Abstract
By studying the loci that contribute to human longevity, we aim to identify mechanisms that contribute to healthy aging. To identify such loci, we performed a genome-wide association study (GWAS) comparing 403 unrelated nonagenarians from long-living families included in the Leiden Longevity Study (LLS) and 1670 younger population controls. The strongest candidate SNPs from this GWAS have been analyzed in a meta-analysis of nonagenarian cases from the Rotterdam Study, Leiden 85-plus study, and Danish 1905 cohort. Only one of the 62 prioritized SNPs from the GWAS analysis (P < 1 × 10−4) showed genome-wide significance with survival into old age in the meta-analysis of 4149 nonagenarian cases and 7582 younger controls [OR = 0.71 (95% CI 0.65–0.77), P = 3.39 × 10−17]. This SNP, rs2075650, is located in TOMM40 at chromosome 19q13.32 close to the apolipoprotein E (APOE) gene. Although there was only moderate linkage disequilibrium between rs2075650 and the ApoE ε4 defining SNP rs429358, we could not find an APOE-independent effect of rs2075650 on longevity, either in cross-sectional or in longitudinal analyses. As expected, rs429358 associated with metabolic phenotypes in the offspring of the nonagenarian cases from the LLS and their partners. In addition, we observed a novel association between this locus and serum levels of IGF-1 in women (P = 0.005). In conclusion, the major locus determining familial longevity up to high age as detected by GWAS was marked by rs2075650, which tags the deleterious effects of the ApoE ε4 allele. No other major longevity locus was found.
Keywords: aging, apolipoprotein E, genetics, genome-wide association study, human, longevity
Introduction
Worldwide human populations have shown an increase in mean life expectancy in the past two centuries (Oeppen & Vaupel, 2002). This is mainly because of environmental factors such as improved hygiene, nutrition, and health care. The large variation in healthy lifespan among the elderly has prompted research into the determinants of aging and lifespan regulation. The genetic contribution to human lifespan variation was estimated at 25–30% in twin studies (Gudmundsson et al., 2000; Skytthe et al., 2003; Hjelmborg et al., 2006). The most prominent genetic influence is observed in families in which the capacity to attain a long lifespan clusters (Perls et al., 2000; Schoenmaker et al., 2006). Exceptional longevity can be reached with a low degree of age-related disability (Christensen et al., 2008; Terry et al., 2008), raising the question whether protective mechanisms against disease exist in long-lived subjects.
In most experimentally modified animal model systems, single-gene mutations in many different genes have major life extension effects (Fontana et al., 2010; Kenyon, 2010). However, natural human and animal longevity is presumed to be a complex trait (Finch & Tanzi, 1997). In humans, both candidate gene and genome-wide genetic association approaches have been applied in an attempt to identify longevity loci. The frequency of genetic variants has been typically compared between nonagenarian cases and young controls, revealing loci at which genetic variants may contribute to a higher or lower probability of survival into old age. The initial candidate gene studies aimed at finding human longevity genes were dominated by contradictory results (Christensen et al., 2006). The more consistent evidence obtained by repeated observation in independent cohort studies for association with longevity has so far only been observed for three loci, the apolipoprotein E (APOE) locus (Schachter et al., 1994; Christensen et al., 2006), the FOXO3A locus (Willcox et al., 2008; Flachsbart et al., 2009; Pawlikowska et al., 2009; Soerensen et al., 2010), and the AKT1 locus (Pawlikowska et al., 2009). Thus, despite the expectation that longevity would be influenced by many genetic variants with small effect sizes, the effect of variants has consistently been shown in only three genes.
Hypothesis-free genome-wide approaches have also been undertaken. Genome-wide linkage scans reported evidence for linkage with longevity on chromosome 4q25 (Puca et al., 2001), 3p24-22, 9q31-34, and 12q24 (Boyden & Kunkel, 2010). However, the evidence for these loci is still very weak as the results, obtained in centenarians and their families, could not be replicated in nonagenarian sibling pairs (Beekman et al., 2006) or have yet to be tested in other studies. A meta GWAS of survival to 90 years or older in 1836 cases and 1955 controls did not find any significant genome-wide associations (Newman et al., 2010). Thus far, hypothesis-free approaches have not identified any loci involved in longevity.
In a few studies, such as the Ashkenazi Jewish Centenarian Study and the Leiden Longevity Study (LLS), different generations of long-lived families are being investigated for parameters and pathways contributing to the longevity phenotype (Atzmon et al., 2004; Schoenmaker et al., 2006). The survival benefit of the LLS families is marked by a 30% decreased mortality risk in the survival analysis of three generations, i.e., the parents of the probands in this study (nonagenarian sibling pairs), their unselected additional siblings, and their offspring (Schoenmaker et al., 2006). As compared to their partners, the offspring of nonagenarians siblings have a lower prevalence of type 2 diabetes, myocardial infarction and hypertension (Westendorp et al., 2009), a beneficial glucose, lipid, and thyroid metabolism, and a preservation of insulin sensitivity with age (Rozing et al., 2009, 2010a,b; Vaarhorst et al., 2011; Wijsman et al., 2011). Hence, in middle age, these families display beneficial metabolic profiles.
Because the longevity phenotype is inherited in the LLS families, they offer a route to identify genetic variants that influence human longevity. Previously, we tested whether the absence of GWAS-identified alleles promoting common diseases might explain their familial longevity (Beekman et al., 2010). Longevity was not easily explained by the absence of disease-susceptibility alleles. More likely therefore, the genome of the long-lived harbors longevity-promoting alleles. To identify such loci, we performed a GWAS comparing nonagenarian siblings from the LLS and younger population controls. We subsequently investigated emerging candidate SNPs in nonagenarian cases from the Rotterdam Study, the Leiden 85-plus study, and the Danish 1905 cohort.
Results
GWAS
A GWAS was performed in nonagenarian participants from the LLS and middle-aged controls from the Rotterdam Study (RS). Genotype data for 516,721 SNPs that passed quality control thresholds were analyzed in a comparison of 403 unrelated nonagenarians (94 years on average) and 1670 controls (58 years on average). A flow chart of the consecutive analysis steps is depicted in Fig. 1, and a description of the population samples investigated in the GWAS and subsequent replication studies is given in Table 1. Results of the association analysis of stage 1 are depicted in Fig. S1. None of the SNPs reached genome-wide significance (P < 5 × 10−8).
Fig. 1.
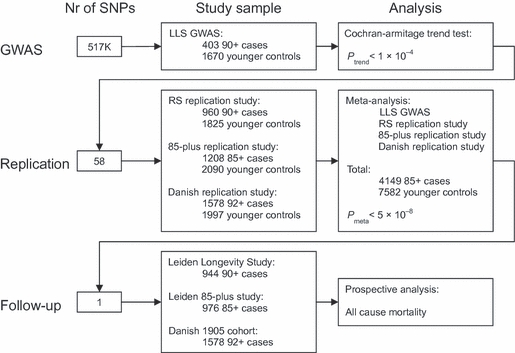
Flow chart of experimental work.
Table 1.
Characteristics of the genotyped samples used for analysis
| Study | SNPs | Samples | Number | Mean age | Age range | Men/women |
|---|---|---|---|---|---|---|
| LLS GWAS | 517K | Cases | 403 | 94 | 89–102 | 137/266 |
| 517K | Controls | 1670 | 58 | 55–59 | 745/925 | |
| RS replication study | 58 | Cases | 960 | 94 | 90–106 | 217/743 |
| 58 | Controls | 1825 | 62 | 60–65 | 805/1020 | |
| Leiden 85-plus replication study | 58 | Cases | 1208 | 92 | 85–109 | 372/836 |
| 58 | Controls | 2090 | 35 | 15–70 | 743/1347 | |
| Danish replication study | 58 | Cases | 1578 | 93 | 92–93 | 430/1148 |
| 58 | Controls | 1997 | 57 | 46–68 | 900/1097 |
LLS, Leiden Longevity Study; RS, Rotterdam Study; GWAS, genome-wide association study.
Replication studies
We prioritized the SNPs that had the most significant association with survival into old age according to the analysis of stage 1 (P < 1 × 10−4, Table S1). For 58 of the 62 selected SNPs, successful genotyping was obtained in the replication cohorts. In stage 2, these 58 SNPs were tested for association comparing 960 RS replication cases (mean age of 93 years), 1208 Leiden 85-plus replication cases (mean age of 92 years), and 1578 Danish replication cases (mean age of 93 years) with appropriate middle-aged population controls (Table 1). Meta-analysis for the 58 SNPs, comprising a total of 4149 nonagenarian cases and 7582 younger controls (from the LLS GWAS, RS replication, Leiden 85-plus replication, and Danish replication studies), was performed.
Rs2075650 on chromosome 19 was the only SNP that was associated with survival into old age at the genome-wide significance level (P = 3.39 × 10−17) (Table S2A). The minor allele was underrepresented among the older cases as compared to middle-aged controls, hence associated with the decreased probability of carriers surviving into old age corresponding to an odds ratio (OR) below unity [OR = 0.71 (95% CI 0.65-0.77)]. This effect is observed in both sexes (Table S2B, C). The remaining 57 SNPs did not show genome-wide significant effects on longevity either in men or women (Table S2B for men and S2C for women). The association of rs2075650 with survival did show some heterogeneity across the four studies (P = 0.0495), which is mainly because of the RS.
rs2075650 and the APOE ε2/ε3/ε4 polymorphism
Rs2075650 is located in the TOMM40 gene, next to the APOE gene (Fig. S2). APOE was previously associated with longevity (Schachter et al., 1994; Christensen et al., 2006). The ApoE protein has three isoforms (ApoE ε2, ApoE ε3, and ApoE ε4) which are defined by two SNPs, rs7412 (Arg136Cys; ε2) and rs429358 (Cys112Arg; ε4). A meta-analysis of rs7412 and rs429358, in the LLS GWAS study, the Leiden 85-plus replication study, and the Danish replication study samples (3189 cases and 5757 controls), showed a significant association of rs429358 with longevity [OR = 0.62 (95% CI 0.56–0.68), P = 1.33 × 10−23], which was comparable to rs2075650 [OR = 0.67 (95% CI 0.61–0.74), P = 9.15 × 10−17]. Rs7412 also showed an association with longevity, with a higher prevalence of the minor allele in nonagenarians [OR = 1.31 (95% CI 1.17–1.46), P = 1.35 × 10−6].
We observed only moderate linkage disequilibrium (LD) between rs2075650 and rs429358 (r2a = 0.553) and low LD between rs2075650 and rs7412 (r2a = 0.014) when analyzing all samples with genotype data of rs2075650, rs429358, and rs7412 (na = 8946). Nevertheless, in a conditional analysis with rs429358 and rs7412 (Model 1, described in the Experimental procedures section), rs2075650 was no longer associated with longevity [OR = 0.93 (95% CI 0.81–1.07), P = 0.337]. The OR increased from 0.67 to 0.93, i.e., the deleterious effect of rs2075650 on longevity diminishes and is statistically non-significant. However, the deleterious effect of rs429358 [OR = 0.64 (95% CI 0.56–0.74), P = 2.68 × 10−9] and the protective effect of rs7412 [OR = 1.20 (95% CI 1.07–1.36), P = 0.002] on longevity remained significant.
To determine whether there was an APOE-independent effect of rs2075650 on survival after 90 years, prospective analysis of rs2075650, adjusted for rs429358 and rs7412, was performed. This analysis showed that carriers of the minor allele of rs2075650 displayed no increased mortality, i.e., a significant hazard ratio (HR) above 1, after 90 years of age independently of APOE in two of the three cohorts analyzed [LLS, HR = 0.99 (95% CI 0.78–1.25), P = 0.914; Leiden 85-plus study, HR = 1.06 (95% CI 0.89–1.27), P = 0.521; Danish 1905 cohort, HR = 1.21 (95% CI 1.01–1.44), P = 0.036) (Table S3A, Fig. S3).
Overall, our results suggest that the association of rs2075650 with longevity is most likely a reflection of the effects of rs429358, caused by the moderate LD between the loci.
Association of rs429358 (ε4) and rs2075650 with serum parameters
As previous studies showed that rs429358 was associated with several metabolic phenotypes (Boerwinkle & Utermann, 1988; Topic et al., 2008; Hubacek et al., 2010), association of this SNP with relevant serum parameters was determined in the offspring of the elderly LLS cases and their partners (na = 2324, Model 2 described in the Experimental procedures section). We replicated the previously reported associations of rs429358 with plasma levels of ApoE (P = 7.42 × 10−28), total cholesterol (P = 0.001), LDL cholesterol (P = 4.91 × 10−5), HDL cholesterol (P = 0.062), and high sensitivity C-reactive protein (hsCRP) (P = 0.028) and with HDL (P = 0.061) and LDL particle size (P = 0.062) (Table 2). In addition, we detected a minor effect on IGF-1 (P = 0.025) and IGFBP3 levels (P = 0.042) (Table 2). The effect on IGF-1 seems to be female-specific (P = 0.005 and P = 0.748, in women and men, respectively) and is still significant after correction for multiple testing. We observed no APOE-independent effect of rs2075650 on these traits, except for an increase of 0.18 mmol L−1 total cholesterol (P = 0.017) and 0.14 mmol L−1 LDL cholesterol (P = 0.014) with each minor allele of rs2075650 (using Model 3 described in the Experimental procedures section).
Table 2.
Association analysis of serum parameters between carriers and non-carriers of rs429358
| All | Males | Females | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serum parameter | N* | Effect† | 95% CI‡ | P-value§ | N* | Effect† | 95% CI‡ | P-value§ | N* | Effect† | 95% CI‡ | P-value§ |
| ApoE (mg dL−1)¶ | 2222 | 0.83 | 0.80 to 0.86 | 7.42 × 10−28 | 1015 | 0.85 | 0.80 to 0.89 | 2.74 × 10−11 | 1207 | 0.81 | 0.78 to 0.85 | 1.13 × 10−22 |
| Total cholesterol (mmol L−1) | 2229 | 0.18 | 0.07 to 0.29 | 0.001 | 1019 | 0.18 | 0.04 to 0.32 | 0.011 | 1210 | 0.18 | 0.02 to 0.33 | 0.024 |
| HDL cholesterol (mmol L−1) | 2228 | −0.04 | –0.07 to 0.00 | 0.062 | 1018 | −0.04 | −0.09 to 0.00 | 0.064 | 1210 | −0.03 | −0.08 to 0.02 | 0.286 |
| LDL cholesterol (mmol L−1) | 2168 | 0.20 | 0.10 to 0.29 | 4.91 × 10−5 | 978 | 0.19 | 0.07 to 0.31 | 0.002 | 1190 | 0.20 | 0.07 to 0.33 | 0.003 |
| HDL Size (nm) | 2219 | −0.04 | −0.08 to 0.00 | 0.061 | 1011 | −0.04 | −0.10 to 0.02 | 0.159 | 1208 | −0.04 | −0.09 to 0.02 | 0.165 |
| LDL Size (nm) | 2219 | −0.06 | −0.13 to 0.00 | 0.062 | 1011 | −0.08 | −0.19 to 0.02 | 0.117 | 1208 | −0.05 | −0.14 to 0.03 | 0.246 |
| hsCRP (mg/L−1)¶ | 2216 | 0.90 | 0.81 to 0.99 | 0.028 | 1014 | 0.84 | 0.73 to 0.94 | 0.005 | 1202 | 0.94 | 0.83 to 1.08 | 0.399 |
| IGF-1 (nmol L−1) | 2223 | −0.49 | −0.92 to −0.06 | 0.025 | 1015 | −0.10 | −0.74 to 0.53 | 0.748 | 1208 | −0.80 | −1.36 to −0.24 | 0.005 |
| IGFBP3 (mg L−1) | 2223 | −0.09 | −0.17 to 0.00 | 0.042 | 1015 | −0.06 | −0.18 to 0.06 | 0.281 | 1208 | −0.10 | −0.21 to 0.01 | 0.062 |
| IGF-1/IGFBP3 | 2223 | −0.03 | −0.11 to 0.04 | 0.384 | 1015 | 0.04 | −0.08 to 0.15 | 0.504 | 1208 | −0.09 | −0.19 to 0.01 | 0.065 |
ApoE, apolipoprotein E; HDL, high-density lipoprotein; LDL, low-density lipoprotein; hsCRP, high sensitivity C-reactive protein; IGF-1, insulin-like growth factor 1; IGFBP3, insulin-like growth factor binding protein 3.
N, Number of samples in the analysis.
Effect; Effect on serum parameter per minor allele of rs429358.
95% CI; 95% Confidence Intervals.
P-value; Nominal P-value obtained from Model 2 (described in the Experimental procedures section).
Natural log transformed serum parameter was used in the association analysis.
No significant effects of rs429358 were observed on glucose (P = 0.388), insulin (P = 0.123), triglyceride (P = 0.203), and fT3 (P = 0.141) levels (Table 3); the phenotypes that have previously been associated, in middle age, with familial longevity in the LLS families (Rozing et al., 2009, 2010a,b; Vaarhorst et al., 2011; Wijsman et al., 2011).
Table 3.
Association analysis of serum parameters previously associated with familial longevity in middle age in the Leiden longevity study families between carriers and non-carriers of rs429358
| All | Males | Females | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serum parameter | N* | Effect† | 95% CI‡ | P-value§ | N* | Effect† | 95% CI‡ | P-value§ | N* | Effect† | 95% CI‡ | P-value§ |
| Glucose (mmol L−1) | 2234 | −0.05 | −0.17 to 0.07 | 0.388 | 1021 | −0.16 | −0.36 to 0.04 | 0.116 | 1213 | 0.03 | −0.11 to 0.18 | 0.660 |
| Insulin (mU L−1)¶ | 2163 | 0.95 | 0.88 to 1.02 | 0.123 | 990 | 0.93 | 0.84 to 1.03 | 0.158 | 1173 | 0.96 | 0.87 to 1.06 | 0.400 |
| HDL cholesterol (mmol L−1) | 2228 | −0.04 | −0.07 to 0.00 | 0.062 | 1018 | −0.04 | −0.09 to 0.00 | 0.064 | 1210 | −0.03 | −0.08 to 0.02 | 0.286 |
| Triglyceride (mmol L−1)¶ | 2229 | 1.03 | 0.98 to 1.08 | 0.203 | 1016 | 1.07 | 0.99 to 1.15 | 0.095 | 1208 | 1.01 | 0.95 to 1.07 | 0.834 |
| HDL Size (nm) | 2219 | −0.04 | −0.08 to 0.00 | 0.061 | 1011 | −0.04 | −0.10 to 0.02 | 0.159 | 1208 | −0.04 | −0.09 to 0.02 | 0.165 |
| LDL Size (nm) | 2219 | −0.06 | −0.13 to 0.00 | 0.062 | 1011 | −0.08 | −0.19 to 0.02 | 0.117 | 1208 | −0.05 | −0.14 to 0.03 | 0.246 |
| fT3 (pmol L−1) | 2223 | 0.05 | −0.02 to 0.12 | 0.141 | 1015 | 0.07 | −0.02 to 0.16 | 0.127 | 1208 | 0.03 | −0.06 to 0.13 | 0.470 |
HDL, high-density lipoprotein; LDL, low-density lipoprotein; fT3, free triiodothyronine.
N, Number of samples in the analysis.
Effect; Effect on serum parameter per minor allele of rs429358.
95% CI; 95% Confidence Intervals.
P-value; Nominal P-value obtained from Model 2 (described in the Experimental procedures section).
Natural log transformed serum parameter was used in the association analysis.
Analysis of Alzheimer's disease SNPs
Rs2075650 has consistently been associated with an increased risk of Alzheimer's disease in several independent GWAS studies (Harold et al., 2009; Lambert et al., 2009; Seshadri et al., 2010). Therefore, we studied the effect of SNPs present in the AlzGene database [http://www.alzgene.org/, (Bertram et al., 2007)], on survival into old age in the LLS GWAS. Apart from rs2075650, none of the 751 measured Alzheimer's disease SNPs showed a significant association after adjustment for multiple testing (Table S4).
Analysis of FOXO3A and AKT1 SNPs
Apart from APOE, two other genes have shown consistent evidence for association with longevity, FOXO3A (Willcox et al., 2008; Flachsbart et al., 2009; Pawlikowska et al., 2009; Soerensen et al., 2010) and AKT1 (Pawlikowska et al., 2009). For the longevity-promoting FOXO3A SNPs previously reported with centenarian longevity, we observed no association with survival into old age in our nonagenarians (Table S5). For AKT1, one of the two measured SNPs, rs2498804, showed a significant association with survival into old age [OR = 0.75 (95% CI 0.63–0.89), P = 0.001] (Table S5).
Discussion
To identify common SNPs contributing to longevity, GWAS analysis of 403 nonagenarian cases and 1670 population controls was performed. Of the 62 top associating SNPs, 58 were tested in a meta-analysis of 4149 nonagenarian cases and 7582 younger controls and we identified one SNP, rs2075650, that associated significantly with survival into old age (P = 3.39 × 10−17). Carriers of the minor allele had a 29% decreased probability of reaching 90 years on average. Although cases and controls originate from different generations, we concluded that there was no substructure to an extent that would affect the observations.
Rs2075650 is located in the TOMM40 gene at chromosome 19q13.32 close to and centromeric of the APOE gene (Fig. S2), which has shown consistent evidence for association with longevity (Schachter et al., 1994; Christensen et al., 2006). The ApoE protein has three isoforms (ApoE ε2, ApoE ε3, and ApoE ε4) that are defined by two SNPs, rs7412 (Arg136Cys; ε2) and rs429358 (Cys112Arg; ε4). ApoE ε4 carriers have an increased risk of cardiovascular disease and Alzheimer's disease, while ApoE ε2 carriers are protected from these diseases (Corder et al., 1993; Eichner et al., 1993; Christensen et al., 2006). Although we detected only moderate LD (r2a = 0.553) between rs2075650 and the ApoE ε4-defining SNP rs429358, we could not detect a significant effect of rs2075650 on longevity independent of rs429358. Several prospective studies, including one with the Danish 1905 cohort (Jacobsen et al., 2010), reported increased mortality for ApoE ε4 carriers, even though there is still much debate about APOE being a ‘frailty gene’ or a ‘longevity gene’ (Gerdes et al., 2000; Christensen et al., 2006; Ewbank, 2007; Jacobsen et al., 2010). The prospective data in the LLS and Leiden 85-plus study support the ‘frailty gene’ hypothesis, as rs429358 affects mortality after 85 years and continues the effect after 90 years [HR = 1.08 (95% CI 1.03–1.13), P = 0.001 and HR = 1.08 (95% CI 1.03–1.12), P = 0.001, respectively] (Table S3B, Fig. S4). In these prospective studies, carriers of the minor allele of rs2075650 showed no increased mortality independent of rs429358, which indicates that the association of rs2075650 with longevity is most likely due to variation in the APOE gene. Although GWAS studies have reported significant associations between rs2075650 and Alzheimer's disease, brain imaging, total cholesterol, and CRP plasma levels (Reiner et al., 2008; Aulchenko et al., 2009; Seshadri et al., 2010; Shen et al., 2010), no analyses were performed to determine whether these associations are APOE independent. We observed no APOE-independent effect on the phenotypes investigated in the LLS offspring and partners except for total and LDL cholesterol.
Previously, rs429358 had been associated with several metabolic phenotypes, such as ApoE, total cholesterol, HDL cholesterol, LDL cholesterol, and hsCRP levels, as well as HDL and LDL particle size (Boerwinkle & Utermann, 1988; Topic et al., 2008; Hubacek et al., 2010) and, here, we have confirmed these findings using serum measurements of the offspring and partners from the LLS. Because the insulin/IGF-1 signaling (ISS) pathway has a lifespan regulating effect in several model organisms (Fontana et al., 2010; Kenyon, 2010) and humans (Suh et al., 2008), we also investigated the effect of rs429358 on serum levels of IGF-1 and IGFBP3, which both play a role in this pathway. Both proteins are involved in the etiology of several age-related diseases. However, up till now, it is not clear whether higher or lower serum levels are beneficial for longevity. Low IGF-1 serum levels associate to a decreased risk of cancer, but an increased risk of cardiovascular disease and neurodegenerative disease (Juul, 2003). Previously, we showed in the Leiden 85-Plus Study cohort that genetic variants known to associate to lower IIS activity and IGF-1 serum levels at younger age associated with better survival at ages above 85 years (van Heemst et al., 2005). However, the effect of these genetic variants on IGF-1 serum levels was not tested in the Leiden 85-Plus Study cohort. In addition, we showed previously that neither IGF-1 and IGFBP3 levels nor their ratio differed between partners and offspring from the LLS (Rozing et al., 2009), which indicates that IGF-1 serum levels are, in middle age, not a marker for longevity, whereas a decreased risk of metabolic diseases was evident at that age in long-lived families (Westendorp et al., 2009). In the current study, we found that the minor allele of rs429358 associates with lower IGF-1 levels in middle-aged women, which to our knowledge has not previously been reported. Like low IGF-1 levels, ApoE ε4 was previously associated with an increased risk of developing cardiovascular disease and neurodegenerative disease (Corder et al., 1993; Eichner et al., 1993; Christensen et al., 2006). Thus, the mechanism behind the increased risk of female ApoE ε4 carriers of developing cardiovascular and/or neurodegenerative diseases might involve serum levels of IGF-1 or other aspects of IIS activity reflected by these levels. Apart from lipid metabolism, the parameters determining the longevity phenotype in middle age in the LLS, such as glucose metabolism, insulin sensitivity, and thyroid hormone metabolism (Rozing et al., 2009, 2010a,b; Vaarhorst et al., 2011; Wijsman et al., 2011), were not influenced by the presence of the minor allele of rs429358. This indicates that it is likely that other loci could explain the differences in these phenotypes between LLS offspring and partners.
The strength of this study is that, by using a GWAS, we were able to replicate the previously reported association of the APOE locus with longevity (Schachter et al., 1994; Christensen et al., 2006) as the major locus. This was not observed in the previously published meta genome-wide association study of Newman et al. (Newman et al., 2010), possibly because of differences in the study design and population control selection between the studies. While Newman et al. used nonagenarian cases in a population-based design, we made use of a family-based design in which the families are genetically enriched for longevity. In addition, Newman et al. used population controls from the same cohort which had died before the age of 80. Between 60 and 80 years however, there might already have been a selection on survival, decreasing the frequency of ApoE ε4 carriers in the control group. In contrast, we made comparisons to a younger population group (55–60 years) from a different cohort (RS).
As we previously reported that long-lived individuals carry the same number of disease risk alleles for cardiovascular disease, cancer, and type 2 diabetes as young controls (Beekman et al., 2010), we expected to primarily find longevity-promoting alleles. However, although most of the 58 prioritized SNPs (na = 43) from the LLS GWAS showed a longevity-promoting effect ranging from 36 to 168%, none of them could be replicated in additional study populations of nonagenarian singletons. The only replicated locus is APOE, which is a mortality locus that has previously been reported to be the major locus responsible for Alzheimer's disease (Harold et al., 2009; Lambert et al., 2009; Seshadri et al., 2010), a well-known age-related disease. Nevertheless, none of the other Alzheimer's disease loci showed an association with survival to 90 years, which indicates that the remaining genetic variation in longevity in the LLS could not be explained by the genetic variation which contributes to Alzheimer's disease. In addition to APOE, we also observed evidence for association at the previously reported AKT1 locus (Pawlikowska et al., 2009) with survival into old age in the LLS GWAS, although the effect of this SNP is relatively small (25% decreased probability of becoming 90 years) compared to the effect of rs429358 (51%). The previously reported longevity-promoting effect of the FOXO3A locus could not be replicated in this study. This is probably due to the relatively low number of centenarians in the LLS GWAS case group, in which the effect of SNPs in FOXO3A on longevity seems to be most prominent. The still unexplained genetic variation in longevity might be attributable to rare variants or variants with small effects, which has previously been reported for other complex traits, such as Alzheimer's disease. These loci could not be identified in this study because of the relatively small number of cases in the LLS GWAS, the heterogeneity of factors influencing lifespan within populations, and the difference in the design of the studies used for replication. One way to identify variants with small effects would be to increase the initial sample size of the GWAS study and perform replication in other studies of nonagenarians. Given the higher heritability of longevity at older ages (Tan et al., 2004), one may also limit the study population to centenarians or supercentenarians. In addition to common variants with small effects, rare variants with large effects might play a role in longevity. By whole-genome/exome sequencing of long-lived subjects and their families, rare variants can be identified and associated with human longevity.
In conclusion, we have shown that the deleterious effect of the ApoE ε4 allele, tagged by rs2075650, is the single major hit in our GWAS study for longevity, indicating that no other major longevity locus was present among these nonagenarians. We confirmed the previously reported associations of the ApoE ε4 allele with lipid metabolism parameters and report an additional effect on IGF-1 signaling in women. To identify genetic variants with smaller and protective effects on human lifespan, a meta-GWAS for longevity with a larger sample size is merited.
Experimental procedures
Study populations
Leiden longevity study
For the LLS, long-lived siblings of European descent were recruited together with their offspring and the partners of the offspring. Families were included if at least two long-lived siblings were alive and fulfilled the age criterion of 89 years or older for men and 91 years or older for women, representing <0.5% of the Dutch population in 2001 (Schoenmaker et al., 2006). In total, 944 long-lived proband siblings were included with a mean age of 94 years (range 89–104), 1671 offspring (61 years, 39–81), and 744 partners (60 years, 36–79). DNA from the LLS was extracted from samples at baseline using conventional methods (Beekman et al., 2006). For the GWAS, 403 unrelated LLS siblings (one sibling from each sibling pair) were included (LLS GWAS cases).
Rotterdam study
The Rotterdam Study (RS) is a prospective population-based study of people aged 55 years and older, which was designed to study neurological, cardiovascular, locomotor, and ophthalmological diseases (Teichert et al., 2009). The study consists of 7983 participants from the baseline cohort (RS-I) and 3011 participants from an independent extended cohort formed in 1999 (RS-II) from which DNA was isolated between 1990 and 1993 (RS-I) or between 2000 and 2001 (RS-II). For the GWAS, 1731 participants from the combined cohort who were below 60 years of age and for whom GWAS data were available were included as controls (RS GWAS controls). For the replication study, 960 cases above 90 years at time of recruitment (RS replication cases) and 1825 controls between 60 and 65 years at baseline (RS replication controls) from the combined cohorts, for whom GWAS data were also available, were included.
Leiden 85-plus study
In the Leiden 85-plus study, two prospective population-based cohorts were recruited from inhabitants of Leiden (Weverling-Rijnsburger et al., 1997; der Wiel et al., 2002). Between 1987 and 1989, 673 subjects aged 85 years and older were enrolled in a prospective study (Cohort 1). Between 1997 and 1999, 563 subjects were enrolled in the month of their 85th birthday with follow-up (Cohort 2). Subjects were visited at their home, and there were no exclusion criteria related to health. DNA was available from the combined cohorts consisting of 1208 subjects aged 85 years and older (Leiden 85-plus replication cases).
Netherlands twin registry
From the Netherlands Twin Registry (NTR), 2090 unrelated participants of European descent for whom DNA was available were selected as control samples (Boomsma et al., 2008) (Leiden 85-plus replication controls). The substructure in the NTR has been reported before (Sullivan et al., 2009), and in this study, we included samples aged between 15 and 70 years at the time of blood sampling, without known family relations (i.e., those without any substructure).
Danish 1905 cohort
The participants in this study are from the Danish 1905 birth cohort recruited in 1998 (Nybo et al., 2003) when they were aged 92–93 years. From this cohort, 3,600 subjects were still alive, of whom 2262 participated in the study. Participants were subjected to a home-based interview on health and lifestyle parameters, physical and cognitive function tests, and the collection of biological material. The current genetic study comprises a total of 1578 of these individuals (Danish replication cases). Survival was followed up until January 2010. Ninety-nine percent (1561 subjects) of subjects died in the 12 years of follow-up. Control samples were 1997 twins (one twin for each pair) between 46 and 68 years of age collected from all over Denmark (Danish replication controls).
The cases in all three replication cohorts originate from population-based cohort studies from a genetic background similar to the LLS (Heath et al., 2008). All the participants in these studies have signed an informed consent.
Genotyping
Genome-wide association study (GWAS)
Leiden Longevity Study GWAS cases were genotyped using Illumina Infinium HD Human660W-Quad BeadChips (Illumina, San Diego, CA, USA). The RS-I and RS-II cohorts were genotyped using Illumina Infinium II HumanHap 550K Beadchips and Illumina Infinium II HumanHap550-Duo BeadChips (Illumina), respectively (Teichert et al., 2009).
For the GWAS, we selected 551 606 SNPs for analysis because these were genotyped in both the LLS GWAS cases and (some of) the RS GWAS controls. Of these 551 606 SNPs, 34 885 SNPs were excluded on the basis of the following criteria: SNP call rate <0.95 or MAF <0.01 in RS GWAS controls or LLS GWAS cases (n = 8908 and n = 24 586, respectively), and PHWE < 10−4 in RS GWAS controls (n = 1355). In addition, SNPs with a between-chip effect in the RS GWAS controls were removed using a genotype trend test comparing the RS GWAS controls from RS-I with RS-II (n = 36), leaving 516,721 SNPs for statistical analysis. The Illumina clusterplots of the SNPs with P < 1 × 10−4 (n = 71) were visually inspected to confirm high-quality genotyping, and 9 SNPs were excluded on the basis of bad clustering in the LLS GWAS cases or RS GWAS controls.
Genotype data were used to confirm gender and family relationships. Two RS GWAS control samples were excluded because of abnormalities in the sex chromosome (both samples had Triple X Syndrome). Latent clustering of genotypes because of population substructure was assessed by pairwise identity-by-state (IBS) distance using Graphical Relationship Representation (GRR) [http://bioinformatics.well.ox.ac.uk/GRR, (Abecasis et al., 2001)]. LLS GWAS cases showed no relationship errors. From the RS GWAS controls, 59 samples were excluded because of high IBS. In total, 403 LLS GWAS cases and 1670 RS GWAS control samples with a sample call rate >0.95 were analyzed. Because cases and controls originate from different generations, we investigated whether substructure in these cohorts could have influenced the observed associations. IBS estimates for all pairs of subjects in the data set were computed on a randomly selected set of 10% of the SNPs that passed quality control thresholds, using the –genome, –cluster, and –mds-plot 4 commands in PLINK [http://pngu.mgh.harvard.edu/purcell/plink, (Purcell et al., 2007)]. The first two resulting principal components (C1 and C2) were plotted against each other, which gives a representation of the data in two dimensions. In the resulting scatter plot, each point represents an individual (green = LLS GWAS case and blue = RS GWAS control) (Fig. S5). If there had been substructure, one would see multiple clusters in one plot. However, because all our samples seem to be in one cluster, we concluded that there was no substructure to an extent that would affect the observations.
Replication studies
For the RS replication study, we used the existing GWAS data in the Rotterdam Study after the quality control screening described by Teichert et al. (Teichert et al., 2009). For the Leiden 85-plus and Danish replication studies, genotyping was performed using the Sequenom MassARRAY iPLEX Gold and TaqMan SNP Genotyping assays. Of the 62 prioritized SNPs, 58 could be designed for replication studies using Sequenom, of which 56 were successfully genotyped in >95% of the samples displayed in Table 1. The average genotype call rate for SNPs genotyped with Sequenom was 98.40%, and the average concordance rate with GWAS data among the LLS GWAS cases was 99.97%. For 2 of the 6 SNPs that could not be genotyped with Sequenom, rs2075650 and rs642990, pre-designed TaqMan SNP genotyping assays (C___3084828_20 and C___2206314_20, respectively) were used for genotyping, following the manufacturer's instructions. The average genotype call rate for the SNPs genotyped with TaqMan was 99.04%, and the average concordance rate with GWAS data among the LLS GWAS cases was 100%.
APOEε2/ε3/ε4 polymorphism
The APOEε2/ε3/ε4 defining SNPs, rs429358 (Cys112Arg; ε4) and rs7412 (Arg136Cys; ε2), were genotyped in the LLS GWAS cases, Leiden 85-plus replication study, and Danish replication study controls using pre-designed TaqMan SNP genotyping assays (C___3084793_20 and C____904973_10, respectively). For the RS GWAS controls and Danish replication study cases, previously measured data were used (Slooter et al., 1998; Jacobsen et al., 2010).
Measurement of serum parameters
All standard serum measurements were performed using fully automated equipment.
Glucose, total cholesterol, high-density lipoprotein cholesterol (HDL-C), and triglyceride levels were measured using the Hitachi Modular P 800 (Roche, Almere, the Netherlands) (Rozing et al., 2009). Low-density lipoprotein cholesterol (LDL-C) was calculated using the Friedewald formula (Friedewald et al., 1972).
LDL and HDL particle sizes were measured using proton NMR spectroscopy (LipoScience Inc, Raleigh, NY, USA) (Heijmans et al., 2006).
Insulin-like growth factor-1 (IGF-1), insulin-like growth factor-binding protein 3 (IGFBP3), and insulin levels were measured using the Immulite 2500 (DPC, Los Angeles, CA, USA) (Rozing et al., 2009).
Free triiodothyronine (fT3) was measured using the Modular E170, and hsCRP was measured using Cobas Integra 800 (both from Roche) (Rozing et al., 2010b).
The level of ApoE was determined in serum samples using a human ApoE-specific sandwich ELISA (van Vlijmen et al., 1994; Mooijaart et al., 2006).
Statistical analysis
GWAS and replication studies
For the association analysis of the GWAS data, we applied a Cochran-Armitage trend test (Cochran, 1954; Armitage, 1955). For X-linked SNPs, the genotypes of the men were considered as homozygous genotypes. SNPs with a P-value <1 × 10−4 (na = 62) were selected for replication. Odds ratios were estimated and the corresponding 95% confidence intervals were computed. For meta-analyses, a fixed effect approach was used. Scores and their variances were computed within each study and combined across the four studies to obtain a single meta-statistic. P-values below 5 × 10−8 were considered as genome-wide significant (Pe'er et al., 2008). The between-study variance was calculated to determine heterogeneity across the four studies. All these analysis were performed using Bioconductor R [http://www.bioconductor.org, (Gentleman et al., 2004)].
The quantile–quantile plot (Fig. S6), constructed using Bioconductor R [http://www.bioconductor.org, (Gentleman et al., 2004)], showed that the P-value distribution of stage 1 conformed to a null distribution at all but the extreme tail. The genomic inflation factor (λ), which measures over-dispersion of test statistics from association tests indicating population stratification, was 1.027 and we therefore decided not to adjust for population stratification.
Linkage disequilibrium between rs2075650 and the APOEε2/ε3/ε4 polymorphism
Pairwise linkage disequilibrium (LD) between rs2075650 and the APOEε2/ε3/ε4 polymorphism determining SNPs rs7412 and rs429358 was calculated in 8946 individuals using the –ld command in PLINK (http://pngu.mgh.harvard.edu/purcell/plink, (Purcell et al., 2007)).
APOE -independent association of rs2075650 with longevity
To determine whether the association of rs2075650 with longevity was independent of the APOEε2/ε3/ε4 polymorphism, a logistic regression model with adjustment for rs429358, rs7412, and an interaction term for ε2/ε3 with ε3/ε4 was tested (Ronald et al., 2009):
Logit (Pstatus = 1) = β0 + β1*rs2075650 + β2*rs429358 + β3*rs7412 + β4*(rs429358*rs7412)+β5*Study (Model 1)
Status was coded as 0 (control) or 1 (long-lived case), Study was coded as 0 (LLS GWAS), 1 (Leiden 85-plus replication study), or 2 (Danish replication study), and the genotypes of rs2075650, rs429358, and rs7412 were coded as 0 (homozygous for the common allele), 1 (heterozygous), or 2 (homozygous for the rare allele). STATA/SE 11.1 (StataCorp LP, College Station, TX, USA) was used for this analysis.
Prospective analysis
Prospective analysis of rs2075650 and rs429358 was performed with 944 nonagenarian siblings from the LLS, 976 octogenarians and nonagenarians from the Leiden 85-plus study, and 1578 nonagenarians from the Danish 1905 cohort.
After a mean follow-up time of 5.7 years (LLS), 14.8 years (Leiden 85-plus study), and 11.4 years (Danish 1905 cohort), 73.2% (na = 691) (LLS), 84.8% (na = 828) (Leiden 85-plus study), and 98.9% (na = 1561) (Danish 1905 cohort) of the individuals had died.
Mortality analyses were performed with STATA/SE 11.1 (StataCorp LP) using a sex-adjusted, left-truncated Cox proportional hazards model to adjust for late entry into the data set according to age.
Association of rs429358 (ε4) and rs2075650 with serum parameters
To determine the association of rs429358 and the APOE-independent association of rs2075650 with serum parameters in the offspring and their partners from the LLS, the following regression models were tested:
Serum parameter = β0 + β1*Age + β2*Sex + β3*(Age*Sex) + β4*Group + β5*rs429358 (Model 2)
Serum parameter = β0 + β1*Age + β2*Sex + β3*(Age*Sex) + β4*Group + β5*rs2075650 + β6*rs429358 + β7*rs7412 + β8*(rs429358*rs7412) (Model 3)
Age was coded in years. Sex was coded as 1 (male) or 2 (female), Group was coded as 0 (partner) or 1 (offspring). Robust standard errors were used to account for sibship relations. STATA/SE 11.1 (StataCorp LP) was used for these analyses.
Acknowledgments
We thank all participants of the Leiden Longevity Study, Rotterdam Study, Leiden 85-plus study, Netherlands Twin Register, and the Danish 1905 cohort. This study was supported by a grant from the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology, and the Netherlands Consortium for Healthy Ageing (Grant 050-060-810), all in the framework of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NWO) and by Unilever Colworth.
The generation and management of GWAS genotype data for the Rotterdam study is supported by the Netherlands Organisation for Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2) and the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. The Rotterdam Study is funded by the Erasmus Medical Center and Erasmus University, Rotterdam, the Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam.
The data collection of the Danish 1905 cohort is supported by the US National Institute on Aging, research grant NIA-P01-AG08761, and the Danish Aging Research Center is supported by a grant from the VELUX foundation.
The data collection of the Netherlands Twin Register is supported by NWO (904-61-090; 904-61-193; 480-04-004; 400-05-717; SPI 56-464-14192), Center for Medical Systems Biology (NWO Genomics; Centre for Neurogenomics and Cognitive Research (CNCR-VU); the EU (EU/QLRT-2001-01254); Genome scan for neuroticism (NIMH R01 MH059160); the Geestkracht program of ZonMW (10-000-1002), and institutes involved in NESDA (VU University Medical Center, Leiden University Medical Center, GGZ Buitenamstel-Geestgronden, Rivierduinen, University Medical Center Groningen, GGZ Lentis, GGZ Friesland, GGZ Drenthe). The genotyping of the samples was provided through the Genetic Association Information Network (GAIN).
Author contributions
Authors contributed as follows JD, MB, BTH, RGJW, PES; Data analysis, drafting manuscript, study design Leiden Longevity Study H-WU, QH, JJH-D; Statistical data analysis DK, RB, HEDS, NL, EBA, WMP, DH; Data collection and data analysis Leiden Longevity Study MK, LC, HT, AJC, FR, EJdG, DIB, AGU, KC, CMD; Data analysis and data collection GWAS controls and replication cohorts MP; DNA isolations Leiden Longevity Study FJO, DAG; Data collection Leiden Longevity Study GWAS cases.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1 Manhattan plot presenting the −log10P from the Cochran-Armitage trend test for the 516,721 SNPs that passed the quality control thresholds in the LLS GWAS.
Fig. S2 Genomic region surrounding rs2075650 (obtained from the UCSC genome browser (http://genome.ucsc.edu/). The physical distances between rs2075650 and rs429358 and between rs2075650 and rs7412 are 16.32 kb and 16.46 kb, respectively.
Fig. S3 Kaplan-Meier curves showing the survival rate over years to follow-up of ApoE ε3ε3 carriers with zero (solid line) or one (large dashed line) minor allele(s) of rs2075650 in the LLS, Leiden 85-plus study and Danish 1905 cohort.
Fig. S4 Kaplan-Meier curves showing the survival rate over years to follow-up of carriers of zero (solid line), one (large dashed line), or two (small dashed line) ε4 allele(s) of rs429358 in the LLS and Leiden 85-plus study.
Fig. S5 C1 values plotted against the C2 values, both resulting from the multidimensional scaling analysis, of the 403 LLS GWAS cases (green) and the 1670 RS GWAS controls (blue).
Fig. S6 Quantile–quantile plot of expected vs. observed chi-square values for the test statistic from the Cochran-Armitage trend test for 516,721 SNPs that passed the quality control thresholds in the LLS GWAS. The slope of the dashed line represents the genomic inflation factor (λ = 1.027). The shaded region represents the 95% confidence band.
Table S1 SNPs (n = 62) selected for replication analysis, associating at P < 1 × 10−4 with survival into old age in the analysis of the LLS GWAS.
Table S2 (A) Results of the association analysis with survival into old age of the 58 prioritized SNPs from the LLS GWAS in the RS replication study, Leiden 85-plus replication study, Danish replication study, and the meta-analysis. (B) Results of the meta-association analysis with survival into old age of the 58 prioritized SNPs in male cases compared to all controls. (C) Results of the meta-association analysis with survival into old age of the 58 prioritized SNPs in female cases compared to all controls.
Table S3 (A) Results of the prospective analysis of rs2075650 adjusted for rs429358 (ε4) and rs7412 (ε2). (B) Results of the prospective analysis of rs429358 (ε4).
Table S4 Association of LLS GWAS SNPs selected from the Alz-Gene database [http://www.alzgene.org/, (Bertram et al., 2007)] with survival into old age.
Table S5 Association of LLS GWAS SNPs within a 10 Kb window around FOXO3A and AKT1 with survival into old age.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- Atzmon G, Schechter C, Greiner W, Davidson D, Rennert G, Barzilai N. Clinical phenotype of families with longevity. J. Am. Geriatr. Soc. 2004;52:274–277. doi: 10.1111/j.1532-5415.2004.52068.x. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Doring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman M, Blauw GJ, Houwing-Duistermaat JJ, Brandt BW, Westendorp RG, Slagboom PE. Chromosome 4q25, microsomal transfer protein gene, and human longevity: novel data and a meta-analysis of association studies. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:355–362. doi: 10.1093/gerona/61.4.355. [DOI] [PubMed] [Google Scholar]
- Beekman M, Nederstigt C, Suchiman HE, Kremer D, van der Breggen R, Lakenberg N, Alemayehu WG, de Craen AJ, Westendorp RG, Boomsma DI, de Geus EJ, Houwing-Duistermaat JJ, Heijmans BT, Slagboom PE. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18046–18049. doi: 10.1073/pnas.1003540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Boerwinkle E, Utermann G. Simultaneous effects of the apolipoprotein E polymorphism on apolipoprotein E, apolipoprotein B, and cholesterol metabolism. Am. J. Hum. Genet. 1988;42:104–112. [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D, Kluft C, Smit G, Nolen WA, Zitman FG, Smit JH, Hoogendijk WJ, van Dyck R, de Geus EJ, Penninx BW. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur. J. Hum. Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- Boyden SE, Kunkel LM. High-density genomewide linkage analysis of exceptional human longevity identifies multiple novel Loci. PLoS ONE. 2010;5:e12432. doi: 10.1371/journal.pone.0012432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat. Rev. Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, McGue M, Petersen I, Jeune B, Vaupel JW. Exceptional longevity does not result in excessive levels of disability. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13274–13279. doi: 10.1073/pnas.0804931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran WG. Some methods for strengthening the common χ2 tests. Biometrics. 1954;10:417–451. [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Eichner JE, Kuller LH, Orchard TJ, Grandits GA, McCallum LM, Ferrell RE, Neaton JD. Relation of apolipoprotein E phenotype to myocardial infarction and mortality from coronary artery disease. Am. J. Cardiol. 1993;71:160–165. doi: 10.1016/0002-9149(93)90732-r. [DOI] [PubMed] [Google Scholar]
- Ewbank DC. Differences in the association between apolipoprotein E genotype and mortality across populations. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:899–907. doi: 10.1093/gerona/62.8.899. [DOI] [PubMed] [Google Scholar]
- Finch CE, Tanzi RE. Genetics of aging. Science. 1997;278:407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes LU, Jeune B, Ranberg KA, Nybo H, Vaupel JW. Estimation of apolipoprotein E genotype-specific relative mortality risks from the distribution of genotypes in centenarians and middle-aged men: apolipoprotein E gene is a “frailty gene,” not a “longevity gene”. Genet. Epidemiol. 2000;19:202–210. doi: 10.1002/1098-2272(200010)19:3<202::AID-GEPI2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Gudmundsson H, Gudbjartsson DF, Frigge M, Gulcher JR, Stefansson K. Inheritance of human longevity in Iceland. Eur. J. Hum. Genet. 2000;8:743–749. doi: 10.1038/sj.ejhg.5200527. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, de Deyn PP, van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, Fabianova E, Foretova L, Georges M, Janout V, Kabesch M, Krokan HE, Elvestad MB, Lissowska J, Mates D, Rudnai P, Skorpen F, Schreiber S, Soria JM, Syvanen AC, Meneton P, Hercberg S, Galan P, Szeszenia-Dabrowska N, Zaridze D, Genin E, Cardon LR, Lathrop M. Investigation of the fine structure of European populations with applications to disease association studies. Eur. J. Hum. Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Beekman M, Mooijaart SP, Heijmans BT, Brandt BW, Zwaan BJ, Slagboom PE, Westendorp RG. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4:79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Beekman M, Houwing-Duistermaat JJ, Cobain MR, Powell J, Blauw GJ, van der Ouderaa F, Westendorp RG, Slagboom PE. Lipoprotein particle profiles mark familial and sporadic human longevity. PLoS. Med. 2006;3:e495. doi: 10.1371/journal.pmed.0030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmborg JV, Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, Kaprio J, Pedersen NL, Christensen K. Genetic influence on human lifespan and longevity. Hum. Genet. 2006;119:312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- Hubacek JA, Peasey A, Pikhart H, Stavek P, Kubinova R, Marmot M, Bobak M. APOE polymorphism and its effect on plasma C-reactive protein levels in a large general population sample. Hum. Immunol. 2010;71:304–308. doi: 10.1016/j.humimm.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen R, Martinussen T, Christiansen L, Jeune B, Andersen-Ranberg K, Vaupel JW, Christensen K. Increased effect of the ApoE gene on survival at advanced age in healthy and long-lived Danes: two nationwide cohort studies. Aging Cell. 2010;9:1004–1009. doi: 10.1111/j.1474-9726.2010.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juul A. Serum levels of insulin-like growth factor I and its binding proteins in health and disease. Growth Horm. IGF Res. 2003;13:113–170. doi: 10.1016/s1096-6374(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, de Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Mooijaart SP, Berbee JF, van Heemst D, Havekes LM, de Craen AJ, Slagboom PE, Rensen PC, Westendorp RG. ApoE plasma levels and risk of cardiovascular mortality in old age. PLoS. Med. 2006;3:e176. doi: 10.1371/journal.pmed.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AB, Walter S, Lunetta KL, Garcia ME, Slagboom PE, Christensen K, Arnold AM, Aspelund T, Aulchenko YS, Benjamin EJ, Christiansen L, D'Agostino RB, Sr, Fitzpatrick AL, Franceschini N, Glazer NL, Gudnason V, Hofman A, Kaplan R, Karasik D, Kelly-Hayes M, Kiel DP, Launer LJ, Marciante KD, Massaro JM, Miljkovic I, Nalls MA, Hernandez D, Psaty BM, Rivadeneira F, Rotter J, Seshadri S, Smith AV, Taylor KD, Tiemeier H, Uh HW, Uitterlinden AG, Vaupel JW, Walston J, Westendorp RG, Harris TB, Lumley T, van Duijn CM, Murabito JM. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J. Gerontol. A Biol. Sci. Med. Sci. 2010;65:478–487. doi: 10.1093/gerona/glq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybo H, Petersen HC, Gaist D, Jeune B, Andersen K, McGue M, Vaupel JW, Christensen K. Predictors of mortality in 2,249 nonagenarians–the Danish 1905-Cohort Survey. J. Am. Geriatr. Soc. 2003;51:1365–1373. doi: 10.1046/j.1532-5415.2003.51453.x. [DOI] [PubMed] [Google Scholar]
- Oeppen J, Vaupel JW. Demography. Broken limits to life expectancy. Science. 2002;296:1029–1031. doi: 10.1126/science.1069675. [DOI] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Perls T, Shea-Drinkwater M, Bowen-Flynn J, Ridge SB, Kang S, Joyce E, Daly M, Brewster SJ, Kunkel L, Puca AA. Exceptional familial clustering for extreme longevity in humans. J. Am. Geriatr. Soc. 2000;48:1483–1485. [PubMed] [Google Scholar]
- Puca AA, Daly MJ, Brewster SJ, Matise TC, Barrett J, Shea-Drinkwater M, Kang S, Joyce E, Nicoli J, Benson E, Kunkel LM, Perls T. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Barber MJ, Guan Y, Ridker PM, Lange LA, Chasman DI, Walston JD, Cooper GM, Jenny NS, Rieder MJ, Durda JP, Smith JD, Novembre J, Tracy RP, Rotter JI, Stephens M, Nickerson DA, Krauss RM. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J, Rajagopalan R, Ranchalis JE, Marshall JK, Hatsukami TS, Heagerty PJ, Jarvik GP. Analysis of recently identified dyslipidemia alleles reveals two loci that contribute to risk for carotid artery disease. Lipids Health Dis. 2009;8:52. doi: 10.1186/1476-511X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozing MP, Westendorp RG, Frolich M, de Craen AJ, Beekman M, Heijmans BT, Mooijaart SP, Blauw GJ, Slagboom PE, van Heemst D. Human insulin/IGF-1 and familial longevity at middle age. Aging (Albany. NY) 2009;1:714–722. doi: 10.18632/aging.100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozing MP, Westendorp RG, de Craen AJ, Frolich M, de Goeij MC, Heijmans BT, Beekman M, Wijsman CA, Mooijaart SP, Blauw GJ, Slagboom PE, van Heemst D. Favorable glucose tolerance and lower prevalence of metabolic syndrome in offspring without diabetes mellitus of nonagenarian siblings: the Leiden longevity study. J. Am. Geriatr. Soc. 2010a;58:564–569. doi: 10.1111/j.1532-5415.2010.02725.x. [DOI] [PubMed] [Google Scholar]
- Rozing MP, Westendorp RG, de Craen AJ, Frolich M, Heijmans BT, Beekman M, Wijsman C, Mooijaart SP, Blauw GJ, Slagboom PE, van Heemst D. Low serum free triiodothyronine levels mark familial longevity: the Leiden Longevity Study. J. Gerontol. A Biol. Sci. Med. Sci. 2010b;65:365–368. doi: 10.1093/gerona/glp200. [DOI] [PubMed] [Google Scholar]
- Schachter F, Faure-Delanef L, Guenot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- Schoenmaker M, de Craen AJ, de Meijer PH, Beekman M, Blauw GJ, Slagboom PE, Westendorp RG. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur. J. Hum. Genet. 2006;14:79–84. doi: 10.1038/sj.ejhg.5201508. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr, Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Jr, Weiner MW, Saykin AJ. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skytthe A, Pedersen NL, Kaprio J, Stazi MA, Hjelmborg JV, Iachine I, Vaupel JW, Christensen K. Longevity studies in GenomEUtwin. Twin Res. 2003;6:448–454. doi: 10.1375/136905203770326457. [DOI] [PubMed] [Google Scholar]
- Slooter AJ, Cruts M, Kalmijn S, Hofman A, Breteler MM, van Broeckhoven C, van Duijn CM. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch. Neurol. 1998;55:964–968. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, Christiansen L. Replication of an Association of Variation in the FOXO3A Gene with Human Longevity Using Both Case-control and Longitudinal Data. Aging Cell. 2010;9:1010–1017. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, Arolt V, Baune BT, Blackwood D, Cichon S, Coventry WL, Domschke K, Farmer A, Fava M, Gordon SD, He Q, Heath AC, Heutink P, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hu Y, Kohli M, Lin D, Lucae S, Macintyre DJ, Maier W, McGhee KA, McGuffin P, Montgomery GW, Muir WJ, Nolen WA, Nothen MM, Perlis RH, Pirlo K, Posthuma D, Rietschel M, Rizzu P, Schosser A, Smit AB, Smoller JW, Tzeng JY, van Dyck R, Verhage M, Zitman FG, Martin NG, Wray NR, Boomsma DI, Penninx BW. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol. Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Zhao JH, Iachine I, Hjelmborg J, Vach W, Vaupel JW, Christensen K, Kruse TA. Power of non-parametric linkage analysis in mapping genes contributing to human longevity in long-lived sib-pairs. Genet. Epidemiol. 2004;26:245–253. doi: 10.1002/gepi.10304. [DOI] [PubMed] [Google Scholar]
- Teichert M, Eijgelsheim M, Rivadeneira F, Uitterlinden AG, van Schaik RH, Hofman A, de Smet PA, van Gelder T, Visser LE, Stricker BH. A genome-wide association study of acenocoumarol maintenance dosage. Hum. Mol. Genet. 2009;18:3758–3768. doi: 10.1093/hmg/ddp309. [DOI] [PubMed] [Google Scholar]
- Terry DF, Sebastiani P, Andersen SL, Perls TT. Disentangling the roles of disability and morbidity in survival to exceptional old age. Arch. Intern. Med. 2008;168:277–283. doi: 10.1001/archinternmed.2007.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topic A, Spasojevic KV, Zeljkovic A, Spasojevic-Kalimanovska V, Zeljkovic A, Vekic J, Jelic-Ivanovic Z. Gender-related effect of apo E polymorphism on lipoprotein particle sizes in the middle-aged subjects. Clin. Biochem. 2008;41:361–367. doi: 10.1016/j.clinbiochem.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Vaarhorst AA, Beekman M, Suchiman EH, van Heemst D, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE, Heijmans BT. Lipid metabolism in long-lived families: the Leiden Longevity Study. Age (Dordr.) 2011 doi: 10.1007/s11357-010-9172-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vlijmen BJ, van den Maagdenberg AM, Gijbels MJ, van der Boom H, HogenEsch H, Frants RR, Hofker MH, Havekes LM. Diet-induced hyperlipoproteinemia and atherosclerosis in apolipoprotein E3-Leiden transgenic mice. J. Clin. Invest. 1994;93:1403–1410. doi: 10.1172/JCI117117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorp RG, van Heemst D, Rozing MP, Frolich M, Mooijaart SP, Blauw GJ, Beekman M, Heijmans BT, de Craen AJ, Slagboom PE. Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic nonagenarians: The Leiden Longevity Study. J. Am. Geriatr. Soc. 2009;57:1634–1637. doi: 10.1111/j.1532-5415.2009.02381.x. [DOI] [PubMed] [Google Scholar]
- Weverling-Rijnsburger AW, Blauw GJ, Lagaay AM, Knook DL, Meinders AE, Westendorp RG. Total cholesterol and risk of mortality in the oldest old. Lancet. 1997;350:1119–1123. doi: 10.1016/s0140-6736(97)04430-9. [DOI] [PubMed] [Google Scholar]
- der Wiel AB, van Exel E, de Craen AJ, Gussekloo J, Lagaay AM, Knook DL, Westendorp RG. A high response is not essential to prevent selection bias: results from the Leiden 85-plus study. J. Clin. Epidemiol. 2002;55:1119–1125. doi: 10.1016/s0895-4356(02)00505-x. [DOI] [PubMed] [Google Scholar]
- Wijsman CA, Rozing MP, Streefland TC, le Cessie S, Mooijaart SP, Slagboom PE, Westendorp RG, Pijl H, van Heemst D. Familial longevity is marked by enhanced insulin sensitivity. Aging Cell. 2011;10:114–121. doi: 10.1111/j.1474-9726.2010.00650.x. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


