Abstract
Nematode spermatozoa are highly specialized cells that lack flagella and, instead, extend a pseudopod to initiate motility. Crawling spermatozoa display classic features of amoeboid motility (e.g. protrusion of a pseudopod that attaches to the substrate and the assembly and disassembly of cytoskeletal filaments involved in cell traction and locomotion), however, cytoskeletal dynamics in these cells are powered exclusively by Major Sperm Protein (MSP) rather than actin and no other molecular motors have been identified. Thus, MSP-based motility is regarded as a simple locomotion machinery suitable for the study of plasma membrane protrusion and cell motility in general. This recent focus on MSP dynamics has increased the necessity of a standardized methodology to obtain C. elegans sperm extract that can be used in biochemical assays and proteomic analysis for comparative studies. In the present work we have modified a method to reproducibly obtain relative high amounts of proteins from C. elegans sperm extract. We show that these extracts share some of the properties observed in sperm extracts from the parasitic nematode Ascaris including Major Sperm Protein (MSP) precipitation and MSP fiber elongation. Using this method coupled to immunoblot detection, Mass Spectrometry identification, in silico prediction of functional domains and biochemical assays, our results indicate the presence of phosphorylation sites in MSP of Caenorhabditis elegans spermatozoa.
Keywords: Major Sperm Protein, phosphorylation, cell motility, spermatozoa, C. elegans
Introduction
Nematode spermatozoa are highly specialized cells that lack a flagellum and must extend a pseudopod in order to become motile and fertilize an egg. In nematodes, the Major Sperm Protein (MSP), rather than actin as in typical amoeboid cells, is responsible for sperm locomotion [1]. The MSP-based motility system is analogous to actin-based motility in terms of filament formation and the generation of a protrusive force at the leading edge of the pseudopod, although this process is based on a set of very different molecules [2]. MSP is an immunoglobulin (Ig)-like 15 kDa protein that constitutes approximately 15% of the total sperm protein in nematodes and is synthesized prior to the differentiation of primary spermatocytes [3]. In fully differentiated spermatozoa, MSP forms filaments exclusively involved in locomotion by polymerizing dimers into helical subfilaments [4]; these subfilaments intertwine to form helical filaments that ultimately assemble into fibers [5]. Polymerization of MSP into filaments and fibers produces a protrusive force along the leading edge of the pseudopod, while depolymerization at the base of the pseudopod promotes retraction of the cell body. These two forces, coupled with pseudopod attachment to the substrate, confer movement to the sperm cell [6]. In addition to its role in conferring sperm motility through pseudopod extension, MSP has also been shown to serve as a signal for oocyte maturation and gonad sheath contraction required for ovulation [7]. Thus, MSP is a multifunctional protein that must nucleate filaments in the vicinity of the plasma membrane, elongate MSP fibers along the pseudopod to confer cellular motility and signal for oocyte maturation. This multifunctional identity of MSP could be explained by the the presence of ∼ 50 msp genes and pseudogenes in the genome of C. elegans [8], from which 28 MSP protein sequences have been annotated in GeneBank and at least three MSP protein isoforms have been detected by isoelectric focusing in C. elegans sperm [9]. However, posttranslational modifications that would explain MSP multifunctionality have not been identified.
MSP’s dual role as both a signaling and a cytoskeletal protein involved in locomotion have attracted a great deal of attention since proteins carrying the MSP Domain have been found in taxonomically diverse eukaryotic kingdoms, including protists, fungi, plants, and animals [10]. MSP Domain Proteins (MDPs) contain the MSP Ig-like domain that mediates protein-protein interactions and can be recognized in two categories: cytoskeletal MSPs and VAPs (VAMP-associated proteins, which are integral membrane proteins involved in linking membrane with cytosolic protein complexes) [11]. Among animals, MDPs have been identified in ∼ 20 species of nematodes (including C. elegans), the flatworm Schistosoma japonicum, the mollusk Aplysia californica, the model organisms Drosophila melanogaster, Xenopus laevis, Danio rerio, Mus musculus, and a wide variety of vertebrates including humans.
In nematode sperm, previous studies have suggested a role of MSP in the regulation of cytoskeletal protrusions at the plasma membrane necessary for signals that lead to both oocyte maturation in C. elegans and pseudopod protrusion in Ascaris. In the case of C. elegans, MSP assembly is thought to generate the protrusive force that leads to vesicle budding since MSP is concentrated at the sites of bud formation [12]. In the case of the parasitic nematode Ascaris suum, previous studies have shown that MDPs play a crucial role in the regulation of MSP filament dynamics [13] and that phosphorylation and dephosphorylation events of cytoskeletal accessory proteins are necessary for the assembly of the MSP cytoskeleton at the leading edge of the pseudopod [14]. Furthermore, the role for an activated MSP isoform (MSP*) in Ascaris that would polymerize rapidly under physiological conditions in the vicinity of the plasma membrane has been previously hypothesized [15]. However, to date, no direct phosphorylation events that would explain the multifunctionality of MSP have been reported either in Ascaris or Caenorhabditis sperm. Here we have modified previously published methods [3, 16] to reproducibly obtain relative high amounts of proteins from C. elegans sperm extracts. Using this method of sperm isolation (coupled to biochemical assays) we show that the C. elegans sperm extract share some of the properties observed in sperm extract of Ascaris such as MSP precipitation and fiber elongation. Furthermore, the use of immunoblot detection, Mass Spectrometry identification, and in silico prediction of functional domains indicates the presence of potential phosphorylation sites in MSP of C. elegans spermatozoa.
Materials and methods
Genetics and strains
The strain CB1489: him-8(e1489) IV, was maintained at 20oC on NGM plates seeded with Escherichia coli OP50 as described by [17]. Klass and Hirsh (1981) suggested to use this strain for male separation and sperm isolation since it produces ∼ 40% male progeny compared to ∼0.1% males produced by wild type [18] and their sperm are cytologically indistinguishable from wild-type sperm [19]. This strain was provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Worm synchronization and male separation
Culture synchronization and male separation were performed as described by [16] with some modifications. Briefly, worms were allowed to grow to saturation on plates for three days and the cultures were synchronized using the alkaline hypochlorite method (35 ml of a bleach/ NaOH mixture). Embryos were allowed to hatch for 24 hours in M9 buffer (22mM KH2PO4, 22mM Na2HPO4, 85mM NaCl, 1mM MgSO4) supplemented with cholesterol and spotted on seeded NGM plates allowing the worms to grow for three days at 20oC. Nitex filters (Tetko) were used for worm and spermatid separation. Plates with adult worms were washed using M9 buffer and filtered using a 35-µm Nitex filter. The filtrate containing males and hermaphroditic juveniles was transferred onto a 25-µm Nitex filter and washed using M9 buffer to eliminate juvenile worms. A worm population of ∼ 95 % males was collected from the top of the 25-µm filter and poured over a 30 % Hypaque solution (Hypaque™ 76; Diatrizoate Meglumine and diatrizoate sodium from Amersham Health) to remove bacteria and debris. Male worms were centrifuged at 1000g for 10 min and recovered from the interface between the Hypaque solution and the M9 buffer.
Large-scale sperm isolation method
C. elegans sperm were isolated using previously described methods [3, 16] with some modifications. Male worms were collected and washed in Modified Sperm Medium (MSM) to prevent activation of spermatids. MSM contained 50 mM HEPES, 70 mM Choline Chloride, 5 mM CaCl2, 5mM Dextrose, Polyvinylpyrrolidone (PVP) 10 mg/ml, pH adjusted to 6.5 and supplemented with 10 µl/ml of protease inhibitor cocktail (Sigma), 10 mM Na-Fluoride and 1 mM Naorthovanadate to prevent protein lysis and phosphatase activity. Worms were centrifuged at 6000 rpm using a benchtop centrifuge. The pellet of packed males was transferred to the bottom of a glass Petri dish and worms were minced using a stainless steel razor blade for approximately 5 min on top of an ice block. The minced worms were then washed from the Petri dish using MSM and passed through a 15µm Nitex filter. The filtrate was then poured through a 5µm Nitex filter and this second filtrate was poured on top of 2ml of Percoll 10 % and centrifuged at 1000g for 10 min. The supernatant was discarded and the pellet resuspended in 1 ml of MSM. Spermatids were counted using a hemocytometer, separated in aliquots of ∼4 x 106 cells/ml (∼ 2 mg/ml of protein concentration) and centrifuged again at 1000g for 10 min at 4º C. The pellet was resuspended in 1 ml of Homogenization buffer (20 mM Tris, 20 mM HEPES, 30 mM Mannitol, 1 mM EDTA, 1 mM EGTA, pH adjusted to 7.4) and laid on ice for sonication. The sample was sonicated using a Branson Sonifier 250 (VWR Scientific) using 4 pulses of 15 seconds each and resting on ice for 1 min between pulses. The sample was cleaned from cellular debris and nuclear chromatin by centrifugation at 10 000 g for 5 min at 4º C. The supernatant was then concentrated using a microcon tube with a Molecular Weight Cut Off (MWCO) of 3 kDa (Millipore) and either resuspended in 100 µl of PBS and Laemmli sample buffer, or used for crystal precipitation and/or fiber formation assays.
MSP precipitation and fiber formation
The concentrated extract isolated from C. elegans him-8 male spermatids (after sonication and centrifugation) was used for MSP precipitation or fiber formation assays as described by [20]. For crystal precipitation, the concentrate was diluted to a protein concentration of 1mg/ ml using KPM buffer (10 mM potassium phosphate, 0.5 mM MgCl2, pH 6.8). Aliquots of 10µl were placed in eppendorf tubes and an equal volume of Polethylene glycol (PEG, average Mr 20,000) was added to produce concentrations of 15% after addition to MSP. MSP precipitation was observed using a phase-contrast microscope with a 20X objective. For fiber formation, the protocol from [21] was followed. Briefly, the concentrate was diluted in KPM buffer and incubated with either 1 mM Na orthovanadate (Sigma), 500 nM staurosporine (Sigma), 150 µM IC261 (Sigma), 10 µM YOP (from Yersenia enterocolitica; Sigma) and/or 50 mM ATP (all final concentrations after addition to MSP). For fiber formation at different pH, HKB was used instead (50 mM HEPES, 25 mM KCl, 10 mM NaCL) at pH 5, 7 and 9. Fibers were observed using a phase-contrast Nikon Labophot microscope with a 100X objective and images captured using a Hamamatsu C8484-05G digital CCD camera.
Immunofluorescence Microscopy
MSP fibers were assembled in vitro on a cover-slip chamber and immunolabeled as previously described [22]. Fibers were fixed by perfusion with 1.25% glutaraldehyde in KPM buffer for 30 min and washed three times with PBS. Fibers were treated three times for 20 min with 20 mM NaBH4 to quench unreacted aldehydes and blocked in PBS/0.1% BSA for 2 hrs. Primary (4A5 anti-MSP in mouse) and secondary antibodies (AlexaFluor 488-conjugate in goat) were used at a concentration of 5µg/ml and incubated for 4 hr each. In control assays the primary antibody was omitted. Immunofluorescence labeling was examined under a 100X objective on a Nikon Labophot equipped with appropriate excitation and barrier filters for FITC.
SDS-PAGE and Western blotting
SDS-PAGE was performed using 15 % gels according to the method of [23] and stained with 1% Coomasie brilliant blue. Gels for immunoblotting were transferred to nitrocellulose membranes (Invitrogen) as described previously [24]. Blocking of the membrane was performed in TBS-T (0.1 % Tween-20, 137 mM NaCl, 20 mm Tris, pH 7.6) with 1% bovine serum albumin (BSA) for 1 hr at room temperature and probed with primary antibody for 4 hours. The membrane was washed with TBS-T and probed using a secondary HRP-conjugated antibody for 4 hrs. Bands were observed using the Supersignal West Femto kit (Pierce).
Antibodies and antiserum
Monoclonal antibodies clone PSR-45 (antiphos-phoserine; Sigma) and monoclonal 42H4 (antiphosphothreonine; Cell Signaling) were used as primary antibodies. The monoclonal antibody 4A5 anti-MSP developed by [12] was obtained from the Developmental Studies Hybridoma Bank under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. The polyclonal anti-MPAK antibody developed by [14] was kindly provided by Dr. Thomas Roberts (Florida State University, Tallahassee). Secondary antibodies used were: goat anti -mouse IgG, H&L, conjugated to horseradish peroxidase (EMD Biosciences), goat anti-rabbit IgG also conjugated to horseradish peroxidase (Santa Cruz Biotechnology), and goat anti-mouse conjugated to AlexaFluor 488 (Invitrogen). All primary antibodies were diluted 1:1,000 and the secondary antibodies were diluted 1:10,000.
Mass Spectrometry analysis
Gels were stained with Coomasie Brilliant Blue R250 showing a prominent band at ∼ 16 kDa. This band was excised and sent for identification to the W.M. Keck Proteomics Laboratory at the Center for Plant Cell Biology, University of California, Riverside. Peptides were trypsin digested and identified using Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (LC/MS/MS) and the data generated was submitted to the MASCOT database for protein identification allowing for variable modifications such as Acetyl (K), Acetyl (N-term), Formyl (N-term), Gln->pyro-Glu (N-term Q), Glu->pyro-Glu (N-term E), Oxidation (M), Phospho (ST).
Prediction of functional domains in MSP
All available C. elegans annotated MSP proteins (containing 127 aminoacids) were downloaded from Genbank and aligned using ClustalW [25], available at www.ebi.ac.uk/Tools/clustalw2/index.html. Functional MSP domains were predicted using PROSITE [26], available at http://ca.expasy.org/prosite/. Settings for the search included patterns with a high probability of occurrence. The 3D model of MSP filament structure was obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB), available at www.rcsb.org/pdb/home/home.do.
Results and discussion
MSP from C. elegans sperm extract precipitates and elongates fibers
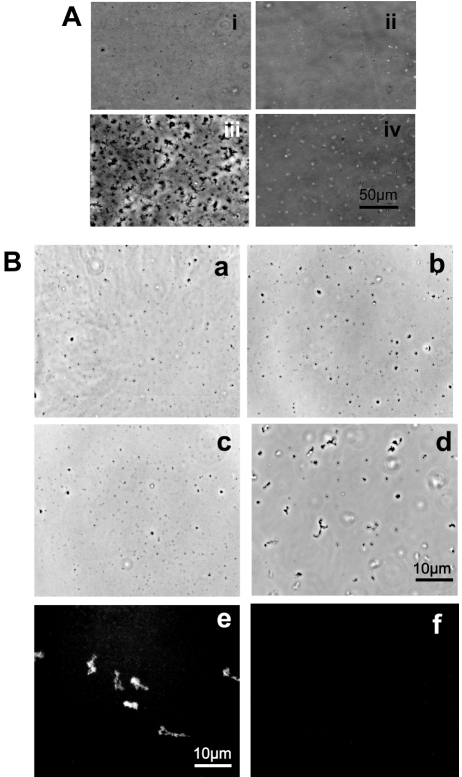
The complexity of performing large-scale isolation of C. elegans sperm has represented a disadvantage to widely spread the use of biochemical assays in the study of MSP-based motility in this model organism. Although a method for biochemical analysis of proteins helped identify MSP from C. elegans [3], only a couple of research groups have used this technique resulting in the identification of proteins involved in sperm activation and the role of MSP to induce oocyte maturation [7, 27, 28]. Other groups have opted for the purification of recombinant MSP to identify amino acid residues that mediate MSP filament elongation [29] or the use of whole-male protein lysates to perform large-scale proteomic analyses [30, 31]. On this regard, the improvement of the method to isolate C. elegans sperm in a large-scale provides an opportunity to use biochemical assays and proteomic analyses (coupled to the genetic tools that C. elegans offers) in sperm enriched extracts to understand MSP-based motility. The modified method for large-scale sperm isolation used for this work yielded ∼ 4x106 spermatids/ ml corresponding to ∼ 2 mg/ml of total protein. According to Klass and Hirsh (1981), 15% of the total protein from C. elegans sperm is MSP, thus we estimated that ∼ 0.3 mg/ml of protein in sperm extracts should be MSP. To test for the presence of MSP proteins, first, the sperm extracts from C. elegans males (him-8 strain) were precipitated with 15% Polyethylene glycol (PEG) since it has been shown that PEG precipitates MSP from Ascaris sperm extracts (King, et al 1992) as well as C. elegans MSP expressed in bacteria (recombinant MSP) [32]. Protein precipitation (similar to that of recombinant MSP) was completed within a few seconds (Figure 1A, iii). A control using BSA (1mg/ml) and 15% PEG did not result in protein precipitation (Figure 1A, iv), suggesting this is due to the proteins present in the C. elegans sperm extracts.
Figure 1.
Protein extracts from C. elegans sperm precipitate into MSP crystals and form fiber-like structures. Sperm extracts isolated using a large-scale method were tested for characteristic properties of MSP, including protein precipitation and fiber elongation. A. Sperm extracts at a concentration of 1 mg/ml were treated with 15% PEG and precipitated proteins in vitro. (i) The extract itself did not contained precipitates, (ii) the same was true for 15% PEG. (iii) Addition of 15% PEG to sperm extracts precipitated proteins instantaneously. (iv) In contrast, a control using BSA (1mg/ml) and 15% PEG did not result in protein precipitation. B. Treatment of sperm extracts with the tyrosine phosphatase YOP, followed by addition of ATP, formed MSP fibers in vitro. (a) Sperm extracts at a concentration of 1 ml/mg did not contain fiber-like structures. (b) Addition of 50 mM ATP was not sufficient to promote fiber formation. (c) Treatment of 10 µM YOP for 20 min did not induce in vitro fiber formation. (d) Fiber formation was induced after treatment of the sperm extract with 10 µM YOP for 10 min, followed by addition of 50 mM ATP. (e) The identity of the MSP fiber was corroborated using a specific C. elegans anti-MSP antibody and immunofluorescence microscopy. (f) The specificity of the anti-MSP antibody was tested by incubating MSP fibers only with a fluorescently tagged secondary antibody resulting in the lack of fluorescent signal.
It has been shown previously that addition of ATP to sperm extracts from Ascaris promotes the elongation of MSP fibers in a pH-dependent manner [21, 22]. Thus, by following the same protocols, we tested whether addition of ATP to the extracts from C. elegans sperm initiated fiber elongation. However, incubation of the extracts in 50 mM ATP at pH 5, 7 or 9 alone did not induced fiber formation. In Ascaris sperm extracts, fiber elongation (through the addition of ATP) is due to the initiation of phosphorylation and dephosphorylation events in accessory proteins involved in the polymerization of MSP filaments as shown by the use of kinase and phosphatase inhibitors [21]. Following the same protocol, we tested whether treatment with either 500 nM staurosporine (a broad kinase inhibitor), 150 µM IC261 (a casein kinase inhibitor), 1 mM Na orthovanadate (a tyrosine phosphatase inhibitor) or 10 µM YOP (a tyrosine phosphatase from Y. enterocolitica) induced fiber formation in C. elegans sperm extracts. However, none of these inhibitors alone induced the formation of fibers. The formation of fiberlike structures was induced only after the extracts were incubated with 10 µM of the tyrosine phosphatase YOP for 10 min, followed by an incubation of 50 mM ATP for 10 min (Figure 1B, d). This suggests that fiber formation requires a tyrosine dephosphorylation event in proteins present in the sperm extracts (either MSP or accessory proteins) prior to the phosphorylation induced by ATP. The formation of fiber-like structures was not immediate (as compared to protein precipitation), nevertheless, we were not able to observe the dynamic elongation of fibers using time-lapse microscopy; instead, we observed that fully formed fiber-like structures deposited on the cover slip 10 min after ATP addition. In Ascaris sperm extracts, the identification of MSP as the monomer that forms fibers was corroborated by immunofluorescence microscopy using an anti-MSP primary antibody [22]. Thus, we also employed immunofluorescence microscopy using a C. elegans anti-MSP monoclonal antibody (see materials and methods) to confirm that fiber-like structures in C. elegans sperm extracts were formed by MSP (Figure 1B, e). A control, in which only the fluorescently tagged secondary antibody was used, demonstrated the specificity of the anti-MSP antibody by the lack of fluorescent signal (Figure 1B, f).
Evidence for MSP phosphorylation using immunoblotting and Mass Spectrometry
Since phosphorylation and dephosphorylation events can induce in vitro fiber formation in Ascaris sperm [21, 22], and our results suggest this might also be the case in C. elegans sperm, we decided to identify phosphorylated proteins by immunoblotting the sperm extracts. The him-8 male sperm extracts at a concentration of 2 mg/ml were run on a 15 % SDS-PAGE gel. The gel stained with Coomassie blue contained a prominent band at ∼16 kDa, a mobility similar to the reported Mr for MSP [3, 20], (Figure 2A, red arrow). This prominent band from the acrylamide gel (total fraction) was cut and subjected to mass spectrometry for protein identification. The results from MS/MS and MASCOT sequence analysis identified 23 peptides that correspond to 9 different isoforms of MSP with a 91 to 93% of amino acid coverage, including 8 peptides specific for 7 MSP isoforms (Table 1). Two of these peptides corresponding to amino acids 70-92 in 8 MSP isoforms; and amino acids 67-89 for MSP-33 showed a putative serinethreonine (ST) phosphorylation site and no tyro-sine phosphorylation sites were present, suggesting that the tyrosine dephosphorylation event observed in fiber formation might correspond to accessory proteins that in turn could phosphorylate MSP. A previous study on the phosphoproteome of C. elegans (based on whole worm protein extraction) showed an enrichement of phosphosites in developmental and sex determination proteins [31], however, phosphorylated MSP peptides were not found in these complex samples. In the present work we used sperm protein extract for MS/MS analyis ensuring that identified peptides are exclusively present (and enriched) in spermatids of C. elegans.
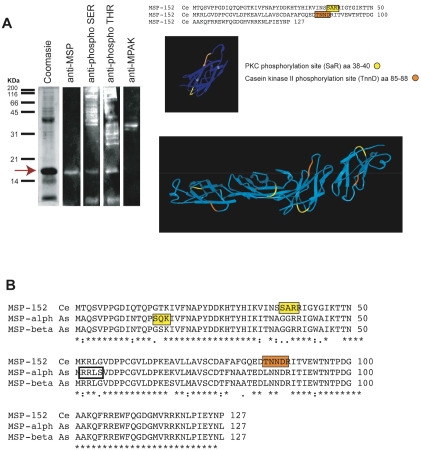
Figure 2.
MSP phosphorylation is suggested by immunoblotting and in silico prediction of functional sites. The sperm extract was subject to immunoblotting and MS/MS identification that revealed putative phosphorylation sites in the MSP sequence. A. MSP is evident as a prominent band ∼ 16 kDa (red arrow) and recognized by an anti-MSP monoclonal antibody. Anti-phosphoserine and phosphothreonine antibodies reacted to the same band identified as MSP. An antibody against MPAK, a Casein kinase from Ascaris sperm, was used as a control for non-specific binding and it cross-reacted with a band ∼ 38 kDa. MS/MS analysis identified the prominent band as C. elegans MSP containing a peptide with a putative phosphorylation site. Using in silico analysis, two predicted phosphorylation sites were identified: A Protein Kinase C phosphorylation site (SaR) in amino acids 38-40 (yellow); and a Casein Kinase II phosphorylation site (TnnD) in amino acids 85-88 (orange). The 3-D model of MSP localized the predicted phosphorylation sites in MSP (pdb id: 1grw) outside the protein-protein interaction domain and exposed for kinase phosphorylation as shown in a filament of MSP. In this figure, the sequence of C. elegans MSP-152 is used as an example. B. Differences on the MSP phosphorylation sites between Ascaris and C. elegans sperm are shown in the alignment of a representative protein sequence of MSP from C. elegans (MSP-152 Ce; NP_494901), and the two Ascaris isoforms: alpha (MSP-alph As; P27439) and beta (MSP-beta As; P27440). Predicted phosphorylation sites are: PKC phosphorylation sites (yellow); Casein kinase phosphorylation site (orange); and PKA phosphorylation site (white).
Table 1.
List of peptide sequences identified as MSP by MS/MS (91 to 93% of amino acid coverage) with their respective amino acid residue and putative modification corresponding to the full-length protein sequences of 9 different MSP isoforms
| MS/MS peptide sequence | Corresponding amino acids residue |
|---|---|
| AQSVPPGDIQTQPGTK; Acetyl (N-term) | 2-17; b,d,e,f,g,h |
| AQSVPPGDIQTQPNAK; Acetyl (N-term) | 2-17; c |
| AHSAQSVPPGDIQTQPGTK; Acetyl (N-term) | 2-20; a |
| IVFNAPYDDK | 21-30; a,b,c,d,e,f,g,h,i |
| IVFNAPYDDKHTYHIK | 21-36; a,b,c,d,e,f,g,h,i |
| HTYHIK | 31-36; a,b,c,d,e,f,g,h,i |
| VINSSAR | 37-43; a,b,c,d,e,f,g,h,i |
| RIVYGIK | 41-47; f |
| RIGYGIK; Acetyl (N-term) | 44-50; a,b,c,d,e,g,h,i |
| IGYGIK | 45-50; a,b,c,d,e,g,h,i |
| LGVDPPCGVLDPK | 57-69; a,b,c,d,e,g,h,i |
| EAVFLAVSCDAFAFGQEDTNNDR; Phospho (ST) | 67-89; d |
| EAVLLAVSCDAFAFGQEDTNNDR; Phospho (ST) | 70-92; a,b,c,e,f,g,h,i |
| ITVEWTNTPDGAAR | 90-103; b |
| ITIEWTNTPDGAAK | 90-103; h |
| ITVEWTNTPDGAAK | 93-106; a,c,d,e,f,g,i |
| ITVEWTNTPDGAAKQFR | 93-109; a,c,d,e,f,g,i |
| REWFQGDGMAR; Oxidation (M) | 107-117; g |
| EWFQGDGMAR; Oxidation (M) | 108-117; g |
| REWFQGDGMVR; Oxidation (M) | 110-120; a,b,c,d,e,f,h,i |
| EWFQGDGMVR; Oxidation (M) | 111-120; a,b,c,d,e,f,h,i |
| KNLPIEYNP | 122-130; a,b,c,d,e,f,g,h,i |
| NLPIEYNP | 123-130; a,b,c,d,e,f,g,h,i |
MSP isoform; Gene ID; NCBI accession No
predicted MSP; Y59E9AR.1; NP 500755
MSP-3; F26G1.7; NP 494858
MSP-10; K07F5.2; NP 501760
MSP-33; R05F9.8; NP 494888
MSP-49; C34F11.6; NP 494970
MSP-55; C09B9.6; NP 500711
MSP-77; F32B6.6; NP 501781
MSP-78; T13F2.11; NP 501742
MSP-152; ZK5456.6; NP 494901
We next corroborated the identity of the prominent band as MSP by immunoblot employing the same C. elegans specific anti-MSP antibody used in the immunofluorescence assay. However, this antibody (which recognizes the amino acid residues 106-126 from the C-terminus of C. elegans MSP) labeled only ∼ 40% the area of the total band from the Coomasiestained gel, suggesting that it recognized only a subfraction of MSP present in the crude extract. This result may be in agreement with the fact that at least three MSP protein isoforms have been detected by isoelectric focusing in C. elegans sperm [9] and can be supported by our MS/MS results in which at least one amino acid of MSP- 77 differs from the other 8 isoforms in the residues 106-126. Thus the possibility of diverse post-translationally modified isoforms of MSP is a logical possibility.
Lastly, we used specific antibodies directed against phospho-serine and phospho-threonine residues (see materials and methods) to search for protein phosphorylation in the sperm extracts. Both antibodies labeled the identical band previously identified as the MSP subfraction (Figure 2A). An antibody against the MSP Polymerization-Activating Kinase (MPAK, a Ser/ Thr Casein kinase involved in filament elongation at the leading edge of the pseudopod in Ascaris sperm) was used as a control for nonspecific binding. This antibody cross-reacted with a ∼ 38 kDa band, in concordance with the predicted molecular weight of SPE-6, a C. elegans Casein kinase homologue of MPAK [14] involved in both MSP filament assembly during spermatogenesis [33] and the suppression of pseudopod extension in spermatids [34]. Although the identification of SPE-6 in sperm extracts was not pursued, this method of large-scale sperm isolation coupled to MS/MS and biochemical assays will give us the opportunity to identify this as well as other accessory proteins involved in MSP filament formation in C. elegans sperm.
In silico prediction of MSP phosphorylation sites
Presence of phosphorylation sites in known MSP protein sequences of C. elegans was predicted by PROSITE (see materials and methods). For all 28 MSP sequences available in Gene-Bank, the program returned two predicted phosphorylation sites: A Protein Kinase C phosphorylation site (SaR) in amino acids 38-40; and a Casein Kinase II phosphorylation site (TnnD) in amino acids 85-88 (Figure 2A). The second predicted site is in agreement with the above mentioned involvement of SPE-6 in both MSP filament assembly and the suppression of sperm motility [34]. In addition, two N-myristoylation sites are predicted in the MSP sequences: (GIktTN) in amino acids 45-50 and (GQedTN) in amino acids 81-86, together with a previously reported N-glycosylation site (NSSA) [35] in amino acids 36-39 (not shown). Alignment of the 28 MSP protein sequences indicates no amino acid variation at any of these sites, suggesting conservation (data not shown). The 3-D model of MSP filaments available at the Protein Data Bank (PDB) was used to localize the predicted phosphorylation sites in MSP (pdb id: 1grw). Based on the Molecular Biology Toolkit (MBT) Protein Workshop application [36], the predicted phosphorylation sites are localized outside the protein-protein interaction domain and thus appear to be available for kinase phosphorylation (Figure 2A). Earlier attempts to label spermatids in vitro using [32P] orthophosphate (before or after activation with Triethanolamine) failed to detect any phosphorylation modifications associated with MSP, suggesting that MSP is not directly phosphorylated during pseudopod extension [9]. In this regard, it is important to point out that MSP phosphorylation by SPE-6 is necessary for filament formation and proper segregation of membranous organ-elles (MOs) into the developing spermatocyte [33], thus, posttranslational modification must occur prior to the differentiation of primary sper-matocytes in the gonad, immediately after MSP synthesis [3]. In this scenario, the complete set of posttranslationally modified MSP isoforms must be packed into the Fibrous Body-Membranous Organelle (FB-MO) complex that ensures the delivery of membrane and cytoplasmic proteins into the developing spermatid and their proper localization in the fully differentiated spermatozoon [37]. Thus, the transcriptionally inactive sperm of C. elegans [38] must rely on posttranslational modifications of proteins to coordinate proper acquisition of motility and subsequent fertilization events, processes that seems to be conserved among both amoeboid and flagellated sperm (for review see [39]).
Although the direct interaction between MSP and SPE-6 has been previously hypothesized during filament nucleation and assembly [33], from our results we cannot conclude whether SPE-6 directly phosphorylates MSP in C. elegans or if phosphorylation occurs exclusively at a specific amino acid residue. Thus, additional biochemical assays will be necessary to elucidate the role of accessory proteins and MSP phosphorylation in C. elegans sperm.
Comparative in silico analysis of MSP phosphorylation sites
In contrast to C. elegans, biochemical and structural studies performed in Ascaris sperm have ruled out phosphorylation sites in MSP from this nematode. Since MSPs from Ascaris and C. elegans sperm share 83% amino acid identity [20, 29], we decided to screen MSP sequences from these two nematodes to search for differences in the pattern of predicted phosphorylation sites. In Ascaris sperm only two isoforms of MSP (alfa and beta) have been identified and, in contrast with results given for all C. elegans MSPs, no phosphorylation sites were predicted for the most abundant isoform, beta-MSP (accession no. P27440), while the alfa-MSP amino acid sequence (accession no. P27439) contained a Protein Kinase C (PKC) phosphorylation site (SqK) in amino acids 14-16 and a cGMP-dependent protein kinase (PKA) phosphorylation site (RRlS) in amino acids 51-54 (Figure 2B). It is interesting to note that, according to King et al. (1992), the two isoforms differ only in four amino acid residues that render alfa-MSP more basic and with properties that enable it to form filaments in vitro at both lower and precipitant concentrations. These characteristics are thought to allow alfa-MSP to nucleate filament assembly or to lengthen existing filaments rapidly in the plasmalemmal end of the fiber, making it a good candidate for the hypothesized activated MSP isoform (MSP*) responsible for the rapid polymerization of filaments in the vicinity of the plasma membrane [15]. Furthermore, PROSITE did not return any significant Casein kinase phosphorylation domain for the Ascaris MSP sequences; this is also in agreement with the fact that MPAK phosphorylates the accessory protein MFP2 (MSP Fiber Protein 2) and not MSP directly in Ascarsis sperm [14] and would support a model in which subtle changes in the amino acid composition of MSP proteins leads to changes in their ability to undergo posttranslational modifications with an impact on their physiological role during fertilization. Nevertheless, to further understand the evolutionary conserved processes that orchestrate sperm motility and reproduction in nematodes, biochemical and proteomic comparative analyses among parasitic and free-living species with different modes of reproduction (gonochoristic, hermaphroditic and parthenogenetic) will be needed. Thus, we believe that the use of this modified large-scale sperm isolation method will enable us to follow up on the phosphorylation of MSP in nematode sperm and the evolutionary conserved mechanisms that allow for successful nematode sperm activation and fertilization.
In summary, we have successfully standardized a methodology to reproducibly obtain relative high amounts of proteins from C. elegans sperm extract that can be used in biochemical assays. The use of this methodology (coupled to immunoblotting, Mass Spectrometry, and in silico prediction of phosphorylation sites) indicates a tyrosine dephosphorylation event (putatively in cytoskeletal accessory proteins) that in turn promotes the phosphorylation of serine and threonine residues in a subfraction of MSP from C. elegans sperm extract. These results together with previous findings [35], raise the possibility of differential posttranslational modifications in MSP isoforms that would explain the multiple roles of this protein both in sperm motility (by differentially localizing MSP at the plasma membrane for filament nucleation or in the cytosol for filament elongation) and as a signaling molecule that promotes oocyte maturation through sperm vesicle budding [7]. The multifunctional characteristics of MSP are in agreement with the structural characteristics of MDPs as integral membrane proteins that provide a scaffold for protein-protein interaction. Thus, future research on this topic will also lead to a better understanding of MDPs as proteins involved in linking membrane and cytosolic components during membrane fusion events.
Conclusions
Amoeboid cell motility has been extensively studied in actin-based systems in vivo and in vitro, and the molecular pathways that lead to pseudopod extension have been fully dissected [40]. Current proteomic approaches are providing new insights on a diverse range of cellular functions (such as novel regulations and interactions of signaling proteins with the actin cytoskeleton) based on the analysis of complex biological samples [41-43]. Recently, these approaches have also been used for the identification of evolutionary conserved proteins in spermatozoa that may ultimately serve as putative targets for male contraception [30, 44-47]. Thus, given the advantages that C. elegans offers as a model organism (and complementing the genetic information available with proteomics, biochemistry, molecular biology and imaging analysis) we believe there is an opportunity to elucidate both the role of MSP post-translational modifications on the cytoskeletal dynamics of amoeboid sperm and the analogous signaling pathways that lead to the acquisition of cell motility between MSP- and actin-based system.
Acknowledgements
We thank Dr. Tom Roberts (Florida State University, Tallahassee) for useful comments in the preparation of this manuscript and for sharing an aliquot of MPAK antibody. We are also thankful to Dr. Kexi Yi for technical advice in the protocol for fiber formation and immunofluorescence.
References
- [1].Nelson GA, Roberts TM, Ward Caenorhabditis elegans spermatozoan locomotion: Amoeboid movement movement with almost no actin. Journal of Cell Biology. 1982;92:121–131. doi: 10.1083/jcb.92.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rohtagi R, Ma L, Miki H, lopez M, Kircchausen T, Takenawa, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- [3].Klass MR, Hirsh D. Sperm isolation and biochemical analysis of the Major Sperm Protein from Caenorhabditis elegans. Developmental Biology. 1981;84:299–312. doi: 10.1016/0012-1606(81)90398-5. [DOI] [PubMed] [Google Scholar]
- [4].Stewart M, King KL, Roberts TM. The motile Major Sperm Protein (MSP) of Ascaris suumforms filaments constructed from two helical subfilaments. Journal of Molecular Biology. 1994;243:60–71. doi: 10.1006/jmbi.1994.1630. [DOI] [PubMed] [Google Scholar]
- [5].King KL, Stewart M, Roberts TM. Supramolecular assemblies of the Ascaris suum major sperm protein (MSP) associated with amoeboid cell motility. Journal of Cell Science. 1994;107:2941–2949. doi: 10.1242/jcs.107.10.2941. [DOI] [PubMed] [Google Scholar]
- [6].Roberts TM, Stewart M. Acting like actin: The dynamics of the nematodeMajor Sperm Protein (MSP) cytoskeleton indicate a push-pull mechanism for amoeboid cell motility. Journal of Cell Biology. 2000;149:7–12. doi: 10.1083/jcb.149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- [8].Ward S, Burke DJ, Sulston JE, Coulson AR, Albertson DG, Ammons D, Klass M, Hogan E. Genomic organization of the Major Sperm Protein genes and pseudogenes in the nematode Caenorhabditis elegans. J Mol Biol. 1988;199:1–13. doi: 10.1016/0022-2836(88)90374-9. [DOI] [PubMed] [Google Scholar]
- [9].Burke D, Ward S. Identification of a large multigene family encoding the major sperm protein of Caenorhabditis elegans. Journal of Molecular Biology. 1983;171:1–29. doi: 10.1016/s0022-2836(83)80312-x. [DOI] [PubMed] [Google Scholar]
- [10].Tarr DE, Scott AL. MSP domain proteins. TRENDS in Parasitology. 2005;21:224–231. doi: 10.1016/j.pt.2005.03.009. [DOI] [PubMed] [Google Scholar]
- [11].Laurent F, Labasse G, de Wit P. Molecular cloning and partial characterization of a plant VAP33 homologue with a Major Sperm Protein domain. Biochemical and Biophysical Research Communications. 2000;270:286–292. doi: 10.1006/bbrc.2000.2387. [DOI] [PubMed] [Google Scholar]
- [12].Kosinski M, McDonald K, Schwartz J, Yamamoto I, Greenstein D. C. elegans sperm bud vesicles to deliver a meiotic maturation signal too distant oocytes. Development. 2005;132:3357–3369. doi: 10.1242/dev.01916. [DOI] [PubMed] [Google Scholar]
- [13].Buttery SM, Ekman GC, Seavy M, Stewart M, Roberts TM. Dissection of the Ascaris sperm motility machinery identifies key proteins involved in major sperm protein-based amoeboid locomotion. Molecular Biology of the Cell. 2003;14:5082–5088. doi: 10.1091/mbc.E03-04-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yi K, Buttery SM, Stewart M, Roberts TM. A ser/thr kinase required for membrane-associated assembly of the major sperm protein motility apparatus in the amoeboid sperm of Ascaris. Molecular Biology of the Cell. 2007;18:1816–1825. doi: 10.1091/mbc.E06-08-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roberts TM, Salmon ED, Stewart M. Hydrostatic pressure shows that lamellipodial motility in Ascaris sperm requires membrane-associated major sperm protein filament nucleation and elongation. Journal of Cell Biology. 1998;140:367–375. doi: 10.1083/jcb.140.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].L’Hernault SL, Roberts TM. Cell biology of nematode sperm. Methods in Cell Biology. 1995;48:273–301. doi: 10.1016/s0091-679x(08)61392-8. [DOI] [PubMed] [Google Scholar]
- [17].Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hodgkin J, Horvitz HR, Brenner S. Nondis-junction mutants of the nematode Caeonorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ward S, Argon Y, Nelson GA. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. Journal of Cell Biology. 1981;91:26–44. doi: 10.1083/jcb.91.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].King KL, Stewart M, Roberts TM, Seavy M. Structure and macromolecular assembly of two isoforms of the major sperm protein (MSP) from the amoeboid sperm of the nematode, Ascaris suum. Journal of Cell Science. 1992;101:847–857. doi: 10.1242/jcs.101.4.847. [DOI] [PubMed] [Google Scholar]
- [21].Miao L, Yi K, Mackey JM, Roberts TM. Re-constitution in vitro of MSP-based filopodium extension in nematode sperm. Cell Motility and the Cytoskeleton. 2007;64:235–247. doi: 10.1002/cm.20177. [DOI] [PubMed] [Google Scholar]
- [22].Italiano JE, Roberts TM, Stewart M, Fontana C. Reconstitution in vitro of the motile apparatus from the amoeboid sperm of Ascaris shows that filament assembly and bundling move membranes. Cell. 1996;84:105–114. doi: 10.1016/s0092-8674(00)80997-6. [DOI] [PubMed] [Google Scholar]
- [23].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [24].Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedures and some applications. PNAS. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Larkin M, Blackshields G, Brown NP, Chenna R, McGettigan PA, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- [26].Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Research. 1991;19:2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nance J, Minniti A, Sadler C, Ward S. spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics. 1999;152:209–220. doi: 10.1093/genetics/152.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Washington NL, Ward S. FER-1 regulates Ca2+-mediated membrane fusion during C elegans spermatogenesis. Journal of Cell Science. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- [29].Smith HE, Ward S. Identification of protein-protein interactions of the Major Sperm Protein (MSP) of Caenorhabditis elegans. J Mol Biol. 1998;279:605–619. doi: 10.1006/jmbi.1998.1793. [DOI] [PubMed] [Google Scholar]
- [30].Chu DS, Liu H, Nix P, Wu TF, Ralston EJ, Yates JR, III, Meyers BJ. Sperm chromatin proteomics identifies evolutionary conserved fertility factors. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zielinska DF, Gnad F, Jedrusik-Bode M, Wisniewski JR, Mann M. Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. Journal of Proteome Research. 2009;8:4039–4049. doi: 10.1021/pr900384k. [DOI] [PubMed] [Google Scholar]
- [32].del Castillo-Olivares A, Smith HE. Critical contact residues that mediate polymerization of nematode Major Sperm Protein. Journal of Cellular Biochemistry. 2008;104:477–487. doi: 10.1002/jcb.21636. [DOI] [PubMed] [Google Scholar]
- [33].Varkey JP, Jansma PL, Minniti AN, Ward S. The Caenorhabditis elegans spe-6 gene is required for Major Sperm Protein assembly and shows second site non-complementation with an unlinked deficiency. Genetics. 1993;133:79–86. doi: 10.1093/genetics/133.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Muhlrad PJ, Ward S. Spermiogenesis initiation in Caenorhabditis elegans involves a Casein kinase 1 encoded by the spe-6 gene. Genetics. 2002;161:143–155. doi: 10.1093/genetics/161.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Klass MR, Kinsley S, Lopez LC. Isolation and characterization of a sperm-specific gene family in the nematode Caenorhabditis elegans. Mol Cell Biol. 1984;4:529–537. doi: 10.1128/mcb.4.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Moreland J, Gramada A, Buzko OV, Zhang Q, Bourne PE. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Roberts TM, Pavalko FM, Ward S. Membrane and cytoplasmic proteins are transported in the same organelle complex during nema-tode spermatogenesis. J Cell Biol. 1986;102:1787–1796. doi: 10.1083/jcb.102.5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pavalko FM, Roberts TM. Posttranslational insertion of a membrane protein on Caenorhabditis elegans sperm occurs without de novo protein synthesis. Journal of Cellular Biochemistry. 1989;41:57–70. doi: 10.1002/jcb.240410203. [DOI] [PubMed] [Google Scholar]
- [39].Fraire-Zamora JJ, Cardullo RA. The physiological acquisition of amoeboid motility in nematode sperm: Is the tail the only thing the sperm lost? Mol Rerpod Dev. 2010;77(9):739–50. doi: 10.1002/mrd.21193. [DOI] [PubMed] [Google Scholar]
- [40].Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- [41].Liu N, Academia K, Rubio T, Wehr t, Yeck T, Jordan L, Hamby K, Paulus A. Actin deficiency induces cofilin phosphorylation: Proteome analysis of HeLa cells after beta-actin gene silencing. Cell Mot Cyto. 2007;64:110–120. doi: 10.1002/cm.20169. [DOI] [PubMed] [Google Scholar]
- [42].Vandermoere F, Yazidi-Belkoura IE, Demont Y, Slomianny C, Antol J, Lemoine J, Hondermarck H. Proteomics exploration reveals that actin is a signaling target of the kinase Akt. Mol Cell Prot. 2007;6:114–124. doi: 10.1074/mcp.M600335-MCP200. [DOI] [PubMed] [Google Scholar]
- [43].Akgül B, Zigrino P, Frith D, Hanrahan S, Storey A. Proteomic analysis reveals the actin cytoskeleton as cellular target for the human papillomavirus type 8. Virology. 2009;386:1–5. doi: 10.1016/j.virol.2009.01.036. [DOI] [PubMed] [Google Scholar]
- [44].Karr TL. Fruit flies and the sperm proteome. Hum Mol Genet. 2007;16:R124–R133. doi: 10.1093/hmg/ddm252. [DOI] [PubMed] [Google Scholar]
- [45].Aitken RJ, Baker MA. The role of proteomics in understanding sperm cell biology. Int J Androl. 2008;31:295–302. doi: 10.1111/j.1365-2605.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- [46].Oliva R, de Mateo S, Estanyol JM. Sperm cell proteomics. Proteomics. 2009;9:1004–1017. doi: 10.1002/pmic.200800588. [DOI] [PubMed] [Google Scholar]
- [47].Brewis IA, Gadella BM. Sperm surface proteomics: from protein list to biological function. Mol Hum Reprod. 2010;16:68–79. doi: 10.1093/molehr/gap077. [DOI] [PubMed] [Google Scholar]