Abstract
Recombineering techniques have been developed to modify bacterial artificial chromosomes (BACs) via bacterial homologous recombination systems, simplifying the molecular manipulations of large DNA constructs. However, precise modifications of a DNA fragment larger than 2-3 kb by recombineering remain a difficult task, due to technical limitations in PCR amplification and purification of large DNA fragments. Here, we describe a new recombineering strategy for the replacement of large DNA fragments using the commonly utilized phage/Red recombination host system. This approach involved the introduction of rare restriction enzyme sites and positive selection markers into the ends of a large DNA fragment, followed by its release from the donor BAC construct and integration into an acceptor BAC. We have successfully employed this method to precisely swap a number of large DNA fragments ranging from 6 to 40 kb between two BAC constructs. Our results demonstrated that this new strategy was highly effective in the manipulations of large genomic DNA fragments and therefore should advance the conventional BAC recombineering technology to the next level.
Keywords: Bacterial artificial chromosome, recombineering, homologous recombination, chimeric BACs, large DNA fragments
Introduction
The Human Genome Project yielded sequence information of complete genomes of human and several other species. These sequence resources laid the foundation for the rapidly developing genomic and epigenomic studies of normal human development and disease processes. In addition, huge selections of constructs containing large genomic segments, in particular the bacterial artificial chromosomes (BACs), have become indispensible tools for the genetic analysis and functional studies of genes as well as their promoters and regulatory elements [1-3]. Moreover, such constructs have been used to generate complex vectors for the creation of genetically engineered animals.
The development of recombination-mediated engineering, also called recombineering, using phage-based Escherichia coli homologous recombination systems has circumvented the needs for restriction enzymes and ligases, allowing the manipulations of large DNA constructs [4]. Since its first publication [5], a number of variations and improvements of this technique have been reported [6-13]. Of the two widely used recombineering systems, one employs episomal plasmids to supply RecE/RecT of the Rac phage (ET cloning system) [5, 6, 14] and the other utilizes a temperature-sensitive repressor to control the expression of recombinases from a replication-defective prophage [7-9].
As a significant improvement of the technique, the two-step schemes of recombineering, involving insertion of markers (galK or kanamycin-resistant gene (Kana) and RpsL+) and their subsequent removal via negative selections, made it possible for precise BAC modifications without leaving behind any unwanted sequences [9, 13]. These methods are generally very efficient for engineering point mutations, insertions or replacement involving sequences of up to 2 kb. However, modifying larger DNA regions can be very challenging, mostly because of the difficulty of obtaining sufficient molar amounts of large substrate DNA fragments by PCR or restriction enzyme digestions from BAC constructs. In addition, significant background colonies arise during counter selection of galK or RpsL+ markers, due to the loss of these markers as a result of intra-molecular recombination of repetitive sequences. Thus, inefficient recombination of large DNA fragments, combined with high background in counter-selection processes, renders the manipulation of large genomic regions difficult.
Recently, Rivero-Müller et al. reported a method for large DNA fragment recombineering using the plasmid-based ET cloning system [12]. In their method, homology arms (HAs) together with homing endonuclease I-SceI sites were first engineered into an acceptor BAC. Following in vivo cleavages of I-SceI sites, a 55-kb fragment containing the human Lhcgr gene was then inserted into its corresponding site of mouse genome in the acceptor BAC. Here, we describe a novel alternative approach that allows large fragment replacement in recombineering hosts with a temperature-sensitive prophage. Using this new stratergy, we have successfully generated a number of chimeric BAC reporters containing both human and mouse genomic sequences of the telomerase reverse transcriptase loci.
Materials and methods
Bacterial stains and BAC clones
Bacterial host SW102, a galK version of DY380, contained a defective prophage with the cI857 repressor and was obtained from the National Cancer Institute [9]. BAC reporter constructs 117B23-cFtR (H(wt)) and 183M22-cFtR (M(wt)) were derived from BAC clones RPCI11-117B23 and RP24-183M22 (Research Genetics), which contained the loci of the human and mouse telomerase reverse transcriptase (TERT), respectively [15]. The human and mouse BAC sequences were based on the NCBI36/hg18 assembly (March 2006) and the NCBI36/mm8 assembly (February 2006), respectively.
Recombineering substrates
GalK and kanamycin resistant gene (Kana) cassettes were PCR amplified using pGalK and pREP4 as the templates, respectively [9]. In earlier experiments, large DNA fragments containing galK were isolated from agarose gels following pulsed field gel electrophoresis (CHEF DR® III System, Bio-Rad). Mixtures of restriction fragments used for later recombineering experiments were obtained by digesting minipreparations of BAC DNAs (∼10 µg) with relevant restriction enzymes, followed by RNase treatment, phenol/chloroform extraction, ethanol precipitation, and resuspension in dH2O.
Recombineering
The recombineering procedure was carried out as previously described [9, 13]. Briefly, 1 ml overnight culture of SW102 harboring a BAC construct was diluted to a fresh culture of 50 ml and grown at 32ºC to the log phase (A600 ≈ 0.5). The culture was then incubated at 42ºC for 15 min to induce the recombinases, followed by chilling to 4ºC. Electro-competent cells were prepared by washing and resuspending the cells in ice-cold 10% glycerol. For positive selections, 50 µl competent SW102 cells were electroporated with a purified substrate fragment (100– 200 ng) or a mixture of restriction fragments (0.5-1 µg total DNAs), plated on kanamycin LB or minimal medium (MM) plates supplemented with 12.5 µg/ml chloramphenicol, and incubated at 32ºC for 1 or 3 days, respectively. For counter-selection against galK, the electroporated cells were diluted to 10 ml SOC medium and incubated for 4.5 h at 32ºC. Following the recovery period, 1 ml culture was spun down, washed twice, and plated on MM plates containing 0.2% (w/v) 2-deoxy-galactose (DOG) [9].
Identification and verification of recombinants
For positive selections, the integrities of modified BACs that underwent specific homologous recombinations were verified by restriction enzyme digestion analyses. For counter selections against the galK marker, single-colony PCR was used as an initial screen for candidate clones that underwent the desired recombination event. The PCR positive clones were further tested by restriction enzyme digestion analysis. Following each recombineering step, single digestions of BAC mini preps with four enzymes were performed to ensure that the resultant BACs suffered no rearrangements other than the desired recombination. To assist analyses, Gene Construction Kit 2.5 (Textco, Inc.) was utilized to predict digestion profiles of the BACs, which were compared to the band patterns on 0.7% agarose gels.
Results and discussion
Replacement of large DNA fragments with a pre -existing homologous end – a 3-step recombineering scheme
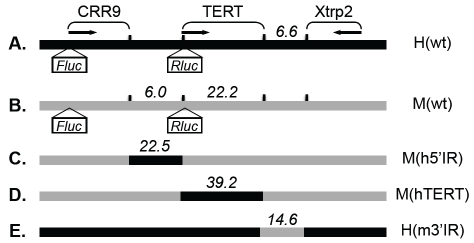
BAC constructs encompassing the entire TERT loci contain all the cis elements required for their developmental regulation [15, 16]. H(wt) and M(wt) are dual-luciferase reporters derived from such BACs that contain a 160-kb human genomic sequence and the matching 135-kb mouse sequence, respectively. In each of these reporters, a Renilla (Rluc) and a Firefly luciferase (Fluc) cassette were inserted downstream of the TERT and CRR9 promoters, respectively, to monitor their activities (Figures 1A and 1B) [15, 16]. In order to study the differential regulation of the human and mouse TERT genes, we set out to construct chimeric BACs (Figure 1C-1E) by swapping large segments of genomic sequences, up to 40 kb, between these two BAC reporters.
Figure 1.
A schematic illustration of BAC constructs. A & B. The wild type forms of human and mouse BAC reporters, H(wt) and M(wt), respectively. C, D, & E. Three chimeric BACs. Genomic positions of the CRR9, TERT, and Xtrp2 loci are shown. Arrows indicate the directions of transcription. Black and grey bars represent human and mouse sequences, respectively. Diagrams are not drawn to scale and lengths of exchanged sequences are indicated in kilobases.
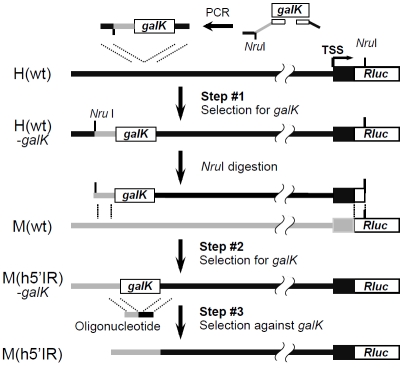
To generate M(h5’IR), in which the 6-kb 5’ intergenic region of the mTERT gene was replaced by the corresponding 22.5-kb human sequence, we designed a 3-step scheme, as illustrated in Figure 2. First, a galK marker cassette was inserted into one end of the donor sequence of H(wt), generating the intermediate product H(wt)-galK (Step #1). The galK gene was amplified from plasmid pGalK using a 150-bp forward primer (25-bp human homology arm (HA) + an overlapping NruI site + 105-bp mouse HA + 20-bp galK forward primer) and a 60-bp reverse primer (20-bp galK reverse primer + 40-bp human HA). The resultant 1.4-kb PCR fragment was transformed into electro-competent SW102 host cells containing H(wt), followed by selection on galactose MM plates. A total of approximately 2500 colonies were obtained and restriction enzyme digestion analyses of four randomly picked colonies indicated that all four clones correctly underwent the desired recombination event (Table 1).
Figure 2.
The 3-step scheme: exchanges of large DNA fragments between BACs using one existing HA. Black and grey bars represent human and mouse sequences, respectively. TSS, transcription start site. galK, E. coli galactose K gene.
Table 1.
Summary of BAC modifications by the three-step procedure
| Chimera BACs | Steps | Substrate BACs | Substrate fragment (bp) | Substrate fragment (ng) | Total # colonies | # collect modifications | ||
|---|---|---|---|---|---|---|---|---|
| Total length | Homology arms | |||||||
| Left | Right | |||||||
| M(h5’IR) | galK insertion | H(wt) | 1,401 | 25 | 40 | 220 | ∼2,500 | 4/4 |
| M(hTERT) | (step #1) | 1,397 | 40 | 26 | 199 | ∼3,200 | 4/4 | |
| M(h5’IR) | Fragment replacement | M(wt) | 24,818 | 105 | 936 | 154 | 3 | 3/3 |
| M(hTERT) | (step #2) | 41,060* | 280 | 100 | 268 | 18 | 4/4 | |
| M(h5’IR) | galK removal | M(h5’IR)-galK | 100 | 50 | 50 | 214 | TMTC | 4/4 |
| M(hTERT) | (step #3) | M(hTERT)-galK | 100 | 50 | 50 | 124 | TMTC | 5/5 |
a mixture of fragments from NruI digestion of H(wt)-galK BAC DNA was used as recombineering substrate. TMTC, too many to count.
Second, the 24.8-kb human intergenic region containing the galK marker was released from H(wt)-galK by NruI digestion and used as a donor fragment for the next recombineering step (Step #2). Because both H(wt) and M(wt) contained a Rluc cassette in the first exon of TERT, the 936-bp Rluc sequence upstream of the 3’ end NruI site was used as the right HA. This nearly 25-kb large NruI fragment was isolated from an agarose gel following pulsed field gel electrophoresis and transformed into SW102 cells containing M(wt). Three colonies were obtained upon selection on galactose MM. Although the efficiency of this second recombination step was much lower than that of Step #1, due to the large size of the substrate fragment, all three clones were correct recombinants of the intermediate product M(h5’IR)-galK, as determined by restriction enzyme digestion analysis (Table 1).
Lastly, the galK marker in M(h5’IR)-galK was removed via recombination with a 100-bp double-stranded oligonucleotide containing a 50-bp mouse HA and a 50-bp human HA, followed by a 3-day counter selection against galK in MM containing 2-deoxy-galactose (DOG) (Step #3). An estimated total of 0.5 to 1 million colonies were yielded from this last recombination step. Because intra-molecular recombinations could also result in the loss of galK marker, single-colony PCR was first carried out, using primers specific for the expected recombinant. Over 50% of PCR screened colonies contained the potential product of precise rearrangement. Restriction enzyme analyses of four randomly picked PCR-positive clones revealed that all four were legitimate M(h5’IR) recombinants (Table 1). Thus, we have successfully replaced the 6.0kb mTERT 5’ intergenic region with the 22.5-kb hTERT 5’ intergenic region employing the 3-step recombineering procedure.
Preparation of large donor DNA fragments for BAC recombineering
Using the 3-step strategy, we constructed M(hTERT) by replacing all exons and introns of the mTERT gene (22.2 kb) in M(wt) with the corresponding 39.2-kb human sequence. The efficiency of each recombination step is summarized in Table 1. In the second step, the 41-kb donor fragment was so large that purifying sufficient molar amount of this fragment became very difficult. To test if it was possible to skip the purification procedure of the DNA fragment for recombineering, the H(wt)-galK construct was digested with NruI and the reaction mixture was extracted by phenol/chloroform, ethanol precipitated, and transformed into SW102 cells containing the acceptor BAC M(wt). As shown in Table 1, this simplified procedure in fact significantly increased the recombineering efficiency, yielding a total of 18 colonies upon galK positive selection. All four colonies examined by restriction enzyme analysis exhibited correctly recombineered fragments but no unintended rearrangements. This result and subsequent experimental data indicated that large substrate DNA fragments could be transformed into host bacteria as a mixture without lessening the recombination efficiencies, thereby omitting the difficult purification step. Thus, in later experiments, mixtures of digested BAC fragments were used as recombination substrates for fragments larger than 10 kb.
Exchange of large DNA fragments without a homologous end – a 4-step scheme
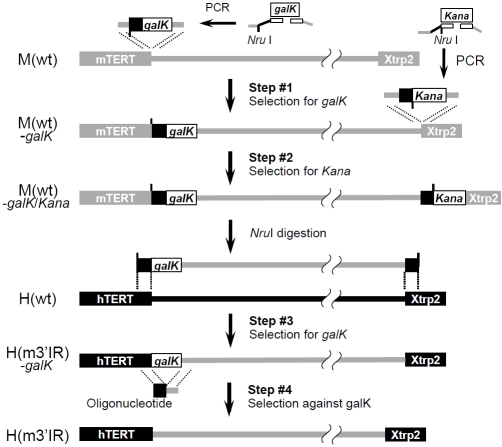
Using the Rluc cassette as a HA simplified the procedures of constructing M(h5’IR) and M(hTERT), as discussed in the 3-step recombineering method. Yet in the case of creating H(m3’IR), in which the 6.6-kb human 3’ intergenic region of H(wt) was replaced by the 14.6-kb mouse counterpart, HAs at both 5’ and 3’ ends needed to be engineered. To accommodate such needs, we designed a four-step scheme, as summarized in Figure 3. First, the galK marker was amplified from pGalK and inserted into the upstream end of the donor sequence in M(wt) using a similar strategy as described in the 3-step approach (Figure 2). In Step #2, the kanamycin-resistant gene (Kana) was amplified from plasmid pREP4 using a 90-bp forward primer (23-bp mouse HA + an overlapping NruI site + 44-bp human HA + 20-bp Kana forward primer) and 60-bp reverse primer (20-bp Kana reverse primer + 40-bp mouse HA), and recombined into the downstream end of the mouse 3’ intergenic region. Although insertion of a positive selection marker was usually very efficient, we obtained only 20 colonies, likely due to the very short HA (23 bp) in this particular case. Nonetheless, all ten randomly picked colonies contained predicted recombineered products as determined by restriction enzyme digestion analyses (Table 2). In Step #3, the 16-kb modified intergenic region including the galK marker was released by NruI digestion and the digested fragment mixture was electroporated into SW102 cells containing H(wt). More than 3000 galK positive colonies were obtained after selection on galactose MM plates. Three randomly picked colonies were examined and all contained the correct replacement of the 3’ intergenic region of H(wt) by the corresponding mouse fragment, as determined by restriction enzyme digestion analyses.
Figure 3.
The 4-step scheme: exchanges of large DNA fragments between BACs without pre-existing HAs. Kana, Kanamycin resistant gene.
Table 2.
Generation of chimera BAC H(m3’IR) by the four-step procedure
| Recombineering steps | Substrate BACs | Substrate fragment (bp) | Substrate fragment (ng) | Total # colonies | # correct modifications | ||
|---|---|---|---|---|---|---|---|
| Total length | Homology arms | ||||||
| Left | Right | ||||||
| galK insertion (step #1) | M(wt) | 1,397 | 24 | 40 | 67 | ∼200 | 5/5 |
| Kana insertion (step #2) | M(wt)-galK | 1,377 | 23 | 40 | 151 | 20 | 10/10 |
| Fragment replacement (step #3) | H(wt) | 15,884* | 105 | 104 | 3,900 | ∼3,500 | 3/3 |
| galK removal (step #4) | H(m3’IR)-galK | 100 | 50 | 50 | 260 | TMTC | 4/4 |
a mixture of fragments from NruI digestion of H(wt)-galK BAC DNA was too many to count.
Finally, the galK marker was removed through recombination with a double-stranded oligonucleotide, resulting in the final product H(m3’IR) (Step #4), as described in the step #3 of the 3-step procedure. Taken together, we precisely substituted the 6.6-kb hTERT 3’ intergenic region with the matching 14.6-kb mTERT sequence by using the 4-step recombination procedure. Because this method did not require any pre-existing sequences, it could be employed to exchange any sequences between two BAC constructs.
Homology arm designing and recombineering efficiency
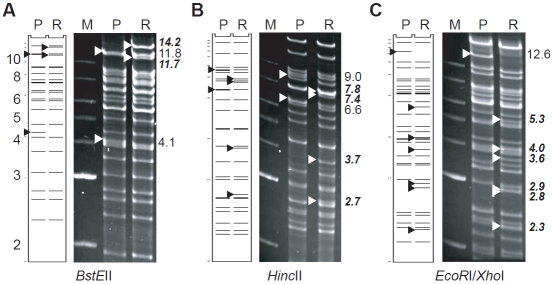
An important feature of this new multistep recombineering strategy is the insertion of HAs, positive selection markers (galK and Kana), and restriction sites (NruI) at the ends of donor fragments. Although this requires extra recombineering steps as compared to the previously published 2-step methods [9, 13], positive selections of recombinants involving large DNA fragments are much more efficient than identifying legitimate recombination products following negative selections. The HAs were designed as parts of PCR primers for amplifying the galK and Kana markers. For insertions of relatively short fragments (< 2-kb) such as galK and Kana markers, a HA of 23-bp appeared to be the minimum length required for generating sufficient recombinant colonies, since one additional base pair enhanced the efficiency by 10-fold (Steps #1 in Table 2). Conversely, recombineering of large fragments required longer HAs (Step #2 in Table 1 and Step #3 in Table 2). Because these recombineering steps involved positive selections of the galK marker and had virtually no background colonies, we found that HAs of about 100 bp were sufficient for swapping long DNA fragments of up to 40 kb (Tables 1 and 2). Therefore, we recommend using oligonucleotides with overall lengths of about 150 bp (consisting of 24-bp human/ mouse HA, 6-bp NruI site, 100-bp mouse/ human HA, and 20-bp galK or Kana primer) for amplifying the galK and Kana genes. The recombineering efficiencies using fragments with these 100-bp HAs and the galK marker were reasonably high, varying from 0.02-179 colonies per ng DNA, considering the large sizes of exchanged DNA fragments (15-40 kb). In fact, the majority of examined colonies from recombineering experiments we have performed thus far using large DNA fragments were correct recombinants (28/30 or 93%, data not shown), as determined by restriction digestion analyses using multiple enzymes. Figure 4 shows an example of restriction enzyme digestion analysis of an H(m3’IR)-galK clone derived from Step #3 of Figure 3, in comparison to its parental H(wt). All fragment sizes remained the same except for the computer-predicted changes in the recombinant.
Figure 4.
Verification of precise modifications by restriction enzyme digestion analysis. The example shown is the recombineering Step #3 in Figure 3. BAC DNAs were digested with BstEII (A), HincII (B), or EcoRI/XhoI (C), and electrophoresed on a 0.7% agarose gel. P, parental BAC H(wt); R, intermediate recombinant H(m3’IR)-galK. M, molecular weight markers in kb (labeled on the left side of the figure). Arrowheads indicate fragments with size changes upon recombineering. The sizes (in kb) of these bands, before (regular) and after (bold italic) recombineering, are shown on the right. In each panel, the computer-predicted band patterns are illustrated on the left and the actual gel patterns are shown on the right. The computer predicted band patterns were similar, but not identical, to the actual gel patterns. The differences were likely caused by polymorphic repetitive sequences within the human and mouse genomes. Other than predicted changes of band sizes, all other restriction fragments remained the same upon recombineering, indicating that no unintended rearrangements had occurred.
The final steps of the multi-step recombineering strategies involved the removal of galK gene by counter selection against the marker, using a 100-bp double-stranded oligonucleotide containing a 50-bp mouse HA and a 50-bp human HA as a substrate. Because sufficient molar amounts of synthetic oligonucleotides were used as recombineering substrates, high recombination efficiencies were obtained from this last step as determined by colony PCR and restriction enzyme analyses. In fact, most of the tested colonies yielded PCR products of correct sizes, using primers flanking the galK marker. Out of the PCR-positive clones, the vast majority displayed expected BAC DNA fragment patterns upon digestion by multiple restriction enzymes. The final products of the multi-step recombineering were further verified by sequencing through the junctions of recombination at both ends.
A novel recombineering strategy for large DNA fragments
Recombineering technology [5, 17] and its modifications [7] allowed the manipulation of large DNA constructs such as BACs, thus becoming an important tool in the construction of targeting vectors for genetic engineering and gene function analysis. By using positive/ negative selection markers, precise modifications, including point mutations, insertion, deletion, and replacement of DNA sequences, could be achieved without leaving behind any unwanted sequences [9, 13]. However, modifying large DNA fragments remained a difficult task until recently. To overcome this barrier, Rivero-Müller et al. recently reported the ALFIRE method (Assisted Large Fragment Insertion with Red/ET-recombination), a modification of previous recombineering techniques by introducing I-SceI recognition sites into recipient BACs. In vivo cleavages of such BAC constructs by I-SceI thus generated double-stranded breaks and greatly increased homologous recombination efficiency, allowing the recombineering of DNA fragments as large as 55 kb. While the ALFIRE method was a modification of the plasmid-based ET cloning system [5, 17], the new strategy reported here was carried out in the widely used temperature-sensitive prophage host system, originally developed by the Copeland laboratory [7, 9], without the need of a bacterial host with inducible I-SceI. Although the 40-kb fragment was the longest recombineerred in this study, the high efficiencies achieved using this new method should allow manipulations of even larger DNA segments.
In summary, we have developed a novel multistep recombineering strategy that is suitable for modifying large DNA sequences. Using this method with either the three- or four-step recombineering schemes, we generated a set of chimeric BACs by swapping large tracks of DNA up to 40 kb between two BAC constructs. The 3-step scheme requires one pre-existing HA. In the experiments reported here, we took advantage of the Rluc cassette present in both human and mouse TERT BAC reporters for the generation of chimeric BACs. Besides, this strategy should also allow the fusion of sequences from two different BACs into one, by using shared sequences in the BAC vector backbones. On the other hand, the 4-step scheme allows the manipulations of any large DNA sequences in BAC constructs without the prerequisite of an existing HA. Therefore, we have demonstrated the feasibility of modifying large DNA sequences in BAC constructs, in the commonly used bacterial host system containing a temperature-sensitive and replication-defective prophage. This efficient method alleviates the long-standing barrier of large DNA fragment modifications in the recombineering field and thus should allow precise rearrangements of DNA segments of any lengths at will.
Acknowledgements
The authors wish to thank Longgui Chen for the technical assistance. We also would like to thank the Macromolecular and Molecular Genetics Core Facilities at Penn State College of Medicine for the excellent services. None of the authors have professional or financial affiliations that can be perceived to bias the presentation of this manuscript.
References
- [1].Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- [2].Tsyrulnyk A, Moriggl R. A detailed protocol for bacterial artificial chromosome recombineering to study essential genes in stem cells. Methods Mol Biol. 2008;430:269–293. doi: 10.1007/978-1-59745-182-6_19. [DOI] [PubMed] [Google Scholar]
- [3].Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP, Dietzel S, Mechtler K, Durbin R, Stewart AF, Peters JM, Buchholz F, Hyman AA. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- [5].Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genetics. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- [6].Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- [7].Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- [8].Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hegde S, Paulson RF. Co-targeting a selectable marker to the Escherichia coli chromosome improves the recovery rate for mutations induced in BAC clones by homologous recombination. Biotechniques. 2004;36:936–938. 940. doi: 10.2144/04366BM03. [DOI] [PubMed] [Google Scholar]
- [11].Sopher BL, La Spada AR. Efficient recombination-based methods for bacterial artificial chromosome fusion and mutagenesis. Gene. 2006;371:136–143. doi: 10.1016/j.gene.2005.11.034. [DOI] [PubMed] [Google Scholar]
- [12].Rivero-Muller A, Lajic S, Huhtaniemi I. Assisted large fragment insertion by Red/ET-recombination (ALFIRE)–an alternative and enhanced method for large fragment recombineering. Nucleic Acids Res. 2007;35:e78. doi: 10.1093/nar/gkm250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol Biotechnol. 2009;42:110–116. doi: 10.1007/s12033-009-9142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Muyrers JP, Zhang Y, Benes V, Testa G, Ansorge W, Stewart AF. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 2000;1:239–243. doi: 10.1093/embo-reports/kvd049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang S, Zhao Y, Leiby MA, Zhu J. Studying human telomerase gene transcription by a chromatinized reporter generated by recombinase-mediated targeting of a bacterial artificial chromosome. Nucleic Acids Res. 2009;37:e111. doi: 10.1093/nar/gkp511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang S, Hu C, Zhu J. Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Molecular Biology of the Cell. 2007;18:669–677. doi: 10.1091/mbc.E06-09-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]