Abstract
The ascomycete Hypocrea jecorina, an industrial (hemi)cellulase producer, can efficiently degrade plant polysaccharides. At present, the biology underlying cellulase hyperproduction of T. reesei, and the conditions for the enzyme induction, are not completely understood. In the current study, three different strains of T. reesei, including QM6a (wild-type), and mutants QM9414 and RUT-C30, were grown on 7 soluble and 7 insoluble carbon sources, with the later group including 4 pure polysaccharides and 3 lignocelluloses. Time course experiments showed that maximum cellulase activity of QM6a and QM9414 strains, for the majority of tested carbon sources, occurred at 120 hrs, while RUT-C30 had the greatest cellulase activity around 72 hrs. Maximum cellulase production was observed to be 0.035, 0.42 and 0.33 µmol glucose equivalents using microcrystalline celluloses for QM6a, QM9414, and RUTC-30, respectively. Increased cellulase production was positively correlated in QM9414 and negatively correlated in RUT-C30 with ability to grow on microcrystalline cellulose.
Keywords: Trichoderma reesei, cellulase, growth characterization, carbon sources
Introduction
The ascomycete Hypocrea jecorina (anamorph Trichoderma reesei) is one of the most studied and industrially important cellulolytic fungi. T. reesei is a saprobic fungus capable of efficiently degrading plant cell wall polysaccharides such as cellulose and hemicelluloses [1]. It was first isolated in Solomon Islands in 1944 by the US Army [2]. The original T. reesei isolate known as T. reesei QM6a soon became an area of great interest, because of its high cellulolytic potential. For many years, random mutagenesis has been applied to improve cellulolytic activity of the strain for its industrial application. The creation of different mutant strains with several-fold increase in the amount of secreted cellulolytic enzymes compared to the wild-type strain has been achieved by both academic and industrial research programs [3, 4].
One of the most hypercellulolytic strains, RUT-C30, originated from T. reesei QM6a by three rounds of mutagenesis including a UV-light treatment followed by N-nitrosoguanidine (NTG) and another round of UV-light treatments [5-7]. Two genetic changes were previously shown in this mutant strain. The first one was a truncation of the cre1 gene encoding CRE1, the carbon catabolite repressor protein that renders the mutant strain carbon catabolite derepressed [8]. The second mutation was a frameshift mutation in the glycoprotein processing β-glucosidase II encoding gene [9]. Recently, massive parallel sequencing of two mutant strains including RUT-C30 and its ancestor NG-14 (originated from T. reesei QM6a) have been reported. These studies have identified higher number of mutagenic events including 223 single nucleotides variants (SNVs), 15 small deletions or insertions, and 18 larger deletions, leading to the loss of more than 100 Kb genomic DNA. These led to mutations in 43 genes mainly involved in nuclear transport, mRNA stability, transcription, protein secretion and metabolism [5-7]. These genetic changes altered the phenotype of RUT-C30 and, for example, it is unable to grow on α-linked oligo- and polysaccharides because of the loss of the maltose permease gene. This alteration makes RUT-C30 an ineffective candidate for cellulolytic enzyme production on carbon sources containing starch and other α-linked glycans [5]. Conversely, RUT-C30 shows an enhanced growth rate on a number of simple carbon sources such as glycerol, D -fructose, D-mannitol and D-mannose which act as catabolite repressing carbon source in T. reesei [5]. This alteration may be caused by the loss of CRE1 function in the mutant strain. Recent studies using array comparative genomic hybridization (aCGH) identified 17 new mutations in RUT-C30 and its ancestor NG-14 [10].
The other well-known hypercellulolytic strain T. reesei QM9414 also originated from T. reesei QM6a; in this case, the two rounds of mutations were included by a particle accelerator [5]. However, the nature of the mutations leading to the mutant strain have never been elucidated [11]. The mutant T. reesei QM9414 strain created by irradiation mutagenesis produces two to four times more cellulases than QM6a. Electrophoretic karyotyping of several T. reesei strains showed that chromosomal rearrangements have occurred in many strains, and in QM9414 the size of the smallest chromosome is different from that of QM6a [10, 12]. Using aCGH, over forty new identified mutations (various types) have been recently reported in QM9414 and/or its ancestor QM9123 [10].
In order to better understand cellulolytic potential of T. reesei, the strain QM6a, with a genome of approximately 34 MB, was subjected to the genome sequencing by US Department of Energy Joint Genome Institute [13]. Although T. reesei is well-known for its hypercellulolytic potential, based on the sequencing results, it had the fewest cellulases among the analyzed fungal genomes. T. reesei produces seven cellulases in total including two exoglucanases and five endoglucanases in addition to a small set of hemicellulases and pectin degrading enzymes [6]. Additionally, T. reesei produces two β-glucosidases to hydrolyze cellobiose (end-product of cellulases) to glucose [14, 15].
The regulation of cellulolytic enzymes expression by hypercellulolytic T. reesei is a complex process and our knowledge is still incomplete. Expression of the cellulolytic enzymes in T. reesei is induced by cellulose and also by some disaccharides such as D-lactose, cellobiose and sophorose whereas the presence of preferred carbon sources such as D-fructose and D-glucose antagonized the induction effect [16, 17]. This is the case because most T. reesei cellulases are adaptive enzymes, meaning their transcripts are not formed during growth on monosaccharides, and their full expression requires the presence of an inducer. Although there are many theories regarding the regulation of cellulases, all of them agree that the actions of cellulases lead to the formation of a cellulase inducer [18]. The need for an inducer to stimulate cellulase gene expression represents tight regulation of the respective promoters. So far, three positive transcriptional activators (XYR1, ACE2 and the HAP2/3/5 complex) as well as two repressors (ACE1 and the carbon catabolite repressor CRE1) have been identified to be involved in the complex regulation of cellulases production by T. reesei [18].
One approach to better understand the biology underlying cellulase hyperproduction is the analysis of improved T. reesei mutant strains as compared to the wild-type T. reesei QM6a. Although the factors that regulate the production of cellulase by T. reesei have been previously studied, we systematically determined the effect of different carbon sources on cellulase production by three important T. reesei strains [19, 20]. Because all the experimental conditions were identical, our results provide a more meaningful comparison between different strains and different carbon sources. In order to expand our understanding of the conditions for the production and activity of cellulases by T. reesei, we investigated the effect of 14 different carbon sources (7 soluble and 7 insoluble including 4 pure polysaccharides and 3 lignocelluloses, Table 1) on three different T. reesei strains including QM6a (wild-type) and its two mutant strains QM9414 and RUT-C30. We also analyzed the appropriate cultivation time for highest cellulase activity for each strain growing on different carbon source. We further investigated the effect of different concentrations of the selected carbon sources on cellulase production. Furthermore, the effect of different carbon sources on the growth of the different strains is reported in this paper.
Table 1.
List of 14 different carbon sources tested for their effect on cellulase production. MA-medium without carbon source was used as the control
| Type | Carbon Source (1%, w/v) | Abbreviation |
|---|---|---|
| Soluble | D-Glucose | Glc |
| D-Xylose | Xyl | |
| D-Lactose | Lac | |
| Cellobiose | CB | |
| Malt extract | ME | |
| Carboxymethyl cellulose | CMC | |
| Potato dextrose a | PD | |
| Insoluble | ||
| Pure polysaccharides | Xylan (oat spelt) | Xylan |
| Cellulose powder (cotton linters) | CP | |
| Microcrystalline cellulose | ||
| Microcrystalline cellulose (J.T. Baker) | MCC | |
| Avicel PH-101(Sigma Aldrich) | Avi | |
| Lignocelluloses | Pine sample | PS |
| Organosolv pretreated pine sample | OrS | |
| Paper-mill sludge | Slu |
Becomes soluble in water when heated.
Materials and methods
Chemicals
All the chemicals and reagents were of analytical grade, and were obtained from Sigma-Aldrich (Sigma-Aldrich Canada Ltd). Microcrystalline cellulose was obtained from J. T. Baker (Phillipsburg, NJ, U.S.A.). Paper mill sludge (obtained from St. Marys Paper Corp., Sault Ste Marie, Canada) was grounded in a blender then dried at 70 °C and kept for later use. Grounded pine sample and organosolv pretreated pine samples were kindly provided by Dr. Charles Xu (Lakehead University, Canada).
Strains and culture conditions
Three different strains of T. reesei including QM6a (wild-type, ATCC13631, kindly provided by Dr. Monika Schmoll, Vienna University of Technology, Austria), and its two hyperproducing mutants QM9414 (ATCC 26921, kindly provided by Dr. Tianhong Wang, Shandong University, China) and RUT-C30 (ATCC 56765, kindly provided by Dr. Xiaobin Yu, Jiangnan University, China) were used in cellulase production experiments. The strains were grown and maintained on potato dextrose agar (PDA) containing 15.0 g/L starch, 20.0 g/L D-glucose, and 18.0 g/L agar [21]. Strains were grown in 250 mL flasks, on a rotary shaker (200 rpm) at 30 °C, and in a 50 mL of medium described by Mandel and Andreotti (MA) [22] with the respective carbon source at a final concentration of 1% (w/v). The media containing the respective carbon sources were autoclaved at 121 °C (15 lb psi) for 15 min.
Inoculum preparation
After 14 days of incubation at 30 °C, the greenish conidia were suspended in 5 mL of sterile saline solution (0.9% w/v, NaCl). The spores were separated from the mycelium by gentle filtration through 12 layers of lens paper (Fisher Scientific, Canada), and spores were counted using a Petroff-Hausser cell counter. The isolated spores were added at 1.0 × 106 (final concentration) to 250 mL flasks containing 50 mL of MA-medium with the respective carbon source at a final concentration of 1% (w/v). Three biological replicates were run for each carbon source.
Carbon source for cellulase production
To test the effect of different types of carbon sources on the mycelia growth and cellulase enzyme production, 14 different carbon sources (Table 1) were used and incorporated into MA-medium at 1% (w/v). To examine total cellulase activity, a time course trial was conducted similar to that of Cianchetta et al (2010) [23]. Specifically, the flasks were incubated at 30 °C in an Innova 44 (New Brunswick Scientific, USA) incubator for a total of 120 hours. Samples of 500 µL each were taken from each of the flasks every 24 hrs in a 1300 Series A2 biosafety cabinet (Thermo Fisher Scientific, Canada) to maintain sterile conditions. These 500 µL samples were centrifuged at 13000 rpm for 5 min, and the supernatant was used as the source of enzyme [21].
Concentration of the carbon source
For each strain, the carbon source with the highest level of reducing sugar production was selected for further study. The concentration of this carbon source was varied from 0.25 to 2% (0.25, 0.5, 0.75, 1.0, 1.25, 1.5, 1.75 and 2.0%), and both the reducing sugar production (Section 2.7), and fungal growth (Section 2.6) were recorded at the time point of maximum activity (obtained from carbon source for cellulase production, Section 2.4).
Determination of fungal growth
To determine mycelia growth, agar plates were inoculated with a small piece of agar in the center of an 8-cm plate, containing MA-agar medium with the respective carbon source at a final concentration of 1% (w/v). The increase in colony diameter was monitored daily, and pictures were taken after 4 days. To measure growth in submerged cultures, dry biomass was recorded at the end of the study. Mycelia were harvested after the experiment was completed —usually 120 hrs—and washing extensively with distilled water. Then they were dried to constant weight in a 70 °C oven. This procedure was carried out for all three biological replicates.
Filter Paper Assay (FPA) and determination of reducing sugar content
A microplate based filter paper assay, similar to the one described by Xiao et al (2004), was carried out to measure the total cellulase activity [24]. The method is 25-fold scale down of the IUPAC protocol for FPA assay [25, 26]. In short, 20 μL of the undiluted cell free culture supernatant was added to 40 μL of 75 mM citrate buffer for a final volume of 60 μL. As noted by Cianchetta et al. (2010), the 75 mM buffer was diluted to the desired 50 mM by the enzyme solution [23]. The final reaction volume, citrate buffer concentration and pH were 60 μL, 50 mM and 4.8, respectively. Each of these samples was placed in a well containing a 6 mm diameter filter paper disk (Whatman No. 1, with average weight of 3.0 mg each, ThermoFisher Scientific, Canada) cut using a standard office hole punch. Reagent blank containing 60 μL of 50 mM citrate buffer and substrate control containing only the filter paper, and 60 μL of 50 mM citrate buffer were also run [27]. To exclude the background of reducing sugars found in the enzyme supernatant from the results, a negative control was run with no filter paper. The absorbance of the no filter paper sets and substrate control were subtracted from the absorbance of the activity assay. A glucose standard curve with a range of 0 to 2 mg/mL was run once, and two standards at 0 and 1.5 mg/mL were run in all subsequent trials. All of the samples and the standards were run in triplicate, while the blanks were run in duplicate.
The microplates were sealed with paraffin wax, covered in a Ziploc bag and incubated at 50 °C in water bath for 60 min. To measure the released reducing sugar, 120 μL of 3,5-dinitrosalicylic acid (DNS reagent) was added, and the plate was resealed with paraffin [28]. Boiling in water bath for 5 min developed the color. A 36 μL aliquot was transferred to a new 96-well flat-bottom microplate containing 160 μL of distilled water, and the absorbance at 540 nm was measured using an xMark Microplate Spectrophotometer (Bio-Rad, Canada). Total reducing sugars generated during the assay was estimated as glucose equivalents. To calculate glucose equivalents, the absorbance of the samples was converted into a concentration using the standard curve, once the negative control and the reagent blank were subtracted. This concentration was converted into glucose equivalents using the assay volume and the molar mass of glucose.
Data processing and statistical analysis
All experimental points are the average values of three independent experiments. The data were collected in a Microsoft Excel spreadsheet where the average and standard error of the mean were determined. A two-way analysis of variance (two-way ANOVA) at a confidence level of 95% (α= 0.05) was carried out with the software PRISM 5 to test the statistical significance of differences between the growth rates of the three different T. reesei strains on each of the carbon sources. A Bonferroni multiple comparisons post hoc test was conducted, also at a 95% confidence level.
Results and discussion
Effect of carbon sources on the growth and total cellulase activity of T. reesei strains
In the current study three different T. reesei strains including QM6a, QM9414 and RUT-C30 were selected to study the effect of carbon sources on cellulases production. Druzhinina et al. (2006) previously examined carbon source utilization of T. reesei QM6a, and some of its cellulase-over producing mutants including QM9414 [29]. In this study, carbon sources were divided into two main groups based on their solubility in water: soluble carbon sources and insoluble carbon sources (the later further divided into two subgroups: pure polysaccharides and lignocelluloses (Table 1) [30]. By measuring the FPA every 24hrs, the effect of incubation time on cellulolytic enzyme production by the T. reesei strains was examined. The effect of each carbon source on the growth of each strain was determined by harvesting the culture, and measuring the dry weight at the end of each experiment.
It was found that the biological variation among the replicated fungal cultures was the main source of the variation observed in FPA studies (Sections 3.1.1, 3.1.2, and 3.1.3) This is in accordance with the other experiments involving T. reesei strains in shake flasks or solid fermentation [23, 31]. Three to four days was chosen by other investigators as the end point for shake flask experiments since at this time the enzyme production reached a plateau phase [23, 32]. This was only the case for RUT-C30, which reached its maximum activity earlier than QM9414 and QM6a. Thus, 5 days was defined as the end point for our shake flask experiment.
Soluble carbon sources
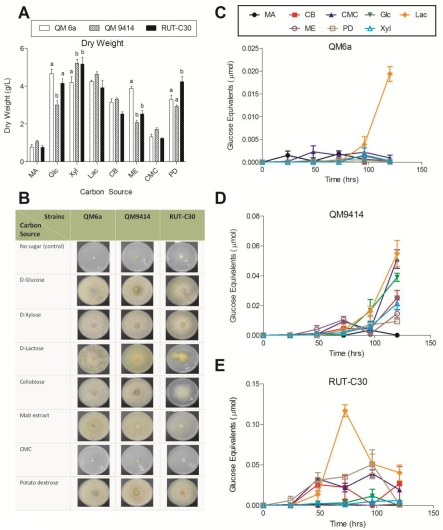
The three T. reesei strains were grown on D-glucose, D-xylose, D-lactose, cellobiose, malt extract, carboxymethyl cellulose (CMC) and potato dextrose in submerged culture in shake flasks and on agar plates. Growth rate of the strains were determined by measuring the dry weight (Figure 1A). The carbon source was found to cause a significant variation in the growth rate of the strains (ANOVA, P< 0.05). All three strains grew less effectively on MA-medium and CMC, as compared to the other soluble carbon sources. The slow growth on MA-medium, suggests that the pure MA-medium contributed very little to the growth of the fungi. The mutant strains QM9414 and RUT-C30 grew significantly slower on malt extract compared to the wild-type QM6a (P< 0.001). RUT-C30 grew significantly faster than QM6a and QM9414 on potato dextrose (P< 0.05). The strains were also grown on the same carbon source on agar plates to visualize the growth (Figure 1B). The similarities between the relative sizes of the agar culture, and the relative dry weight of the mycelium in the liquid culture, suggest that the effect of carbon source on the growth of the fungi is independent from the state of the medium.
Figure 1.
Effect of the different soluble carbon sources on growth rate and total cellulase activity of the three T. reesei strains. (A) Dry weight of the T. reesei strains cultured for 5 days at 30 °C. Letters denote a significant difference between the growth rate of the strains for each carbon source (P< 0.05). Carbon sources that exhibited no significant difference between any of the strains do not have letters; (B) growth comparison of the T. reesei mutant strains QM9414 and RUT-C30 to the wild-type strain QM6a on the different soluble carbon sources after 4 days; (C) total cellulase activity of T. reesei QM6a; (D) QM9414 and (E) RUT-C30 grown on MA-medium supplemented with 1% of the different soluble carbon sources for 24-120 hrs. Error bars denote standard error of the mean.
The effect of the soluble carbon sources on cellulolytic enzymes production by the fungi was determined using a FPA of the liquid culture supernatants (Figure 1C-E). For QM6a strain, the highest total cellulase activity was obtained at 120 hrs using D-lactose as the carbon source with about 0.019 µmol glucose equivalents. This level of activity was significantly larger than the activity induced by any of the other soluble sugars (Figure 1C). For the two mutant strains including QM9414 and RUT-C30, the highest total cellulase activity was also obtained using D -lactose at 120 or 72 hrs with about 0.055 and 0.14 µmol glucose equivalents, respectively (Figure 1D and E). Thus, total cellulase activity of QM9414 and RUT-C30 using D-lactose was, approximately, 3-fold and 7-fold higher than total cellulase activity of the wild-type QM6a, respectively. For QM9414, CMC stimulated high cellulase production at 120 hrs, while cellu-biose, D-xylose, and malt extract exhibited a lower activity. D-glucose was found to be intermediate between these two groupings (Figure 1D). In the case of RUT-C30, cellulase production occurred more quickly than in other strains (Figure 1E). Levels of activity at 48 hrs were found to be within the range of the maximum activity of QM9414, and greater than the activity of QM6a, suggesting that RUT-C30 favours cellulase production.
The background of reducing sugars present in the medium disappeared at different time points for the tested carbon sources. This could be monitored by the amount of the reducing sugars in the no filter paper control. D-xylose and potato dextrose were completely exhausted between 48 to 96 hrs by all the three strains. D-glucose was slowly exhausted from the medium by QM6a and RUT-C30 strains over 120 hrs, and was quickly exhausted by QM9414 after 48 hrs. D-lactose was also slowly exhausted from the medium over 120 hrs by the all three strains. Malt extract and cellobiose were completely exhausted between 48 to 96 hrs by QM6a and QM9414; however, high background sugars were obtained at every time point for RUT-C30. CMC was not exhausted, and the background sugar level stayed the same low level during the experiment which confirmed overall lower T. reesei growth rate using the sugar (Figure 1A and B).
Insoluble, pure polysaccharide carbon sources
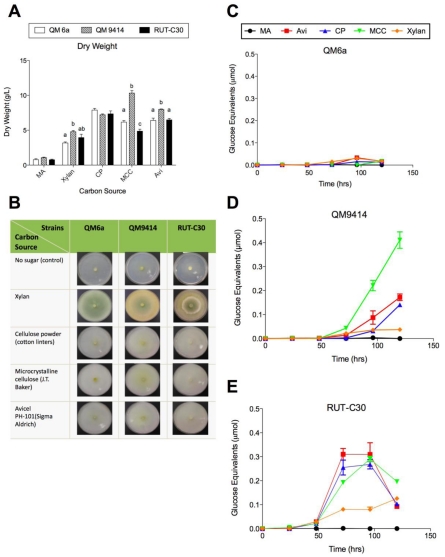
The three T. reesei strains were grown on xylan (oat spelt), cellulose powder (cotton linters) and two brands of microcrystalline cellulose including microcrystalline cellulose (J.T. Baker) and Avicel PH-101(Sigma Aldrich) in submerged culture in shake flasks and on agar plates. Growth rate of the strains were determined by measuring the dry weight (Figure 2A). The carbon source and the strain used were both found to have a significant effect on the growth rate of the mycelium (ANOVA, P< 0.05). All the three strains grew more effectively on cellulose powder (cotton linters), microcrystalline cellulose and Avicel, and less effectively on xylan. The mutant strain QM9414 grew significantly faster on microcrystalline cellulose compared to the wild-type QM6a and RUT-C30 (ANOVA, P< 0.0001). QM6a also produced significantly more biomass than RUT-C30, when grown on microcrystalline cellulose (P< 0.05). This is the only case where the growth rate of all three strains was found to be significantly different, and when compared with the high enzyme production of QM9414 on microcrystalline cellulose, this tentatively suggests that microcrystalline cellulose both stimulates the appropriate inducers of cellulase production and provides an effective source of metabolic substrates for QM9414. The strains were also grown on the same carbon source on agar plates to visualize the growth (Figure 2B).
Figure 2.
Effect of the different insoluble pure polysaccharide carbon sources on growth rate and total cellulase activity of the three T. reesei strains. (A) Dry weight of the T. reesei strains cultured for 5 days at 30 °C. Letters denote a significant difference between the growth rate of the strains for each carbon source (P< 0.05). Carbon sources that exhibited no significant difference between any of the strains do not have letters; (B) growth comparison of the T. reesei mutant strains QM9414 and RUT-C30 to the wild-type strain QM6a on the different insoluble pure polysaccharide carbon sources after 4 days; (C) total cellulase activity of T. reesei QM6a; (D) QM9414 and (E) RUT-C30 grown on MA-medium supplemented with 1% of the different insoluble pure polysaccharide carbon sources for 24-120 hrs. Error bars denote standard error of the mean.
The effect of the insoluble pure polysaccharide carbon sources on cellulolytic enzymes production by the fungi was determined using a FPA of the liquid culture supernatants (Figure 2C-E). For QM6a strain, the highest total cellulase activity was obtained at 96 hrs using Avicel and followed by xylan as the carbon source with about 0.033 and 0.031 µmol glucose equivalents (Figure 2C). For the two mutant strains including QM9414 and RUT-C30, the highest total cellulase activity was obtained using micro-crystalline cellulose and Avicel at 120 or 72 hrs with about 0.41 and 0.31 µmol glucose equivalents, respectively (Figure 2D and E). Thus, total cellulase activity of QM9414 and RUT-C30 using microcrystalline cellulose or Avicel was, approximately, 12-fold and 9.5-fold higher than total cellulase activity of the wild-type QM6a, respectively. In the case of RUT-C30, cellulase production using the tested carbon sources occurred more quickly than in other strains and reached the peak (around 72 or 96 hrs) and (Figure 2E). Similar to QM6a, activity of RUT-C30 was decreased at 120 hrs using most of the tested insoluble carbon sources (Figure 2C and E) whereas QM9414 reached its maximum activity at 120 hrs (Figure 2D). Except for one type of microcrystalline cellulose, the cellulase activity of RUT-C30 was higher than QM9414 among the tested insoluble pure polysaccharide carbon sources (Figure 2D and E). For the most of the insoluble carbon sources tested here the background of reducing sugars present in the medium was not significant. The exception was xylan, which was exhausted after 48.
Insoluble, lignocellulosic carbon sources
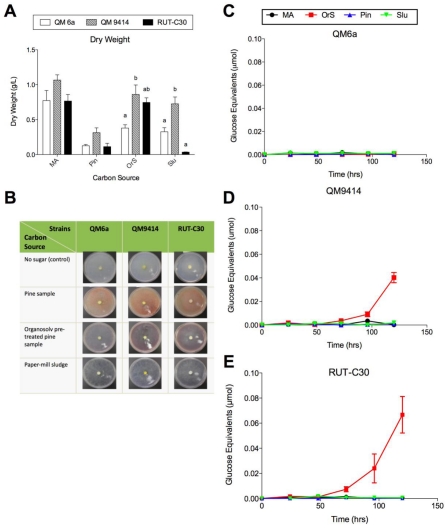
The three T. reesei strains were grown on pine sample, organosolv pretreated pine sample and paper-mill sludge in submerged culture in shake flasks and on agar plates (Figure 3A-E). The dry weight of the mycelium indicates the growth rate of the fungi, and both the carbon source and the strain were found to lead to a significant difference in this rate (ANOVA, P< 0.05) (Figure 3A). All the three strains grew less effectively on all the lignocellulosic carbon sources compared to the other tested carbon sources (soluble and insoluble pure polysaccharides, Figure 1A and Figure 2A). Other studies have found that pre-treated lignocelluloses do not stimulate microbial growth or enzyme production [21]. In the case of the organosolv pre-treated pine sample used in this study, the low activity and fungal growth may be as a result of the inhibitory effects of compounds generated during the pretreatment or the presence of the solvents used for the pretreatment [33]. Also, paper-mill sludge generated by the industrial sources contains a large number of ingredients, some of which are toxic for microbial growth [34]. The lack of microbial growth and enzyme production was also observed using untreated pine sample (Fig 3A-E). This may have been a result of the enzymes inability to access the celluloses found within the wood chips, because of the presence of lignin [35].
Figure 3.
Effect of the different insoluble lignocellulosic carbon sources on growth rate and total cellulase activity of the three T. reesei strains. (A) Dry weight of the T. reesei strains cultured for 5 days at 30 °C. Letters denote a significant difference between the growth rate of the strains for each carbon source (P< 0.05). Carbon sources that exhibited no significant difference between any of the strains do not have letters; (B) growth comparison of the T. reesei mutant strains QM9414 and RUT-C30 to the wild-type strain QM6a on the different insoluble lignocellulosic carbon sources after 4 days; (C) total cellulase activity of T. reesei QM6a; (D) QM9414 and (E) RUT-C30 grown on MA-medium supplemented with 1% of the different insoluble lignocellulosic carbon sources for 24-120 hrs. Error bars denote standard error of the mean.
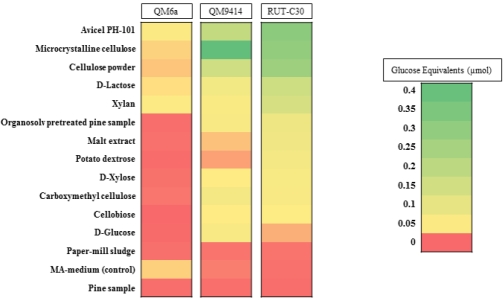
The effect of the insoluble lignocellulosic carbon sources on cellulolytic enzymes production by the fungi was determined using a FPA of the liquid culture supernatants (Figure 3C-E). For QM6a strain, no significant cellulase activity was obtained using all the three different lignocellulosic carbon sources (Figure 3C). For the two mutant strains including QM9414 and RUT-C30, the highest total cellulase activity was obtained using organosolv pretreated pine sample at 120 hrs with about 0.04 and 0.07 µmol glucose equivalents, respectively (Figure 3D and E). The greater activity of RUT-C30, when considered in conjunction with the similarity in dry weight between the two strains, indicates that RUT-C30 is more adept at producing cellulases under these conditions. No reducing sugar background was observed in the medium supplemented with the lignocellulosic carbon sources. Figure 4 summarizes cellulase production by the three fungal strains using all 14 different carbon sources as well as MA-medium (as the control) used in this study.
Figure 4.
An outline of cellulase activity profiles of the wild-type QM6a strain of T. reesei, and its two hypercellulolytic mutants, QM9414 and RUT-C30 on 14 different carbon sources. The colour scale represents the magnitude of the maximum cellulase activity (µmol glucose equivalents) obtained for the strain when grown on each particular carbon source. The legend contains intermediate values within each colour range.
Concentration of carbon source
Based on the carbon source experiments (Figure 1, 2 and 3), Avicel showed the highest activity for QM6a and RUT-C30, whereas micro-crystalline cellulose showed the highest cellulase activity for QM9414. To identify the optimum concentration of carbon source for each T. reesei strain, various concentrations (0.25 to 2%) of Avicel or microcrystalline cellulose were used (Figure 5A-C). The highest cellulase activity for QM6a was obtained using 1% Avicel with all concentrations above this exhibiting repressed cellulase (Figure 5A). T. reesei QM9414 strain showed the highest cellulase activity at about 1.25% microcrystalline cellulose, but stayed almost the same level when the carbon concentration increased up to 2% (Figure 5B). In the case of RUT-C30, cellulase activity using Avicel was gradually increased starting at 0.5%, peaked at about 1.5%, and then decreased when the carbon concentration increased up to 2% (Figure 5C). Growth rate of the strains using the selected carbon sources were also determined by measuring the dry weight (Figure 5A-C). All the three strains showed the same trend with gradually increasing the dry weight when the concentrations of the carbon sources were increased. The maximum dry weight for all the three T. reesei strains was obtained at the highest carbon concentration level (2%). This suggests that an increase in enzyme production is directly caused by an increase in mycelium density, and may be a result of complex inducing factors.
Figure 5.
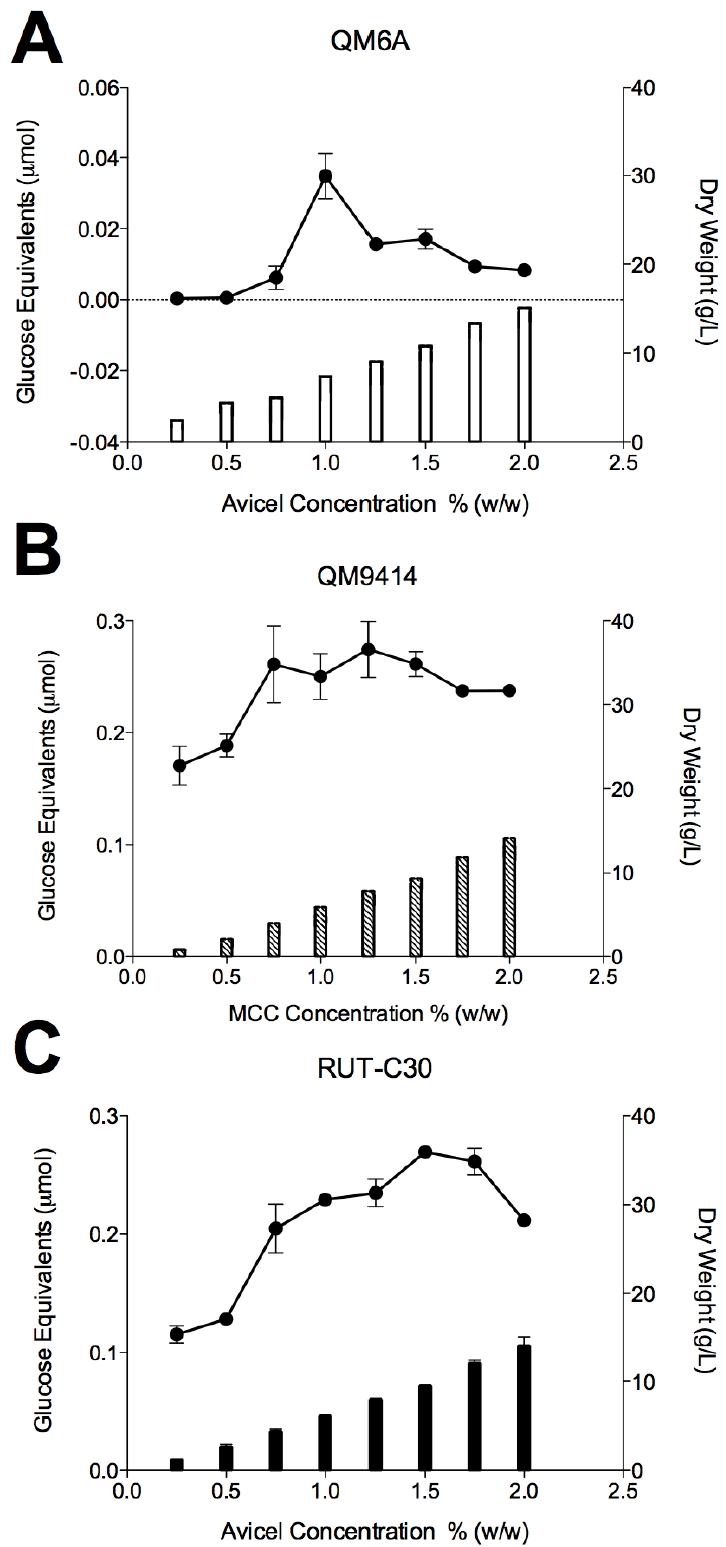
Effect of the selected carbon source concentration (0.25 to 2%) on the growth rate and total cellulase activity of the three T. reesei strains (A) QM6a; (B) QM9414 and (C) RUT-C30. T. reesei strains QM6a and RUT-C30 were grown on MA-medium supplemented with the different concentration of Avicel for 5 or 3 days, respectively. T. reesei strain QM9414 was grown on MA-medium supplemented with the different concentration of microcrystalline cellulose for 4 days. Bars represent dry weight plotted on the right axis, while lines represent glucose equivalents plotted on the left axis. Error bars denote standard error of the mean.
Conclusion
Although the cellulase activity of the T. reesei mutant strains QM9414 and RUT-C30 were appreciably higher than the wild-type QM6a, the carbon source utilization profiles of these strains closely resembled each other. For all the three T. reesei strains maximum cellulase production was observed using two different types of microcrystalline celluloses. The highest cellulase activity for QM6a was obtained using 1% Avicel while RUT-C30 showed the peak at about 1.5%. QM9414 strain showed the highest cellulase activity at about 1.25% microcrystalline cellulose. All the three strains exhibited the same gradual increase in the dry weight of the mycelium, as the concentrations of carbon sources were elevated. This consistency in mycelium growth suggests that the enzyme production is not directly correlated with the growth of the mycelium, and may be a result of complex inducing factors.
Acknowledgement
This work was supported by a scholarship from Ontario Graduate Scholarship (OGS) to M.D. and NSERC-RCD and Ontario Research Chair funding to W.Q.
References
- [1].Dashtban M, Schraft H, Qin W. Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. Int J Biol Sci. 2009;5:578–595. doi: 10.7150/ijbs.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Reese ET. History of the cellulase program at the US army Natick Development Center. Biotechnol. Bioeng. Symp. 1976;6:9–20. [PubMed] [Google Scholar]
- [3].Eveleigh DE, Montenecourt BS. Increasing yields of extracellular enzymes. Adv Appl Microbiol. 1979;25:57–74. doi: 10.1016/s0065-2164(08)70146-1. [DOI] [PubMed] [Google Scholar]
- [4].Durand H, Clanet M, Tiraby G. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microbiol. 1988;10:341–346. [Google Scholar]
- [5].Seidl V, Gamauf C, Druzhinina IS, Seiboth B, Hartl L, Kubicek CP. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics. 2008;9:327. doi: 10.1186/1471-2164-9-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Seidl V, Seiboth B. Trichoderma reesei: genetic approaches to improving strain efficiency. Biofuels. 2010;1:343–354. [Google Scholar]
- [7].Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE, Collett JR, Martin J, Druzhinina IS, Mathis H, Monot F, Seiboth B, Cherry B, Rey M, Berka R, Kubicek CP, Baker SE, Margeot A. Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:16151–16156. doi: 10.1073/pnas.0905848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ilmen M, Thrane C, Penttila M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- [9].Geysens S, Pakula T, Uusitalo J, Dewerte I, Penttila M, Contreras R. Cloning and characterization of the glucosidase II alpha subunit gene of Trichoderma reesei: a frameshift mutation results in the aberrant glycosylation profile of the hypercellulolytic strain Rut-C30. Appl Environ Microbiol. 2005;71:2910–2924. doi: 10.1128/AEM.71.6.2910-2924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vitikainen M, Arvas M, Pakula T, Oja M, Penttila M, Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genomics. 2010;11:441. doi: 10.1186/1471-2164-11-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Morawetz R, Gruber F, Messner R, P KC. Presence, transcription and translation of cello-biohydrolase genes in several Trichoderma species. Curr Genct. 1992;21:31–36. [Google Scholar]
- [12].Mantyla AL, Rossi KH, Vanhanen SA, Penttila ME, Suominen PL, Nevalainen KM. Electrophoretic karyotyping of wild-type and mutant Trichoderma longibrachiatum (reesei) strains. Curr Genet. 1992;21:471–477. doi: 10.1007/BF00351657. [DOI] [PubMed] [Google Scholar]
- [13].Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EG, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, de Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barabote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- [14].Barnett CC, Berka RM, Fowler T. Cloning and amplification of the gene encoding an extracellular beta-glucosidase from Trichoderma reesei: evidence for improved rates of saccharification of cellulosic substrates. Biotechnology (N Y) 1991;9:562–567. doi: 10.1038/nbt0691-562. [DOI] [PubMed] [Google Scholar]
- [15].Takashima S, Nakamura A, Hidaka M, Masaki H, Uozumi T. Molecular cloning and expression of the novel fungal beta-glucosidase genes from Humicola grisea and Trichoderma reesei. J Biochem. 1999;125:728–736. doi: 10.1093/oxfordjournals.jbchem.a022343. [DOI] [PubMed] [Google Scholar]
- [16].Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, Meerman HJ, Mitchell T, Mitchinson C, Olivares HA, Teunissen PJ, Yao J, Ward M. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- [17].Nogawa M, Goto M, Okada H, Morikawa Y. L-Sorbose induces cellulase gene transcription in the cellulolytic fungus Trichoderma reesei. Curr Genet. 2001;38:329–334. doi: 10.1007/s002940000165. [DOI] [PubMed] [Google Scholar]
- [18].Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels. 2009;2:19. doi: 10.1186/1754-6834-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tangnu SK, Blanch HW, Wilke CR. Enhanced Production of Cellulase, Hemicellulase, and β-Glucosidase by Trichoderma reesei (Rut C-30) Biotechnology and Bioengineering. 1981;XXIII:1837–1849. [Google Scholar]
- [20].Ghosh A, Ghosh BK, Trimino-Vazquez H, Eveleigh DE, Montenecourt BS. Cellulase secretion from a hyper-cellulolytic mutant of Trichoderma reesei Rut-C30. Archives of Microbiology. 1984;140:126–133. [Google Scholar]
- [21].Benko Z, Drahos E, Szengyel Z, Puranen T, Vehmaanpera J, Reczey K. Thermoascus aurantiacus CBHI/Cel7A production in Trichoderma reesei on alternative carbon sources. Appl Biochem Biotechnol. 2007;137140:195–204. doi: 10.1007/s12010-007-9051-5. [DOI] [PubMed] [Google Scholar]
- [22].Mandels MM, Andreotti RE. The cellulose to cellulase fermentation. Proc. Biochem. 1978;13:6–13. [Google Scholar]
- [23].Cianchetta S, Galletti S, Burzi PL, Cerato C. A novel microplate-based screening strategy to assess the cellulolytic potential of Trichoderma strains. Biotechnol Bioeng. 2010;107:461–468. doi: 10.1002/bit.22816. [DOI] [PubMed] [Google Scholar]
- [24].Xiao Z, Storms R, Tsang A. Microplate-based filter paper assay to measure total cellulase activity. Biotechnol Bioeng. 2004;88:832–837. doi: 10.1002/bit.20286. [DOI] [PubMed] [Google Scholar]
- [25].Ghose TK. Measurement of cellulase activities. Pure Appl Chem. 1987;59:257–268. [Google Scholar]
- [26].Dashtban M, Maki M, Leung KT, Mao C, Qin W. Cellulase activities in biomass conversion: measurement methods and comparison. Crit Rev Biotechnol. 2010;30:302–309. doi: 10.3109/07388551.2010.490938. [DOI] [PubMed] [Google Scholar]
- [27].Zhang Y-HP, Hong J, Ye X. Cellulase assays. Methods Mol Biol. 2009;581:213–231. doi: 10.1007/978-1-60761-214-8_14. [DOI] [PubMed] [Google Scholar]
- [28].Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- [29].Druzhinina IS, Schmoll M, Seiboth B, Kubicek CP. Global carbon utilization profiles of wild -type, mutant, and transformant strains of Hypocrea jecorina. Appl Environ Microbiol. 2006;72:2126–2133. doi: 10.1128/AEM.72.3.2126-2133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang Y-HP, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv. 2006;24:452–481. doi: 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- [31].King BC, Donnelly MK, Bergstrom GC, Walker LP, Gibson DM. An optimized microplate assay system for quantitative evaluation of plant cell wall-degrading enzyme activity of fungal culture extracts. Biotechnol Bioeng. 2009;102:1033–1044. doi: 10.1002/bit.22151. [DOI] [PubMed] [Google Scholar]
- [32].Kovács K, Megyeri L, Szakacsa G, Kubicekc CP, Galbeb M, Zacchi G. Trichoderma atroviride mutants with enhanced production of cellulase and β-glucosidase on pretreated willow. Enzyme and Microbial Technology. 2008;43:48–55. [Google Scholar]
- [33].Zhao X, Cheng K, Liu D. Organosolv pretreatment of lignocellulosic biomass for enzymatic hydrolysis. Appl Microbiol Biotechnol. 2009;82:815–827. doi: 10.1007/s00253-009-1883-1. [DOI] [PubMed] [Google Scholar]
- [34].Diez MC, Castillo G, Aguilar L, Vidal G, Mora ML. Operational factors and nutrient effects on activated sludge treatment of Pinus radiata kraft mill wastewater. Bioresour Technol. 2002;83:131–138. doi: 10.1016/s0960-8524(01)00204-8. [DOI] [PubMed] [Google Scholar]
- [35].Dashtban M, Schraft H, Syed TA, Qin W. Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol. >2010;1:36–50. [PMC free article] [PubMed] [Google Scholar]