues with 20 exposed to the surface. Our MD simulations (Fig. 3) also confirm this finding—during our 150-ns simulations, the contact number and contact surface area of BFG with SWCNT keep increasing, while other ones reach some plateau. The kinetics curves of protein in FBS adsorbed by SWCNTs and protein adsorption on SWCNTs at different concentrations are shown in Fig. S3. We further analyzed semiquantitatively the amount of “free-proteins” in the supernatant and the “released-proteins” from the protein–SWCNT complex using the SDS-PAGE method (Figs. S4–S6) and confirmed the competitive binding of different blood proteins to SWCNT (see detailed discussions in SI Text).
Toxicity Evaluation of Highly Packed Protein-Coated SWCNTs.
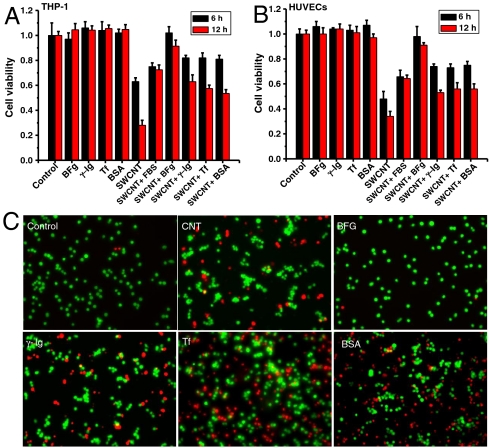
We carried out an additional experiment exploring whether the spontaneous coating of human blood protein influenced the toxicity of SWCNTs in two different human cell lines, human acute monocytic leukemia cell line (THP-1) and human umbilical vein endothelial cells (HUVECs). The results indicated that the highly competitive binding of these proteins on the SWCNT surface can affect the way cells interact and the subsequent cellular responses. Interestingly, we found that any BFG-, BSA-, Tf-, and Ig-coated SWCNTs caused less cytotoxicity than uncoated SWCNTs, whereas BFG-coated SWCNTs were hardly toxic (Fig. 5 A and B). Further fluorescence-based LIVE/DEAD assays showed direct images for dead and live cells after treatment with SWCNTs alone or protein–SWCNT complexes (Fig. 5C), which confirmed the observation in Fig. 5 A and B. In addition, the effects of these SWCNT–protein complexes have similar order to their protein binding capacity, such as BFG > Ig > Tf > BSA, in particular, for THP-1 cells (Fig. 5 A and C).
Fig. 5.
Proliferation and viability of cells in the presence of SWCNTs with/without protein coatings. The differential cytotoxicity of THP-1 (A) and HUVEC cells (B) in 30 μg/mL SWCNTs with/without protein coatings after treatment for 6 and 12 h. (C) The live and dead stains for THP-1 cells after treatment for 12 h (N = 5; error bars are SD).
The reason why BFG-coated SWCNTs have no toxicity might be related to the way the adsorbed proteins rearrange on SWCNTs. Distinct from other proteins, BFG proteins can rearrange themselves on the SWCNT surface with the most compact form and the most layers (five layers as compared to two or three layers for other proteins), which may effectively protect cells from the exposure of SWCNT surfaces. Interestingly, our results also show that cell viability of SWCNTs with fewer BFG proteins adsorbed was obviously less than those with saturated adsorption of BFG (Fig. S7B). When the BFG adsorption was saturated (ca. 12.8 μg/mL BFG), cellular toxicity decreased to levels no longer observable, whereas when the BFG adsorption was nonsaturated (ca. 1.6 μg/mL BFG), the cytotoxicity rises again obviously.
Interestingly, Holt et al. recently found that the dispersed SWCNTs are relatively benign with regard to cytotoxicity (47). The purified and dispersed SWCNTs can induce actin bundling in cells and in purified model actin systems. However, purified and isolated SWCNTs do not induce acute cell death, but cell proliferation is greatly reduced in SWCNT-treated cells with an increase in actin-related division defects, and this subcellular reorganization may cause chronic changes to cellular functions (47). Another very recent work by Holt et al. has shown that BSA can individually disperse SWCNTs at concentrations of up to 0.3 mg mL-1 while retaining SWCNTs’ optical properties; and furthermore, the BSA-dispersed SWCNTs can be uptaken by human mesenchymal stem cells and HeLa cells without apparent acute deleterious cellular effects (48). These important researches have shed light on the biological effects of CNTs at the cellular level. On the other hand, it has been shown previously that the cytotoxicity can depend on different cell lines; for example, Au nanorods have distinct effects on cell viability via killing cancer cells while posing negligible impact on normal cells and mesenchymal stem cells (49).
In general, it is less likely for a protein to be associated and taken up by a cell if it is not recognized by any receptors on the surface of the cell, or if it is bound to nanoparticles without presenting a relevant receptor-binding sequence. After all, cells associate and take up only the proteins they need. A naked particle surface might have much greater nonspecific affinity to the cell surface than a particle hiding behind a corona of bystander proteins (50). However, if a long-lived protein corona that presents the relevant receptor-binding sites is in contact with the cells long enough, they can activate the cell’s uptake machinery in a process known as endocytosis. Effectively, the long-lived proteins give the nanoparticle its biological identity—either by muting the interactions or switching on the cellular machinery (25, 31, 51).
In the complex blood environment, the spontaneous and rapid binding of blood proteins to SWCNTs would have an influence on engulfing and eliminating processes by immune cells in the bloodstream, such as monocytes, platelets, leukocytes, and dendritic cells. The interaction of blood proteins (BSA, Tf, BFG, Ig, etc.) with CNTs may influence their uptake, clearance, distribution, and delivery to the intended target sites, and hence potentially affect their toxicity and biological effects on the organism.
Conclusions
As the blood circulation system is most likely the first entry of nanoparticles in the human body, it is critical to understand how adsorption of blood proteins onto nanoparticles alters the propensity of coated nanoparticles, which in turn associate with other biological interfaces, and thereafter produce cellular responses and other biological effects. Our results demonstrated the interaction processes between SWCNTs and human blood proteins, fibrinogen, immunoglobulin, albumin, transferrin, and ferritin, using both experimental and theoretical approaches. AFM images indicated that the adsorption of Tf and BSA quickly reaches thermodynamic equilibrium in only about 10 min, whereas BFG and Ig gradually packed onto the SWCNT surface in a much longer period of approximately 5 h. Both fluorescence spectroscopy and SDS-PAGE have shown surprising competitive adsorptions among all the blood proteins examined, with a competitive order BFG > Ig > Tf > BSA. CD observation also shows that the protein secondary structure has changed significantly for BFG and Ig, with a decrease in the α-helical content and an increase in the β-sheet structure. In addition, our molecular dynamics simulations on SWCNTs binding with BFG, BSA, Ig, and Tf complexes showed that both the contact residue numbers and binding surface areas exhibited the same order: BFG > Ig > Tf > BSA, in agreement with the experimental findings. Further analysis showed that the π–π stacking interactions between SWCNTs and aromatic residues (Trp, Phe, Tyr) play a critical role in their binding capabilities. The selective binding of these proteins on the SWCNT surface can affect the cellular responses and result in different cytotoxicity. These findings might provide more insight into our understanding of how to preconsider and design the safe surface of nanoparticles in biomedical applications.
Materials and Methods
Materials.
High-purified (purity > 90 wt %) SWCNTs (outer diameter < 2 nm and 5 to approximately 30 µm in bundle length) were purchased from Chengdu Organic Chemical Company, Chinese Academy of Science. BSA was obtained from Solarbio. BFG, Ig, Tf, and ferritin were bought from Sigma-Aldrich. Coomassie brilliant blue R-250 stain was from Bio-Rad. Prestained protein marker was purchased from New England Bio-Labs. Ultrapure water (18.2 MΩ) was obtained from a Milli-Q integral Pure/Ultrapure Water Production unit.
Molecular Dynamics Computational Methods.
The details of the molecular system setup and molecular dynamics simulations are described in the SI Text. In short, we have used NAMD2 to carry out the molecular dynamics simulations on IBM Blue Gene, and used CHARMM force field (c32b1) to describe the protein energy functions. The carbon nanotubes are modeled with (14, 14)-armchair SWCNTs, with carbon atoms having Lennard–Jones parameters εcc = -0.0700 kcal/mol, Rmin/2 = 1.9924 Å. The simulations were performed at constant temperature of 310 K and pressure of 1 atm, with the Particle Mesh Ewald method for long-range electrostatic interactions. The cutoff for the van der Waals interaction was set to 12 Å. All systems were simulated in the NPT (fixed number of atoms N, pressure P, and temperature T) ensemble for more than 120 ns (with time step 2 fs).
In Vitro Cytotoxicity.
Cellular viability was determined by the CCK-8 assay kits (Dojindo Laboratories) as described in specifications. Briefly, about 5,000 cells were plated onto 96-well plates (Corning). After incubation with the indicated dose of phosphatidylserine for different time at 37 °C, reagents of CCK-8 were added following the protocol, and absorbance was recorded at 450 nm. The mean absorbance of nonexposed cells was the reference value for calculating 100% cellular viability. To consistently evaluate the cytotoxicity of pristine SWCNTs and protein-bound SWCNTs, two cell lines, THP-1 and HUVECs (ATCC), were used and cultured in DMEM. Cells were grown to 70% confluence before exposure to each SWCNT sample; each culture plate was incubated in the dark at 37 °C/5% CO2 for 6 or 12 h.
Supplementary Material
Acknowledgments.
We thank the Ministry of Science and Technology of China (2011CB933400 and 2010CB934004), National Natural Science Foundation of China (10975040), and the Chinese Academy of Sciences Knowledge Innovation Program for financial support. R.Z. acknowledges support from the IBM Blue Gene Science Program.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105270108/-/DCSupplemental.
†It should be noted that in the above analysis, we have made some adjustments for BFG and Ig, because we used only one-half of the hexamer for BFG (one of the two sets of three different chains, α, β, and γ) and one-quarter of the tetramer for Ig in our simulations to reduce the computational cost. Therefore, when compared to the experimental results, both the ACRN and ACSA values need to be adjusted, with the BFG values enlarged by a factor of 2 and Ig ones by a factor of 4.
References
- 1.Rutherglen C, Jain D, Burke P. Nanotube electronics for radiofrequency applications. Nat Nanotechnol. 2009;4:811–819. doi: 10.1038/nnano.2009.355. [DOI] [PubMed] [Google Scholar]
- 2.Krauss TD. Biosensors: Nanotubes light up cells. Nat Nanotechnol. 2009;4:85–86. doi: 10.1038/nnano.2008.425. [DOI] [PubMed] [Google Scholar]
- 3.Wilson NR, Macpherson JV. Carbon nanotube tips for atomic force microscopy. Nat Nanotechnol. 2009;4:483–491. doi: 10.1038/nnano.2009.154. [DOI] [PubMed] [Google Scholar]
- 4.Kam NWS, Jessop TC, Wender PA, Dai H. Nanotube molecular transporters: Internalization of carbon nanotube-protein conjugates into mammalian cells. J Am Chem Soc. 2004;126:6850–6851. doi: 10.1021/ja0486059. [DOI] [PubMed] [Google Scholar]
- 5.Liu B, Li X, Li B, Xu B, Zhao Y. Carbon nanotube based artificial water channel protein: Membrane perturbation and water transportation. Nano Lett. 2009;9:1386–1394. doi: 10.1021/nl8030339. [DOI] [PubMed] [Google Scholar]
- 6.De La Zerda A, et al. Carbon nanotubes as photoacoustic molecular imaging agents in living mice. Nat Nanotechnol. 2008;3:557–562. doi: 10.1038/nnano.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Wang H. Nanomedicine: Nanotechnology tackles tumours. Nat Nanotechnol. 2007;2:20–21. doi: 10.1038/nnano.2006.188. [DOI] [PubMed] [Google Scholar]
- 8.Lacerda L, Bianco A, Prato M, Kostarelos K. Carbon nanotubes as nanomedicines: From toxicology to pharmacology. Adv Drug Deliv Rev. 2006;58:1460–1470. doi: 10.1016/j.addr.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Higgins P, Dawson J, Walters M. Nanomedicine: Nanotubes reduce stroke damage. Nat Nanotechnol. 2011;6:121–125. doi: 10.1038/nnano.2011.5. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Xing G, Chai Z. Nanotoxicology: Are carbon nanotubes safe? Nat Nanotechnol. 2008;3:191–192. doi: 10.1038/nnano.2008.77. [DOI] [PubMed] [Google Scholar]
- 11.Jia G, et al. Cytotoxicity of carbon nanomaterials: Single-wall nanotube, multi-wall nanotube, and fullerene. Environ Sci Technol. 2005;39:1378–1383. doi: 10.1021/es048729l. [DOI] [PubMed] [Google Scholar]
- 12.Poland CA, et al. Carbon nanotubes introduced into the abdominal cavity of mice show asbestos-like pathogenicity in a pilot study. Nat Nanotechnol. 2008;3:423–428. doi: 10.1038/nnano.2008.111. [DOI] [PubMed] [Google Scholar]
- 13.Kostarelos K. The long and short of carbon nanotube toxicity. Bioinformatics. 2007;23:2415–2422. doi: 10.1038/nbt0708-774. [DOI] [PubMed] [Google Scholar]
- 14.Ali-Boucetta H, Al-Jamal KT, Kostarelos K. Cytotoxic assessment of carbon nanotube interaction with cell cultures. Methods Mol Biol. 2011;726:299–312. doi: 10.1007/978-1-61779-052-2_19. [DOI] [PubMed] [Google Scholar]
- 15.Lam C, James JT, McCluskey R, Arepalli S, Hunter RL. A review of carbon nanotube toxicity and assessment of potential occupational and environmental health risks. CRC Crit Rev Toxicol. 2006;36:189–217. doi: 10.1080/10408440600570233. [DOI] [PubMed] [Google Scholar]
- 16.Sanhai WR, Sakamoto JH, Canady R, Ferrari M. Seven challenges for nanomedicine. Nat Nanotechnol. 2008;3:242–244. doi: 10.1038/nnano.2008.114. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Sun X, Nakayama-Ratchford N, Dai H. Supramolecular chemistry on water-soluble carbon nanotubes for drug loading and delivery. ACS Nano. 2007;1:50–56. doi: 10.1021/nn700040t. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, et al. Drug delivery with carbon nanotubes for in vivo cancer treatment. Cancer Res. 2008;68:6652–6660. doi: 10.1158/0008-5472.CAN-08-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, et al. Circulation and long-term fate of functionalized, biocompatible single-walled carbon nanotubes in mice probed by Raman spectroscopy. Proc Natl Acad Sci USA. 2008;105:1410–1415. doi: 10.1073/pnas.0707654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng L, et al. Ultralong single-wall carbon nanotubes. Nat Mater. 2004;3:673–676. doi: 10.1038/nmat1216. [DOI] [PubMed] [Google Scholar]
- 21.Moon HK, Lee SH, Choi HC. In vivo near-infrared mediated tumor destruction by photothermal effect of carbon nanotubes. ACS Nano. 2009;3:3707–3713. doi: 10.1021/nn900904h. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Ran T, Li Y, Guo J, Li W. Dependence of the cytotoxicity of multi-walled carbon nanotubes on the culture medium. Nanotechnology. 2006:4668–4674. doi: 10.1088/0957-4484/17/18/024. [DOI] [PubMed] [Google Scholar]
- 23.Chun AL. Toxicology: Food for thought. Nat Nanotechnol. 2006 10.1038/nnano.2006.67. [Google Scholar]
- 24.Wang X, et al. Quantitative techniques for assessing and controlling the dispersion and biological effects of multiwalled carbon nanotubes in mammalian tissue culture cells. ACS Nano. 2010;4:7241–7252. doi: 10.1021/nn102112b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nel AE, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 26.Cedervall T, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linse S, et al. Nucleation of protein fibrillation by nanoparticles. Proc Natl Acad Sci USA. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundqvist M, et al. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci USA. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salvador-Morales C, et al. Binding of pulmonary surfactant proteins to carbon nanotubes; potential for damage to lung immune defense mechanisms. Carbon. 2007;45:607–617. [Google Scholar]
- 30.Zhang B, et al. Functionalized carbon nanotubes specifically bind to α-chymotrypsin’s catalytic site and regulate its enzymatic function. Nano Lett. 2009;9:2280–2284. doi: 10.1021/nl900437n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–47. [Google Scholar]
- 32.Shim M, Kam NWS, Chen RJ, Li Y, Dai H. Functionalization of carbon nanotubes for biocompatibility and biomolecular recognition. Nano Lett. 2002;2:285–288. [Google Scholar]
- 33.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 34.Duan Y, Kollman PA. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 35.Müller DJ, Dufrêne YF. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat Nanotechnol. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 36.Meziani MJ, Sun YP. Protein-conjugated nanoparticles from rapid expansion of supercritical fluid solution into aqueous solution. J Am Chem Soc. 2003;125:8015–8018. doi: 10.1021/ja030104k. [DOI] [PubMed] [Google Scholar]
- 37.Levy Y, Wolynes PG, Onuchic JN. Protein topology determines binding mechanism. Proc Natl Acad Sci USA. 2004;101:511–516. doi: 10.1073/pnas.2534828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snow CD, Nguyen H, Pande VS, Gruebele M. Absolute comparison of simulated and experimental protein-folding dynamics. Nature. 2002;420:102–106. doi: 10.1038/nature01160. [DOI] [PubMed] [Google Scholar]
- 39.Liu P, Huang X, Zhou R, Berne B. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 40.Zhou R, Huang X, Margulis CJ, Berne BJ. Hydrophobic collapse in multidomain protein folding. Science. 2004;305:1605–1609. doi: 10.1126/science.1101176. [DOI] [PubMed] [Google Scholar]
- 41.Zuo G, Huang Q, Wei G, Zhou R, Fang H. Plugging into proteins: Poisoning protein function by a hydrophobic nanoparticle. ACS Nano. 2010:1027–1030. doi: 10.1021/nn101762b. [DOI] [PubMed] [Google Scholar]
- 42.Tu Y, et al. Water-mediated signal multiplication with Y-shaped carbon nanotubes. Proc Natl Acad Sci USA. 2009;106:18120–18124. doi: 10.1073/pnas.0902676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiu P, et al. Manipulating biomolecules with aqueous liquids confined within single-walled nanotubes. J Am Chem Soc. 2009;131:2840–2845. doi: 10.1021/ja804586w. [DOI] [PubMed] [Google Scholar]
- 44.Hummer G, Rasaiah JC, Noworyta JP. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 45.O’Brien EP, Stan G, Thirumalai D, Brooks BR. Factors governing helix formation in peptides confined to carbon nanotubes. Nano Lett. 2008;8:3702–3708. doi: 10.1021/nl8019328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kagan VE, et al. Carbon nanotubes degraded by neutrophil myeloperoxidase induce less pulmonary inflammation. Nat Nanotechnol. 2010;5:354–359. doi: 10.1038/nnano.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holt BD, et al. Carbon nanotubes reorganize actin structures in cells and ex vivo. ACS Nano. 2010;4:4872–4878. doi: 10.1021/nn101151x. [DOI] [PubMed] [Google Scholar]
- 48.Holt BD, Dahl KN, Islam MF. Quantification of uptake and localization of bovine serum albumin-stabilized single-wall carbon nanotubes in different human cell types. Small. 2011;7:2348–2355. doi: 10.1002/smll.201100437. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, et al. Selective targeting of gold nanorods at the mitochondria of cancer cells: Implications for cancer therapy. Nano Lett. 2011;11:772–780. doi: 10.1021/nl103992v. [DOI] [PubMed] [Google Scholar]
- 50.Verma A, et al. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 52.Ge C, et al. Quantitative analysis of metal impurities in carbon nanotubes: Efficacy of different pretreatment protocols for ICPMS spectroscopy. Anal Chem. 2008;80:9426–9434. doi: 10.1021/ac801469b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.