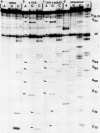
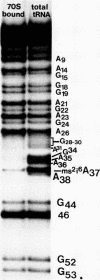
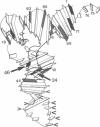
Abstract
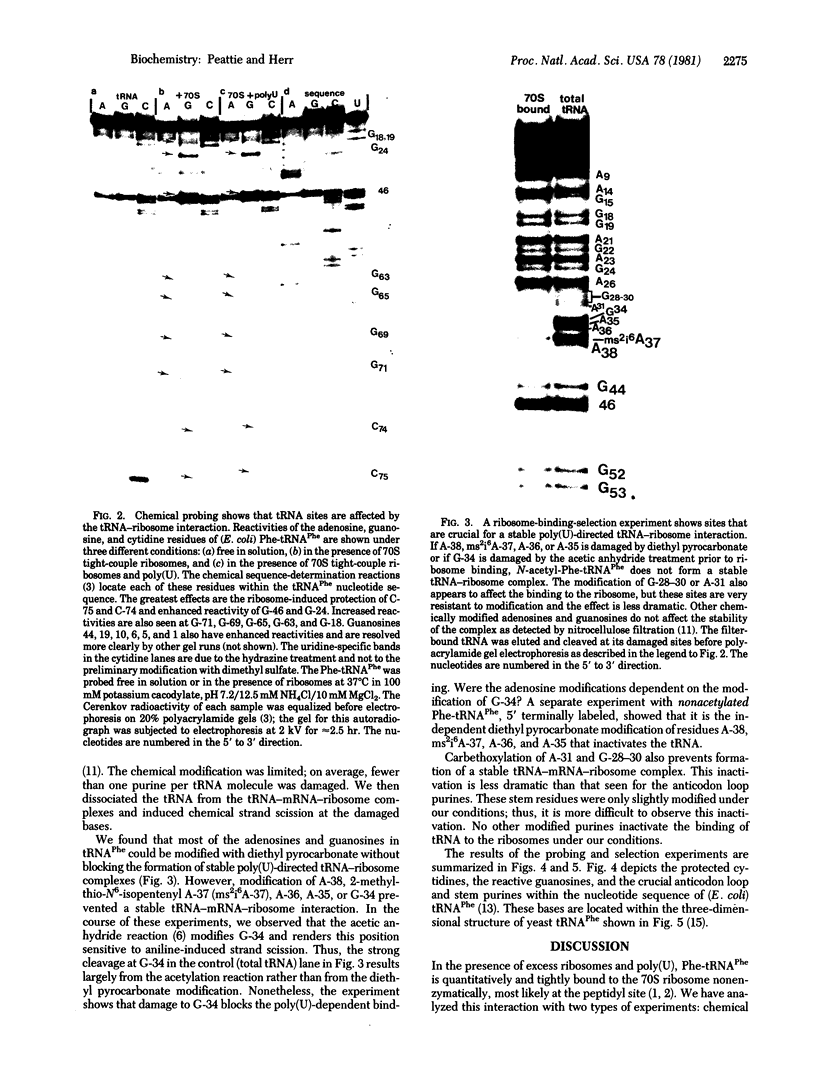
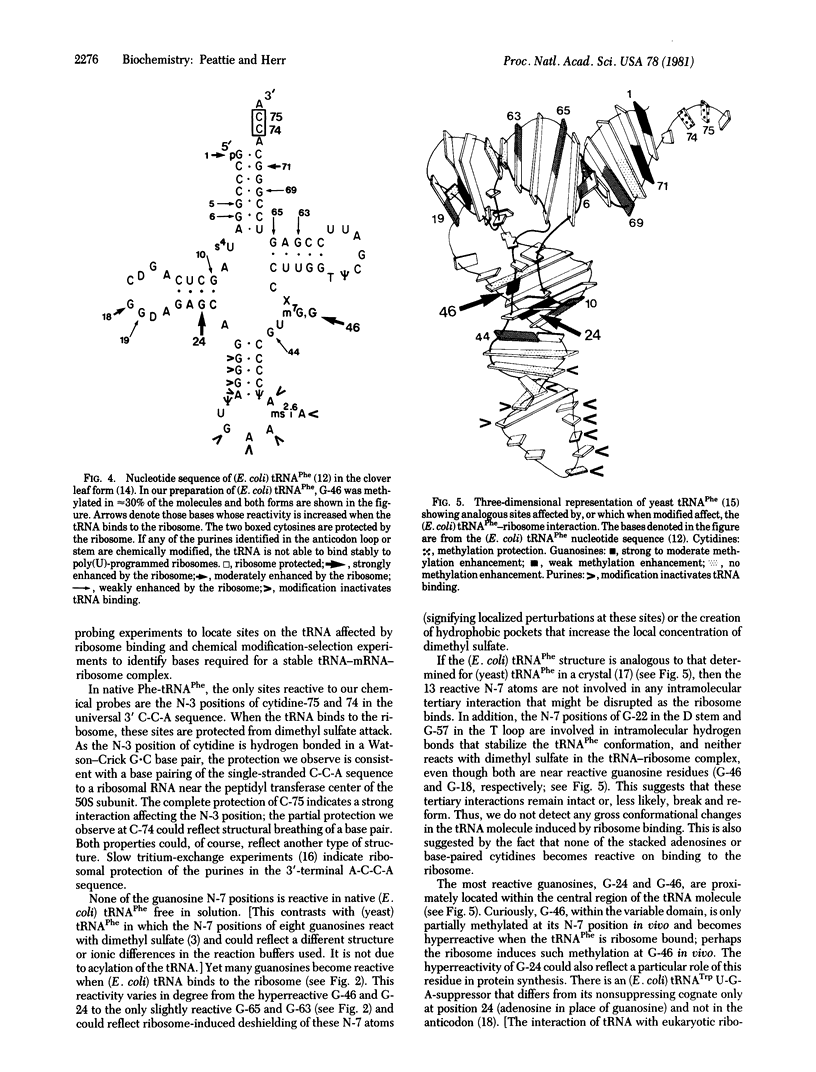
We probed the (Escherichia coli) tRNAPhe--ribosome interaction with the chemical reagents dimethyl sulfate and diethyl pyrocarbonate. This monitored the higher-order structure of the tRNA in this biological complex and identified critical sites in the tRNA molecule involved in binding to the ribosome. The methylation of the N-7 position of guanosine and the N-3 position of cytidine as well as diethyl pyrocarbonate attack on adenosines are sensitive to secondary and tertiary interactions. Here we identify specific bases in E. coli Phe-tRNAPhe affected by the interaction with the ribosome. The 70S ribosome protects the N-3 position of cytidine-74 and 75 in the 3'-terminal C-C-A, suggesting a strong, possibly base pairing, interaction between the ribosome and that universal sequence. The ribosome also induces strong reactivities at the N-7 positions of G-24 and G-46 in the central region of the tRNA molecule near the variable-loop domain as well as less significant reactivities at 11 other guanosines. Two of these, G-10 and G-44, are close to G-24 and G-46 in the center of the molecule; the others (guanosines 1, 5, 6, 18, 19, 63, 65, 69, and 71) are in the coaxial acceptor stem-T stem helix. All of the effects are ribosome induced and occur in the presence or absence of the messenger poly(U). Prior chemical modification of the anticodon bases as well as the two adjacent 3' purines and, less effectively, four purines in the anticodon stem prevent stable poly(U)-directed ribosome binding. Thus, we identify the 3' terminal C-C-A sequence, near the peptidyl transferase site, and the anticodon stem and loop of tRNAPhe as forming critical contacts with the ribosome. Other regions of the molecule become reactive on ribosome binding, but these do not suggest a significant conformational change being more likely due to a change of environment.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrell B. G., Sanger F. The sequence of phenylalanine tRNA from E. coli. FEBS Lett. 1969 Jun;3(4):275–278. doi: 10.1016/0014-5793(69)80157-2. [DOI] [PubMed] [Google Scholar]
- De Groot N., Panet A., Lapidot Y. The binding of purified Phe-tRNA and peptidyl-tRNA Phe to Escherichia coli ribosomes. Eur J Biochem. 1971 Dec 10;23(3):523–527. doi: 10.1111/j.1432-1033.1971.tb01649.x. [DOI] [PubMed] [Google Scholar]
- Fairclough R. H., Cantor C. R., Wintermeyer W., Zachau H. G. Fluorescence studies of the binding of a yeast tRNAPhe derivative to Escherichia coli ribosomes. J Mol Biol. 1979 Aug 25;132(4):557–573. doi: 10.1016/0022-2836(79)90374-7. [DOI] [PubMed] [Google Scholar]
- Farber N., Cantor C. R. Comparison of the structures of free and ribosome-bound tRNAPhe by using slow tritium exchange. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5135–5139. doi: 10.1073/pnas.77.9.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittler F., Hall R. H. Selective modification of yeast seryl-t-RNA and its effect on the acceptance and binding functions. Biochem Biophys Res Commun. 1966 Nov 22;25(4):441–446. doi: 10.1016/0006-291x(66)90225-7. [DOI] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Haenni A. L., Chapeville F. The behaviour of acetylphenylalanyl soluble ribonucleic acid in polyphenylalanine synthesis. Biochim Biophys Acta. 1966 Jan 18;114(1):135–148. doi: 10.1016/0005-2787(66)90261-9. [DOI] [PubMed] [Google Scholar]
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol. 1971 Jun 14;58(2):439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Lucas-Lenard J., Haenni A. L. Release of transfer RNA during peptide chain elongation. Proc Natl Acad Sci U S A. 1969 May;63(1):93–97. doi: 10.1073/pnas.63.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIRENBERG M., LEDER P. RNA CODEWORDS AND PROTEIN SYNTHESIS. THE EFFECT OF TRINUCLEOTIDES UPON THE BINDING OF SRNA TO RIBOSOMES. Science. 1964 Sep 25;145(3639):1399–1407. doi: 10.1126/science.145.3639.1399. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. I. The effect of streptomycin and ribosomal dissociation on 14-C-aminoacyl transfer ribonucleic acid binding to ribosomes. J Biol Chem. 1966 Jan 25;241(2):367–372. [PubMed] [Google Scholar]
- Rich A., Kim S. H. The three-dimensional structure of transfer RNA. Sci Am. 1978 Jan;238(1):52–62. doi: 10.1038/scientificamerican0178-52. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Robertson J. M., Kahan M., Wintermeyer W., Zachau H. G. Interactions of yeast tRNAPhe with ribosomes from yeast and Escherichia coli. A fluorescence spectroscopic study. Eur J Biochem. 1977 Jan 3;72(1):117–125. doi: 10.1111/j.1432-1033.1977.tb11231.x. [DOI] [PubMed] [Google Scholar]
- Ron E. Z., Kohler R. E., Davis B. D. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966 Sep 2;153(3740):1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- Schwarz U., Menzel H. M., Gassen H. G. Codon-dependent rearrangement of the three-dimensional structure of phenylalanine tRNA, exposing the T-psi-C-G sequence for binding to the 50S ribosomal subunit. Biochemistry. 1976 Jun 1;15(11):2484–2490. doi: 10.1021/bi00656a035. [DOI] [PubMed] [Google Scholar]
- Shugart L., Chastain B. m7guanosine in tRNA of Escherichia coli. Int J Biochem. 1979;10(2):155–157. doi: 10.1016/0020-711x(79)90110-1. [DOI] [PubMed] [Google Scholar]
- Thiebe R., Zachau H. G. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem. 1968 Sep 24;5(4):546–555. doi: 10.1111/j.1432-1033.1968.tb00404.x. [DOI] [PubMed] [Google Scholar]
- Wagner R., Garrett R. A. Chemical evidence for a codon-induced allosteric change in tRNALys involving the 7-methylguanosine residue 46. Eur J Biochem. 1979 Jul;97(2):615–621. doi: 10.1111/j.1432-1033.1979.tb13151.x. [DOI] [PubMed] [Google Scholar]
- Watanabe S. Interaction of siomycin with the acceptor site of Escherichia coli ribosomes. J Mol Biol. 1972 Jun 28;67(3):443–457. doi: 10.1016/0022-2836(72)90462-7. [DOI] [PubMed] [Google Scholar]
- Wimmer E., Maxwell I. H., Tener G. M. A simple method for isolating highly purified yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1968 Jul;7(7):2623–2628. doi: 10.1021/bi00847a026. [DOI] [PubMed] [Google Scholar]