Abstract
A large body of literature describes elaborate NF-κB signaling networks induced by inflammatory and immune signals. Decades of research has revealed that transcriptionally functional NF-κB dimers are activated by two major pathways, canonical and non-canonical. Both pathways involve the release of NF-κB dimers from inactive cytoplasmic complexes to cause their nuclear translocation to modulate gene expression programs and biological responses. NF-κB is also responsive to genotoxic agents; however, signal communication networks that are initiated in the nucleus following DNA damage induction are less defined. Evidence in the literature supports the presence of such signaling pathways induced by multiple distinct genotoxic agents, resulting in the activation of cytoplasmic IKK complex. An example is a pathway that involves the DNA damage-responsive kinase ataxia telangiectasia mutated (ATM) and a series of post-translational modifications of NF-κB essential modulator (NEMO) in the nucleus of a genotoxin-exposed cell. Recent evidence also suggests that this nuclear-initiated NF-κB signaling pathway plays significant physiological and pathological roles, particularly in lymphocyte development and human cancer progression. This review will summarize these new developments, while identifying significant unanswered questions and providing new hypotheses that may be addressed in future studies.
Keywords: NF-κB, DNA damage, NEMO, ATM, SUMO, DNA damage response, IKK
Introduction
The NF-κB transcription factor exists in the cytoplasm as a latent, inactive form in most cell types and its activation is induced by the engagement of cell surface receptors by a wide variety of extracellular ligands (reviewed in 1, 2, 3, 4, 5). NF-κB also responds to genotoxic agents that cause damage to nuclear DNA. Unlike membrane receptor-initiated signaling pathways, the critical signal initiation event induced by genotoxic agents that results in NF-κB activation is incompletely understood. This is because many genotoxic agents simultaneously cause both DNA alterations in the nucleus as well as additional cellular stresses (e.g., oxidative stress) in different cellular compartments outside the nucleus. As such, significant research efforts have been devoted to dissect both the mechanisms of activation and the roles of NF-κB signaling induced by genotoxic agents since the discovery of the NF-κB:IκB system in the late 1980s 6, 7, 8. Some of the progress made from these studies over the years has been reviewed in the literature (e.g., 9, 10, 11, 12, 13, 14). More recent studies have revealed a potentially conserved signaling pathway induced by different DNA double strand break (DSB)- and replication stress-inducing agents 15, 16. This review will focus on these newer developments by providing supporting experimental evidence and identifying key unanswered questions that may be pursued in future investigations.
Genotoxic agent-induced NF-κB activation
The role of ataxia telangiectasia mutated (ATM) in DNA damage response
When a eukaryotic cell is exposed to a genotoxic agent, the cell mounts an orchestrated and conserved signaling response, collectively termed the DNA damage response (DDR; reviewed in 17, 18, 19, 20, 21). As a whole, the DDR involves the detection of the specific DNA damage, leading to subsequent activation of DNA damage repair mechanisms, cell cycle checkpoints, and transcriptional responses to facilitate the overall cellular responses to genomic insults. The outcome of the DDR may consist of satisfactory lesion repair and cell survival, while excessive damage may result in persistent DNA damage and activation of apoptotic cell death or induction of senescence. In mammals, the DDR is coordinated by the members of phosphoinositide-3-kinase-related protein kinase (PIKK) family composed of ATM, ATR (ATM and Rad3 related) and DNA-dependent protein kinase (DNA-PK). ATM and DNA-PK are principally activated in response to DSB, while ATR responds primarily to replication stress. On activation, these kinases phosphorylate a myriad of downstream effectors, such as histone H2AX, checkpoint kinase 1 or 2, 53BP1 (p53-binding protein 1), and p53 tumor suppressor, to coordinate the overall cellular response to DNA damage. Dysregulation of activities or genetic alterations in many of the DDR components contribute to human pathologies, such as premature ageing, immunodeficiency, neurodegeneration and cancer, highlighting the importance of these pathways in vivo (reviewed in 21). Of the PIKKs, ATM has surfaced as a central player in mediating NF-κB activation by multiple genotoxic stimuli.
The IKK-NF-κB signaling system
There are five members of the NF-κB family in mammals, RelA (p65), c-Rel, RelB, p50 (NFΚB1), and p52 (NFΚB2; reviewed in 1, 4). The latter two family members are produced as precursor proteins, p105 and p100, respectively, and their processing to the mature forms requires proteasome activity. Maturation of p50 from p105 is co-translationally regulated 22, while that of p52 is signal inducible 23, 24. These five members can form various homo- and heterodimers with one another to control tissue- and signal-specific gene expression programs (reviewed in 25). Dimers containing RelA are kept inactive in the cytoplasm by association with a member of the IκB family of inhibitors, such as IκBα. IκB proteins contain six to seven ankyrin repeat motifs that create a structural element to associate with NF-κB dimers 26, 27. The precursors p105 and p100 also function as IκBs via ankyrin repeat motifs present in their C-termini 28, 29.
The activation of NF-κB dimers is controlled by two major NF-κB signaling mechanisms referred to as the canonical (or classical) and non-canonical (or p100 processing) pathways (reviewed in 1, 4). In the canonical pathway, cell stimulation with different signals, such as the proinflammatory cytokine tumor necrosis factor α (TNFα), causes cascades of intracellular events that culminate in activation of IKΚβ (IκB kinase β) in association with the regulatory subunit IKKγ/NF-κB essential modulator (NEMO). The essential function of NEMO in this context is thought to be as the polyubiquitin binding subunit to recruit IKK to linear or K63-linked polyubiquitin scaffolds that form as a consequence of receptor-initiated signaling events 5. Active IKΚβ then phosphorylates IκBα on two key serine residues, which creates a docking site for a β-transducing repeat containing protein (β-TrCP) ubiquitin ligase complex to cause polyubiquitination and subsequent degradation of IκBα via 26S proteasome. Degradation of IκBα releases free NF-κB, which then translocates into the nucleus and regulates transcription of target genes through binding to decameric κB consensus elements. Canonical NF-κB signaling is induced rapidly within minutes of stimulation without the need for de novo protein synthesis. The non-canonical pathway occurs with much slower kinetics, requires de novo protein synthesis and activation of IKKα without the need for IKΚβ and NEMO. IKKα phosphorylates key C-terminal serine residues of p100; and this causes β-TrCP-mediated polyubquitination and proteasome-dependent processing to generate p52. The newly produced p52 in association with a member of the NF-κB family, such as RelB, translocates to the nucleus to control expression of a subset of NF-κB target genes.
A wide array of structurally and functionally distinct signaling inputs, including many different inflammatory cytokines, bacterial and viral products, and oxidative stress conditions, result in NF-κB activation by the canonical pathway (reviewed in 1, 4, also see http://people.bu.edu/gilmore/nf-kb/). This mechanism is particularly critical for innate and adaptive immune responses and stress responses, as well as embryonic development as evidenced by the embryonic lethality of knockout mice homozygous-null for Rela30, IKKβ 31, 32, 33 or Nemo 34, 35, 36. The non-canonical pathway is activated by a smaller number of inducers, such as lymphotoxin β and B cell activating factor, and plays an important role in B-cell maturation and the formation of secondary lymphoid tissues 37, 38. Improper activation of NF-κB is associated with many types of human pathologies. For example, insufficient activity leads to uncontrolled infection by pathogens or the loss of cells otherwise needed for survival as found during neurodegeneration, whereas chronic activity can contribute to autoimmunity or oncogenesis (see http://people.bu.edu/gilmore/nf-kb/).
A signal transduction paradox
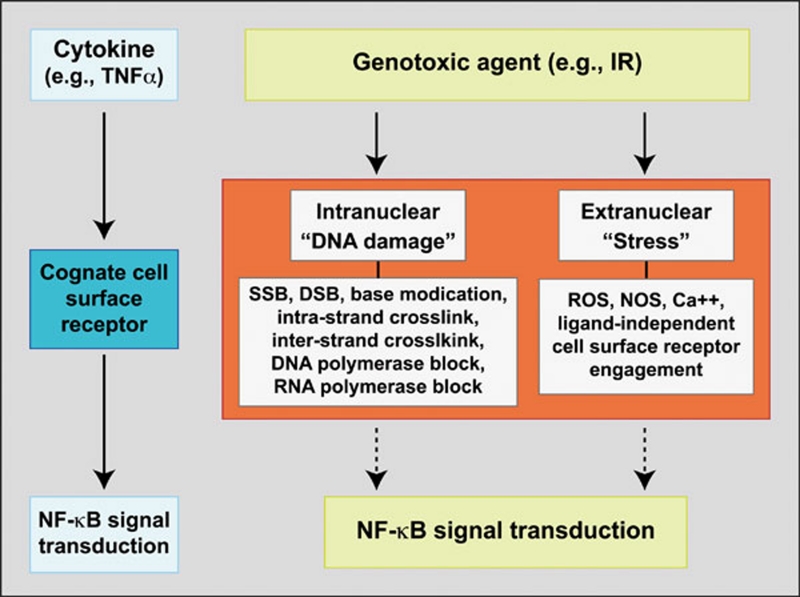
Many different types of genotoxic agents can cause activation of NF-κB via the canonical pathway (reviewed in 9, 10, 14). Whereas a majority of the known inducers of canonical NF-κB activity function exclusively by transducing cell surface and/or cytoplasmic signals to the cytoplasmic IKKβ:NEMO complexes, the site of action of genotoxic agents is located in the nucleus. How nuclear DNA damage induced by genotoxic agents can cause activation of latent NF-κB in the cytoplasm presents an interesting signal transduction paradox. A concept of a nuclear-to-cytoplasmic signaling pathway was postulated to explain, such a signaling mechanism by Stein et al. in 1989 39. However, dissection of NF-κB activation pathways induced by genotoxic agents is complicated by the co-induction of different types of DNA damages as well as other cell stresses, including oxidative stress in different cellular compartments (e.g., 40, 41, 42, 43, 44, 45, Figure 1). Thus, unlike NF-κB signaling induced by cell surface receptors where the signal initiation event can be precisely defined, the necessary and sufficient signal initiation event induced by various genotoxic agents is still being addressed in the literature. Highlighting the complexity of this issue, although ultraviolet (UV) irradiation-induced NF-κB activation led to the conceptualization of nuclear-to-cytoplasmic signaling 39, accumulated evidence over the last two decades demonstrates that nuclear DNA damage is probably not the signal initiation event for immediate activation of NF-κB in this case 46, 47, 48.
Figure 1.
Complexity of signal initiation event(s) in NF-κB signaling induced by gentoxic agents. The ligand-receptor interaction represents the signal initiation event for NF-κB activation induced by a traditional canonical NF-κB inducer, such as TNFα (left). Genotoxic agents, such as IR (ionizing radiation), may induce different types of nuclear DNA damage and stress conditions, as well as induce other cellular stress conditions (right). Different DNA and cell stress conditions listed as examples are not meant to be comprehensive. The fundamental knowledge of the necessary and sufficient events that initiate NF-κB signaling pathways induced by genotoxic agents is currently incomplete. Because of the presence of concurrent molecular events induced by different genotoxic agents, more than one mechanism may be induced to control NF-κB activation with varying dominance and different kinetics.
Importantly, several lines of evidence support the requirement of nuclear DSB in initiating the canonical IKK-NF-κB signaling pathway in response to many different genotoxic agents (reviewed in 9, 10). First, IKK and NF-κB activation by ionizing radiation (IR), topoisomerase (TOP) II-targeting drugs, VP16 (etoposide) and doxorubicin/adriamycin, and a TOP I-targeting drug camptothecin (CPT) in a variety of cell lines also requires the nuclear DSB-responsive ATM kinase activity 49, 50, 51, 52, 53, 54. Second, cyotoplasts resulting from enucleation (physical removal of nucleus from a cell) fail to release NF-κB from IκBα on CPT treatment, but NF-κB can be released by TNFα, thus, demonstrating the essential requirement of intact nucleus for CPT signaling 55. Third, the CEM-C2 human leukemic cell line expressing a CPT-resistant form of nuclear TOP I shows efficient activation of NF-κB by VP16 and IR, but not by CPT 55. Since both cytoplasts and CEM-C2 cells maintain intact mitochondrial TOP I that is susceptible to CPT-induced DNA damage 56, NF-κB is not activated by mitochondrial DNA damage at least at the time points analyzed. Fourth, electroporation of different restriction enzymes, but not heat-inactivated versions, can activate NF-κB via an ATM-dependent pathway involving IKK activation and degradation of IκBα 57. Fifth, exposure of mice to whole body radiation induces canonical IKK activation in wild-type but not in Atm−/− mice, demonstrating the physiological requirement of the DSB responsive ATM in IR-induced NF-κB activation in vivo 49. Sixth, this IR-induced ATM-dependent NF-κB activation is linked to the major pro-survival response in vivo58. Although ATM is not always necessary for NF-κB activation in response to DSB-inducing agents in all cell systems (e.g., 59), these lines of evidence nevertheless highlight the importance of nuclear DSB-induced ATM activation in contributing to the initiation of an IKK-NF-κB activation pathway.
NEMO modifications in genotoxic stress signaling
ATM activation is a frequently necessary, but insufficient intermediate event leading to NF-κB activation (e.g, 52, 57). An insight into the presence of a parallel signaling event that collaborates with ATM in NF-κB signaling in response to genotoxic agents came from the analysis of the NEMO-deficient 1.3E2 mouse pre-B cell line that fails to activate NF-κB by canonical inducers. When wild-type NEMO is stably introduced into these cells, NF-κB activation by genotoxic agents is fully restored, thereby establishing a robust cell system to perform somatic genetic complementation studies with NEMO mutants 52. Through this analysis, it was observed that mutations in the C-terminal zinc finger (ZF) domain were uniquely capable of uncoupling the nuclear signaling role of NEMO in genotoxic stress-induced NF-κB activation from its requirement as the regulatory subunit of the canonical IKK complex in the cytoplasm. This led to further observation of a series of nuclear post-translational modifications (PTMs) of NEMO that are critical for NF-κB activation following genotoxic stress. These include ATM-in dependent SUMO (small ubiquitin-like modifier) 1 modification at lysine residues 277 and 309, and ATM-dependent phosphorylation at serine 85 and monoubiquitination also requiring lysines 277/309 52, 54. Although a small fraction of the total NEMO pool is modified, individual mutations in the acceptor sites for these PTMs completely block NF-κB activation by multiple genotoxic agents, including replication stress inducers 16. In contrast, these mutations of NEMO do not significantly perturb activation by bacterial lipopolysaccharide (LPS), a membrane receptor-triggering canonical stimulus. Thus, these NEMO PTMs are critically required for NF-κB activation by DSB-inducing agents, suggesting the presence of an NF-κB activation pathway shared by multiple DNA-damaging agents. While it is evident that NEMO PTMs in concert with ATM mediate NF-κB activation by DNA-damaging agents in certain in vitro cell systems, the physiological role of these NEMO modifications needs to be evaluated through in vivo models in which specific NEMO modification sites are disrupted.
Modulators of NEMO SUMOylation
After the initial observation of the role of NEMO PTMs, subsequent work has identified additional regulators of NEMO SUMOylation following DNA damage. Mabb et al. 60 determined that PIASy (protein inhibitor of activated STAT y) is the major SUMO E3 ligase for NEMO that is required for NF-κB activation by multiple genotoxic agents. Janssens et al. 61 found that p53-induced death domain protein (PIDD) translocates from the cytoplasm to the nucleus in association with receptor interacting protein 1 (RIP1) 61. RIP1 was previously implicated in both TNFα- and DNA damage-induced NF-κB activation and cell survival 62. This nuclear PIDD-RIP1 complex promoted NEMO SUMOylation and NF-κB activation. While the mechanism of activation remains yet unclear, a recent analysis of multiple cell types obtained from Pidd−/− mice indicates that PIDD does not affect cell death induced by a wide variety of apoptotic agents, including IR and VP16 63. It remains to be determined whether NF-κB activation by DNA damage is modulated in Pidd−/− mice. More recently, Stilmann et al. 64 revealed a critical role for poly-ADP-ribose polymerase 1 (PARP1) in assembling a nuclear signaling complex composed of PARP1, PIASy, NEMO and ATM. On recruitment to DNA damage, PARP1 is activated and causes PAR modification of numerous substrates, as well as automodification of PARP1 itself 65. NEMO is recruited to this complex by direct interaction with PARP-1. PIASy is recruited to PAR-modified PARP1 via two PAR-binding motifs present in PIASy, ultimately promoting SUMOylation of NEMO. The role of PARP1 in NF-κB signaling induced by DNA damage appears to be cell-type-dependent, since PARP1 inhibition does not block NF-κB activation induced by TOP inhibitors in T leukemic cell lines 66 and also does not block IκBα degradation or NF-κB nuclear translocation induced by IR in breast cancer cell lines 67. Further understanding of the regulation of NEMO PTM by SUMOylation requires additional investigation, including a potential regulatory role for deSUMOylation by SUMO proteases.
Nuclear export of NEMO and ATM
While ATM-dependent phosphorylation of NEMO is an essential step in DSB-induced NF-κB activation 54, the requirement for ATM kinase activity in any additional step(s) in the pathway remains unclear. A NEMO-S85A mutant that cannot be phosphorylated by ATM is efficiently SUMO1 modified, but fails to be monoubiquitinated and exported to the cytoplasm following cell exposure to VP16. 1.3E2 cells expressing this mutant NEMO fail to activate IKK or NF-κB in response to multiple genotoxic agents and show hypersensitivity to IR-induced cell death. Surprisingly, if a ubiquitin moiety is fused in frame to the N-terminus of NEMO-S85A mutant, downstream NEMO export, activation of IKK and NF-κB, and survival responses are all restored in a manner still dependent on ATM kinase activity. These observations suggest that NEMO monoubiquitination is critical for NEMO export and ATM has additional downstream role(s) independent of NEMO phosphorylation. Jin et al. 68 recently found that cIAP1 is the K277/K309-specific NEMO ubiquitin ligase involved in the genotoxic stress signaling. In consonance with the hypothesis that ATM mediates additional downstream function(s), a small fraction of activated ATM is exported to the cytoplasm of HEK293 cells in a NEMO-dependent manner, and associates with IKΚβ along with ELKS (a protein rich in glutamic acid, leucine, lysine and serine) 54. ELKS is also thought to play a critical role in canonical NF-κB activation by TNFα stimulation 69. Hinz et al. 70 recently provided evidence that ATM export to the cytoplasm (and to the plasma membrane) is a critical step in DSB-induced NF-κB activation, but is not dependent on NEMO in HepG2 and HeLa cells. Since leptomycin B, a potent inhibitor of the nuclear export receptor CRM1/exportin 1 71, does not prevent the export of NEMO and IKK activation induced by genotoxic stimuli 52, 72, the nuclear export of NEMO and ATM is probably CRM1 independent. Genetic evidence showed that RCC1 (Ran guanine nucleotide exchange factor) is required for NF-κB activation by CPT 72, suggesting that Ran-GTP is likely involved in the NEMO and ATM export step. Consistent with this idea, a complex between NEMO and Ran is transiently induced following VP16 treatment 72. Identification of the nuclear export receptor(s) for ATM and NEMO will shed light on whether ATM export is NEMO-dependent in different cell systems and will also provide a crucial missing piece in the nuclear-to-cytoplasmic signaling puzzle.
ATM-dependent IKK activation in the cytoplasm
In addition to its requirement in NEMO phosphorylation, significant progress has recently been made revealing the role of ATM in regulating TGFβ activated kinase (TAK1), an upstream kinase of IKKβ in the canonical signaling pathway. In addition to identifying cIAP1 as the ligase responsible for NEMO monoubiquitination, Jin et al. 68 further found that TAK1 is required for NF-κB activation by VP16 and CPT, and that cIAP1 is also required for TAK1 activation by these agents. Thus, the authors placed cIAP1-mediated NEMO monoubiquitination upstream of TAK1 activation. The authors also found that another E3 ubiquitin ligase X-linked inhibitor of apoptosis (XIAP) is required for downstream TAK1 and NF-κB activation by CPT and VP16 stimulation. Consistent with this finding, Wu et al. 15 recently found that TAK1 activation by DNA-damaging agents requires XIAP-dependent K63-linked polyubiquitination of ELKS. Surprisingly, while NEMO was required for ELKS ubiquitination, the polyubiquitin binding activity of NEMO was unnecessary for TAK1 activation downstream of ELKS, suggesting NEMO has a unique role in TAK1 activation that is distinct from its well-characterized function as a polyubiquitin-binding protein in the canonical IKK complex 5. In addition, ubiquitination of ELKS promotes subsequent clustering of activated TAK1 and the canonical IKK complex mediated by the ubiquitin-binding subunits, TAK1-binding proteins 2 or 3 and NEMO, respectively. The authors also found that Elks+/− mice (Elks−/− mice are embryonic lethal) are more sensitive to IR-induced lethality, thus, supporting the role of ELKS in physiological response to genotoxic agents in vivo. These studies highlight the two independent roles of NEMO in mediating ATM-dependent TAK1 activation without the need for ubiquitin-binding and downstream TAK1-mediated IKK activation requiring NEMO's ubiquitin-binding activity.
Although the above studies together point to the role of ATM in promoting XIAP-dependent ELKS polyubiquitination in the cytoplasm, Hinz et al. 70 found that ATM is also required for activation of TRAF6 E3 ubiquitin ligase activity. TRAF6 K63-linked polyubiquitination, TRAF6- and cIAP1-dependent monoubiquitination of NEMO at lysine 285, and activation of TAK1 in the cytoplasm were all demonstrated to be necessary intermediate steps in the activation of IKK following exposure to DSB inducers. Interestingly, while knockdown of PIASy or PARP1 prevents NEMO monoubiquitination, ATM-dependent TAK1 activation remains intact. Thus, in the absence of monoubiquitinated NEMO, active TAK1 is somehow unable to activate IKK. From these analyses, the authors hypothesized that SUMO-modified nuclear NEMO induced by PARP1 and PIASy is exported to the cytoplasm to become the substrate for ATM-TRAF6-cIAP1-dependent monoubiquitination at K285 to promote IKK activation.
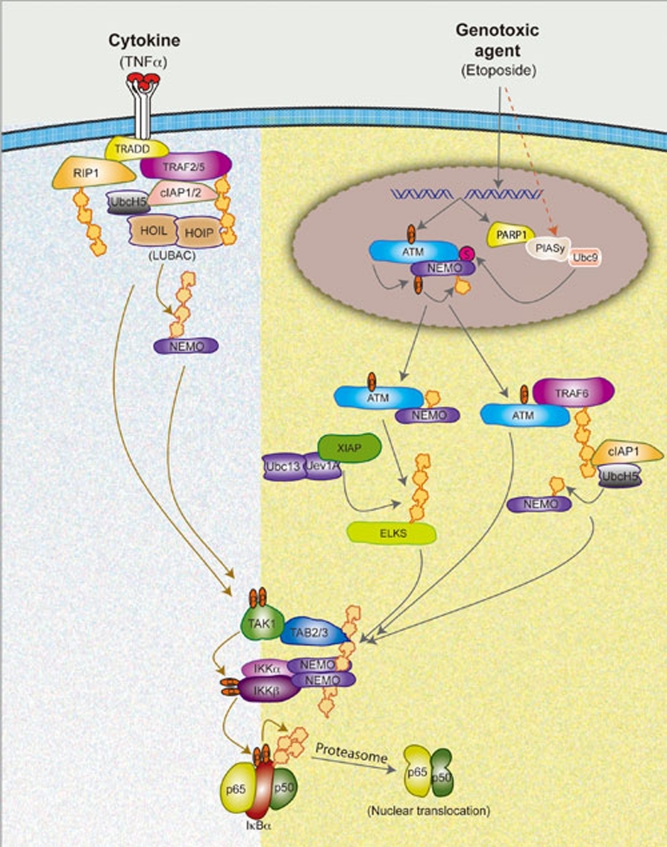
Although some controversies exist in the above studies 15, 68, 70, several consensus points may be drawn regarding the mechanism of NF-κB activation by CPT, VP16 and IR (Figure 2). First, NEMO has a unique upstream role entirely independent of its role as a regulatory component of the canonical IKK complex. Second, ATM activation and NEMO SUMOylation are required for NF-κB activation by these genotoxic agents. Third, ATM and “IKK-free” NEMO are key nuclear-generated signaling molecules that are exported to the cytoplasm to cause a sequential activation of TAK1, IKK and NF-κB. Fourth, ATM is an upstream regulator of TAK1 by assisting K63-linked polyubiquitination in the cytoplasm. Fifth, NEMO ubiquitin-binding activity is dispensable for TAK1 activation, but is necessary for subsequent IKK and NF-κB activation. Finally, like TNFα signaling where several signaling proteins are co-modified by polyubiquitination, multiple proteins may be modified by K63-linked polyubiquitination involving different E3s to coordinate efficient TAK1-dependent IKK activation in the cytoplasm.
Figure 2.
A model depicting distinct and common steps between NF-κB signaling induced by TNFα and DNA damaging agents. TNFα stimulation of TNFR1 engages receptor adaptor proteins (TRADD and RIP1) and results in the recruitment of ubiquitin conjugating enzymes (Ubc13/Uev1A and UbcH5) and E3 ligases (cIAP1/2, TRAF2/5 and HOIL/HOIP) to promote K63-linked, mixed and linear polyubiquitination of multiple target proteins. These polyubiquitin chains form the scaffold on which TAK1/TAB2/3 and IKK/NEMO complexes are formed and TAK1-dependent activation of IKKβ is induced. DNA damaging agents, such as etoposide, cause ATM activation via induction of DSB and SUMOylation of NEMO through a mechanism dependent on PARP1, PIASy and Ubc9. PIASy dependent SUMOylation of NEMO may also be induced by additional stress conditions. ATM then phosphorylates NEMO, which results in cIAP1-dependent monoubiquitination of NEMO. ATM and NEMO are exported to the cytoplasm where K63-linked polyubiquitination of ELKS and TRAF6 via ATM-dependent mechanism, as well as monoubiquitination of NEMO on lysine 285 via cIAP1, are induced. The polyubiquitin scaffolds then activate IKK via TAK1, similar to the mechanism induced by TNFα. Active IKK then phosphorylates IκBα, which then causes K48-linked polyubiquitination and degradation of IκBα by the proteasome to liberate active NF-κB (p50/p65) dimer. Polyubiquitin is represented in repeated yellow units, phosphate is shown in orange oval with “P” and SUMOylation is shown in purple circle with “S”. LUBAC, linear ubiquitin assembling complex.
ATM-NEMO signaling induced by physiological DSB
Does the nuclear ATM-NEMO signaling mechanism revealed above have physiological relevance? A recent study by Bredemeyer et al. 73 provided evidence that an endogenous DSB induced during V(D)J recombination of immunoglobulin loci in differentiating pre-B cells activates NF-κB via an ATM- and NEMO-dependent mechanism. V-abl transformed pre-B cell lines had previously been shown to activate Rag2 synthesis and subsequent recombination on inhibition of v-Abl kinase activity using the specific inhibitor STI571. Using pre-B cells generated from Artemis−/− mice that are deficient in non-homologous end joining, the authors showed that inhibition of v-Abl kinase activity by STI571 produces persistent DSBs at the Igκ gene locus and causes NF-κB activation in an ATM- and NEMO-dependent manner. This activation is DSB-dependent because it is not observed in v-Abl-transformed Rag2−/− pre-B cells that do not produce DSB at the Igκ locus when v-Abl is inhibited. Through a microarray analysis, the authors identified 75 genes whose expression is altered in a DSB-ATM-NF-κB-dependent manner. Changes in some of these genes are also observed in primary pro-B, pre-B and pro-T cells in an ATM-dependent manner, suggesting that physiological V(D)J recombination-induced DSBs activate the ATM-NF-κB signaling pathway in normal developing lymphocytes. The genes activated by this pathway include both cell survival genes and those that play important roles in lymphocyte development, homing and function. The authors speculated, “DNA DSBs generated in other physiological settings, such as immunoglobulin class-switch recombination, meiosis and DNA replication, may have similar effects.” Indeed, a recent study by Sherman et al. 74 has described a new role of DSB-ATM signaling induced by DSB generated during class-switch recombination. This DSB-ATM pathway inactivated CRTC2, a cAMP responsive element-binding protein (CREB) co-activator, thereby preventing CREB-mediated gene regulation and promoting germinal center reaction and plasma cell differentiation. Inhibition of this pathway results in increased germinal center B-cell proliferation, reduced secretion of antibodies and impaired plasma cell differentiation. It is possible that this DSB-ATM pathway also regulates NF-κB as speculated by Bredemeyer et al. Additional studies, including animal models that specifically attenuate the ATM-NEMO signaling pathway, may be informative to test these hypotheses.
ATM-NEMO signaling in pathology
In addition to normal physiological DSBs that can lead to ATM-NEMO-dependent NF-κB activation, a recent study by Grosjean-Raillard et al. 75 provides compelling evidence that ATM-NEMO-NF-κB signaling is constitutively activated in certain acute myeloid leukemia (AML) cell lines and more significantly a high percentage of primary myelodysplastic syndrome (MDS) and AML patient samples. Using the P39 AML cell line, the authors showed that ATM is constitutively active, NEMO and PIDD are constitutively nuclear, and an ATM-NEMO nuclear complex is also detectable. Inhibition of ATM by KU55933 or ATM knockdown results in the loss of nuclear NEMO and PIDD, loss of ATM-NEMO complex, inhibition of NF-κB activity and induction of cell death. Moreover, CD34+ bone marrow mononuclear cells obtained from all high-risk MDS or AML patient samples analyzed stain positive with pS1981-ATM antibody, indicative of constitutive ATM activation. NEMO and PIDD are constitutively found in the nucleus in these primary cells and inhibition of ATM by KU55933 causes their redistribution to the cytoplasm, inhibition of constitutive NF-κB activity, and induction of cell death. Although constitutive activation of NF-κB is frequently observed in many different types of cancer patient samples and in some cases the mechanisms have been revealed 76, 77, 78, 79, 80, in many cases those that maintain NF-κB activity are undefined. Activation of DDR is frequently observed in primary human preneoplastic lesions 81 and DDR can be activated by inappropriate proliferation induced by different oncogenes 82. Thus, it is plausible that the nuclear ATM-NEMO signaling pathway might play a significant role in maintaining constitutive NF-κB activity in different human malignancies, in addition to high-risk MDS and AML.
ATM-NEMO signaling in oxidative stress
In addition to genotoxic agents, molecularly targeted drugs are also employed and being developed to control human malignancies. One class of emerging anticancer drugs is histone deacetylase inhibitors (HDACis). A recent study by Rosato et al. 83 demonstrates that LBH-589 (panobinostat) activates NF-κB via the nuclear ATM-NEMO pathway in different leukemic cell lines. The authors found that LBH-589 causes nuclear localization of NEMO and promotes its interaction with ATM in an oxidative stress-dependent manner. Oxidative stress can activate ATM 84 and cause SUMOylation of NEMO 57. NF-κB activation by LBH-589 is inhibited in cells expressing SUMOylation-defective NEMO-DK (K277A/K309A) mutant 83. Both NEMO nuclear localization and ATM association are prevented in NEMO-DK cells. Knockdown of NEMO or ATM prevents NF-κB activation and sensitizes cells to HDACi-induced cell death. Finally, the authors found that one of the critical NF-κB target genes is manganese superoxide dismutase (Mn-SOD) that counters oxidative stress induced by HDACi and thereby promotes resistance to HDACi-induced cell death. The authors also indicate that other HDACi compounds, Vorinostat (SAHA), LAQ-824 and sodium butyrate, similarly activate NF-κB. Thus, these studies highlight the possibility that HDACi and other oxidative stress-inducing compounds may similarly trigger the nuclear ATM-NEMO SUMOylation signaling pathway to cause NF-κB activation and cancer cell survival.
Significant remaining questions
NEMO structural conundrum
Owing to its essential function as a member of the canonical IKK complex, significant biochemical and structural work has provided mechanistic insight into the role of NEMO in canonical NF-κB activation. Recent structural analyses of NEMO subdomains with or without different binding partners 85, 86, 87, 88, 89, 90 suggest that recombinant NEMO forms an elongated parallel coiled-coil dimer that is predicted to elute in gel filtration with an apparent value of about 270 kDa 90. Experimental evidence provided by Agou et al. 91 showed that the elution profile of recombinant NEMO protein in gel filtration chromatography corresponds to a 500-kDa globular protein with a 73 Å Stokes radius. However, in glycerol gradient centrifugation, it migrated as a globular monomer of ∼50 kDa 92. Indeed, NEMO was reported to exist predominantly as a monomer in IKKα and IKKβ double knockout MEFs 92. In contrast, a recent biochemical analysis indicates that an “apo” form of NEMO1-355 missing the C-terminal ZF domain forms a dimer and a weaker tetramer, arising from self-association of the N-terminal domain, with an apparent Kd of 25.6 μM (dimer-tetramer equilibrium) 93. Moreover, addition of an IKK-derived NEMO-binding peptide eliminates the NEMO tetramer formation in vitro, thus suggesting that IKK prevents NEMO tetramerization. The presence of the ZF domain destabilizes the dimer configuration, causing NEMO to aggregate 93. In cell extracts prepared from HeLa or HEK293 cells, an IKK-free NEMO fraction elutes at [100 kDa in gel filtration chromatography 15. In contrast, a similar IKK-free NEMO fraction in MEF cell extracts elutes at ∼300 kDa in a gel filtration analysis 64. Thus, depending on the cell type the elution profile of “IKK-free” NEMO may be different in gel filtration, suggesting the presence of different free NEMO forms. Therefore, further molecular characterization of free NEMO is required to determine how such a pool of NEMO is differentially regulated in different cell types and how it affects the nuclear ATM-dependent signaling pathway. In this context, the use of the glycerol gradient centrifugation technique may be more informative as demonstrated by Agou and colleagues 92.
Currently available NEMO crystal structures (an extended dimer) also provide a further conundrum with respect to its PTMs induced by genotoxic agents, in particular SUMOylation and phosphorylation. The side chain of one of the SUMOylation sites, K277 present in the CC2 (coil-coil 2) region of NEMO, is pointing interior toward the center of the coil-coil and appears to be inaccessible to the solvent surface 85, 86, 89. This suggests that a regulatory step might be required to induce a conformational change in this CC2 region to increase its accessibility to the SUMOylation machinery (particularly the Ubc9 SUMO conjugating enzyme). Such a change might be induced during the recruitment of NEMO to the PARP1-PIASy complex 64. Alternatively, given the possibility that the free NEMO in cells may be a monomer 92, the SUMOylation lysine sites may be solvent accessible in cells at least some of the time without the need for an active mechanism to induce a conformational switch. Moreover, the covalent attachment of a ∼20 kDa globular SUMO1 molecule to the SUMOylation sites, particularly K277, is expected to significantly distort the coil-coil dimeric structure of NEMO, since it would sterically prevent the movement of the K277 side chain toward the interior of the coil-coil. Addition of a SUMO1 moiety to the side chain of the other SUMOylation site, K309, would also significantly affect the polyubiquitin-binding activity of the NEMO NOA/NUB/UBAN domain, since this residue is located in the middle of the ubiquitin binding patch, IYKADF 85, 86, 89. Thus, SUMOylation of K277 and K309 is likely associated with significant perturbations in the NEMO structure both during and subsequent to the SUMOylation reaction.
Similarly, although the OH group of the S85 residue of NEMO, the site of ATM-induced phosphorylation 54, is pointing somewhat away from the interior of the CC1 domain, the side chain of Q86 appears to be solvent inaccessible in the NEMO N-terminal domain-NBD (NEMO binding domain peptide) complex 88. Since (i) ATM usually (although not absolutely) requires the S/T-Q motif in its substrates for phosphorylation 94 and (ii) Q86N and Q86A point mutations of NEMO completely abrogate NF-κB activation by DNA-damaging agents without affecting activation by LPS 54, it can be concluded that the ATM accessibility of both S85 and Q86 residues is required to mediate S85 phosphorylation and NF-κB activation. Ivins et al. 93 demonstrated that binding of diubiquitin at the NOA/NUB/UBAN domain induced a modest but measurable structural alteration in the N-terminal CC1 domain that is located ∼200 aa away. This suggests the possibility that a binding event in a region away from the S85-Q86 motif may cause a conformational shift to permit efficient ATM accessibility to the SQ motif. Interestingly, FAT domain of ATM can bind the N-terminal region of NEMO that is overlapping with or in the vicinity of S85 16. Such binding could plausibly induce a conformational change to increase the accessibility of the SQ motif for ATM-mediated phosphorylation. Alternatively, constitutive phosphorylation of S68 of NEMO by associated IKKβ in the absence of stimulation has been reported 95. The authors found that this phosphorylation reduces NEMO dimerization and IKKβ association through the N-terminal NEMO domain. It is, therefore, also possible that free NEMO with S68 phosphorylation might be involved in a conformation change that exposes the S85-Q86 motif. Finally, if free NEMO is a monomer, the SQ motif may be solvent accessible. Detailed characterization of the structural and modification status of nuclear IKK-free NEMO is expected to provide significant insight into the role of NEMO PTMs in mediating NF-κB signaling upstream of TAK1 and IKK activation.
Roles of NEMO ZF domain and SUMO1
On the basis of 1.3E2 genetic complementation analysis, the C-terminal ZF domain is essential for NF-κB activation by multiple DSB- and replication stress-inducing agents 52. Signal-inducible SUMOylation of NEMO-ZF mutants is undetectable in cell extracts. When SUMO1 is fused at the N-terminus of such NEMO mutants, NF-κB activation by DNA-damaging agents is fully restored. Thus, in the 1.3E2 cell background, the ZF is required for SUMO1 modification and SUMO1 fusion can fully complement this deficiency. However, the ZF domain is not required for NEMO association with the E3 ligase PIASy, nor is it required for PIASy-mediated SUMOylation in vitro 60. The ZF is suggested to destabilize the dimeric coil-coil structure of full-length NEMO 93. Cordier et al. 96 demonstrated that the NEMO ZF is a ubiquitin-binding module. Moreover, the ZF helps to establish the specificity of the NOA/NUB/UBAN domain toward K63-linked polyubiquitin chains 97. Whether the role of the ZF in promoting NEMO SUMOylation is mechanistically related to the regulation of monomer-dimer states of free NEMO and/or its ubiquitin-binding activity needs to be investigated.
SUMOylation of NEMO induced by genotoxic agents specifically involves SUMO1, and not the related SUMO2 or 3 paralogs 52. Moreover, while the SUMO1 fusion of NEMO-ZF mutants fully complements NF-κB activation by genotoxic agents, SUMO2 and SUMO3 fusions fail to do so (unpublished observations). Since SUMO1 attachment at the N-terminus or at the natural lysine residues permits activation, it appears that SUMO1 functions as a modular domain targeting SUMOylated NEMO to a binding partner or a specific subnuclear location. SUMOylation of several DDR proteins can occur at the DSB site through the recruitment of SUMO paralogs, Ubc9 (SUMO E2), and PIAS1 and PIAS4/y 98, 99. A recent report indicates that NEMO does not localize to the DSB site after VP16 exposure 64. Currently, information regarding detailed NEMO sub-nuclear localization before and after DNA damage is not available. A high-resolution analysis is thus required to determine where in the nucleus the IKK-free unmodified and SUMOylated NEMO species are localized during the activation process.
Additional NF-κB signaling by genotoxic agents
In contrast to the canonical IKK-NF-κB activation, there is very limited information regarding non-canonical NF-κB activation by genotoxic agents. In primary prostate cancer patients, nuclear localization of RelB positively correlates to a patient's Gleason score 100, suggesting that non-canonical activation of RelB may play a significant pathological role in this cancer type. Interestingly, Xu et al. 101 found that nuclear localization of RelB is detected within 10 min of IR exposure, and this localization is maintained for a prolonged duration (up to 12 h) in PC3 prostate cancer cells. The authors also found that p52 protein is found on the Mn-SOD gene after 10 min of IR treatment by chromatin immunoprecipitation analysis. A SN52 peptide that harbors the nuclear localization sequence of p52 blocks nuclear p52:RelB complex appearance and the induction of Mn-SOD gene, and sensitizes cells to apoptosis induced by IR. Although the mechanism underlying the induction of p100 processing to p52 is not clear 102, these studies demonstrate that rapid p52:RelB activation is possible in prostate cancer cells following IR exposure. Similarly, Barre and Perkins 103 found that p100 processing to p52 was induced by treatment of U2OS osteosarcoma cells with cisplatin or UV with slower kinetics (4-12 h). The authors indicate that this p100 processing is ATM and IKKα dependent. Since the presence of nuclear IKKα has been frequently reported in the literature (e.g., 104, 105), and Liao and Sun 106 previously demonstrated that nuclear localization of p100 promotes its processing to p52, it is plausible that ATM activates IKKα in the nucleus to promote the p100-processing pathway. Given that p100 processing requires its prior SUMOylation 107, it is possible that genotoxic agent-induced SUMOylation may also play a role in non-canonical NF-κB activation. Furthermore, Renner et al. 108 showed that DSB-inducing agents cause SUMOylation of nuclear IKKɛ to promote RelA phosphorylation and its transcriptional activity. Thus, SUMOylation may play a broader role in regulating genotoxic stress-induced NF-κB signaling at multiple levels.
Cell type dependence
NF-κB activation by genotoxic agents varies considerably in different cell systems. For example, activation of NF-κB as measured by electrophoretic mobility shift assay by DSB inducers is readily detectable in various cell lines, such as HEK293 embryonic kidney fibroblasts, HeLa cervical cancer, U2OS osteosarcoma, CEM T leukemic and 70Z/3 pre-B leukemic cells, among others 52, 55, 57, 60, 64, 66, 68. In contrast, NF-κB activation by VP16 or IR in primary MEFs and MEF lines, or many human normal cell types, is frequently undetectable or only weakly detected even at relatively high doses, despite efficient ATM activation 11, 55, 59, 64, 68. Most normal tissues in mice, except for the bone marrow, spleen, lymph nodes and intestine, also do not activate NF-κB in response to 8.5 Gy of whole body radiation 109, 110. Activation of IKK and NF-κB may be achieved in kidney and lung tissues after a relatively high dose (20 Gy) of radiation 49. The reason for different efficiencies of NF-κB activation by genotoxic agents in different cancer and normal cell types is unclear but is not likely related to ATM, since ATM activation is readily observed in all cases.
Because NF-κB activation through the nuclear ATM-NEMO pathway depends on the export of these molecules to the cytoplasm, differences in nuclear-cytoplasmic volume ratios could have a significant impact on the efficiency of NF-κB activation by genotoxic agents. A normal hepatocyte, e.g., has an estimated volume ratio between the cytoplasm and nucleus of 20:1. When a signaling molecule moves from the cytoplasm to the nucleus – a classic signaling situation – it may gain a concentration advantage by 20-fold, and may promote a switch-like response (reviewed in 111). In contrast, when ATM and NEMO move to the cytoplasm, their concentrations will be greatly reduced by the much larger cytoplasmic volume thereby potentially reducing the efficiency of signaling. Moreover, the amount of free NEMO in the nucleus may be greater with larger nuclear volume. Thus, by altering nuclear-cytoplasmic volume ratios, it might be possible to significantly alter the efficacy of the nuclear-to-cytoplasmic NF-κB signaling capacity of a cell. Interestingly, a large body of literature demonstrates that mean nuclear volume (MNV) of tumor cells strongly correlates to the progression stage and/or prognosis in many human cancer types (e.g., 112, 113, 114, 115, 116, 117, 118). The difference of MNV can be quite significant; e.g., MNV is 77 and 292 μm3 for normal bladder epithelial cells and bladder carcinoma in situ, respectively 112. The mechanism by which more malignant cells with larger MNV are selected during tumor evolution is currently unknown; however, Ozer et al. 119 found a correlation between larger MNV in breast carcinoma and the presence of mutant p53. One possible mechanism is that cancer cells with progressively higher efficiency of nuclear-initiated survival pathways, such as the NF-κB pathway, because of the increased MNV may be selected. Consistent with this hypothesis, NF-κB activation by DSB inducers is readily detectable in highly progressed, hormone-independent MDA-MB-231 breast cancer and DU145 and PC3 prostate cancer cells, but much less so in hormone-dependent MCF7 breast and LNCaP prostate cancer cells and undetectable in normal diploid counterparts (unpublished observation). In all of these cases, both TNFα-induced NF-κB activation and genotoxic agent-induced ATM activation are readily detected. Thus, differences in MNV might be a critical contributor for cell type differences in NF-κB activation by nuclear DNA damage and possibly may drive the evolution of more aggressive tumor cells.
Concluding remarks
The advancement in our understanding of the complexity of the NF-κB signaling network induced by inflammatory and immune signals over the last two-and-half decades, since its discovery is remarkable. We now know that different NF-κB dimers can be activated by two genetically distinct canonical and non-canonical signaling pathways to control diverse physiological and pathological processes. Similarly, our understanding of the signaling pathways induced by genotoxic agents is also significantly expanding; however, because of the complexity of molecular and stress events imparted by these agents in a cell, the fundamental knowledge of the necessary and sufficient events that initiate NF-κB signaling pathways induced by virtually all genotoxic agents is currently incomplete. Moreover, because of the presence of concurrent molecular events induced by different genotoxic agents, more than one pathway may be simultaneously operating to control NF-κB signaling in the damaged cell with varying dominance and different kinetics. Current strategies to dissect signaling mechanisms employ genetic approaches (e.g., RNA interference, stable reconstitution of deficient cells), chemical inhibitors and molecular strategies that can take considerable effort. More efficient identification of NF-κB signaling pathways in different cell systems under various genotoxic stress conditions mandates the development of unique tools, such as additional chemical inhibitors and cell-permeable peptide competitors that specifically attenuate individual pathways in a high throughput format.
There is considerable experimental support for the presence of a conserved nuclear NEMO- and ATM-dependent NF-κB signaling pathway. This pathway is induced by many different exogenous DSB- and replication stress-inducing agents, and appears to be also induced during normal physiological process (e.g., V(D)J recombination in developing lymphocytes), in certain human malignancies (e.g., AML), and in response to chemotherapeutics that induce oxidative stress (e.g., HDAC inhibitors). SUMOylation of NEMO plays a crucial nuclear role in this signaling pathway; however, there are many unanswered questions regarding this ATM-NEMO-induced NF-κB signaling pathway. Foremost, why is activation of ATM (and PARP1) insufficient to activate NF-κB in many different normal and cancer cell types? The reason for this observation remains unclear but could be at least in part related to amount and/or status of free nuclear NEMO. Although nuclear IKK-free NEMO is targeted for a series of post-translational modifications to mediate this signaling pathway, recent crystal structure analyses indicate that some of these modification sites of NEMO may not be readily solvent accessible. Thus, we do not yet know whether a conformational switch is required for NEMO modifications or whether the structure of free NEMO in the nuclear environment is distinct from that predicted from the analysis of recombinant NEMO protein segments. We also do not know whether free NEMO is a monomer or multimer (e.g., dimer), or in the “apo” form or associated with some partner protein (e.g., a chaperone). Characterization of the free NEMO structure, modification status and associated factors, as well as determination of sub-nuclear localization of free NEMO, could hold answers as to why ATM is insufficient to activate nuclear NEMO-mediated NF-κB signaling pathway.
Finally, because of the involvement of a unique signal transfer step from the nucleus to the cytoplasm to mediate ATM-dependent NF-κB activation, nuclear-cytoplasmic volume ratios may have a considerable impact on the efficiency of NF-κB activation induced by different genotoxic stimuli. A potential increase in the efficiency of a nuclear-to-cytoplasmic NF-κB survival pathway may in part play a role in the evolution of aggressive cancers with progressively larger mean nuclear volume. With the remarkable progress in our understanding of the NF-κB signaling network, answers to these and additional questions are likely forthcoming in the near future. Increased understanding of the NF-κB signaling network and the development of new tools to attenuate or enhance specific signaling mechanisms could aid rational development of drugs for treatment and prevention of specific human diseases, including metastatic cancers.
Acknowledgments
We thank the members of the Miyamoto laboratory for critical reading and for many suggestions, Shelly Wuerzberger-Davis, Kevin McCool and Shawn Jackson for the illustration, and Fabrice Agou for discussions on NEMO structure. The author apologizes to those whose contributions were not referred because of space limitation.
References
- Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hayden MS. New regulators of NF-κB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. J Immunol. 1986;2006;46177:705–716. 7485–7496. [PubMed] [Google Scholar]
- Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D. IκB: a specific inhibitor of the NF-κB transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Miyamoto S. Many faces of NF-κB signaling induced by genotoxic stress. J Mol Med. 2007;85:1187–1202. doi: 10.1007/s00109-007-0227-9. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tschopp J. Signals from within: the DNA-damage-induced NF-κB response. Cell Death Differ. 2006;13:773–784. doi: 10.1038/sj.cdd.4401843. [DOI] [PubMed] [Google Scholar]
- Criswell T, Leskov K, Miyamoto S, Luo G, Boothman DA. Transcription factors activated in mammalian cells after clinically relevant doses of ionizing radiation. Oncogene. 2003;22:5813–5827. doi: 10.1038/sj.onc.1206680. [DOI] [PubMed] [Google Scholar]
- Salminen A, Suuronen T, Huuskonen J, Kaarniranta K. NEMO shuttle: a link between DNA damage and NF-κB activation in progeroid syndromes. Biochem Biophys Res Commun. 2008;367:715–718. doi: 10.1016/j.bbrc.2007.11.189. [DOI] [PubMed] [Google Scholar]
- Ahmed KM, Li JJ. ATM-NF-κB connection as a target for tumor radiosensitization. Curr Cancer Drug Targets. 2007;7:335–342. doi: 10.2174/156800907780809769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y, Piette J. NF-κB activation by double-strand breaks. Biochem Pharmacol. 2006;72:1132–1141. doi: 10.1016/j.bcp.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Wong ET, Shi Y, et al. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell. 2010;40:75–86. doi: 10.1016/j.molcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Miyamoto S. Induction of a pro-apoptotic ATM-NF-κB pathway and its repression by ATR in response to replication stress. EMBO J. 2008;27:1963–1973. doi: 10.1038/emboj.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010;584:3675–3681. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, DeMartino GN, Greene WC. Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, et al. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- Basak S, Hoffmann A. Crosstalk via the NF-κB signaling system. Cytokine Growth Factor Rev. 2008;19:187–197. doi: 10.1016/j.cytogfr.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-κB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- Jacobs MD, Harrison SC. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- Rice NR, MacKichan ML, Israel A. The precursor of NF-κB p50 has IκB-like functions. Cell. 1992;71:243–253. doi: 10.1016/0092-8674(92)90353-e. [DOI] [PubMed] [Google Scholar]
- Mercurio F, DiDonato JA, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-κB-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- Beg AA, Baltimore D. An essential role for NF-κB in preventing TNF-a-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Fuentes ME, Yamaguchi K, et al. Embryonic lethality, liver degeneration, and impaired NF-κB activation in IKK-b-deficient mice. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- Li ZW, Chu W, Hu Y, et al. The IKKb subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Estepa G, Memet S, Israel A, Verma IM. Complete lack of NF-κB activity in IKK1 and IKK2 double-deficient mice: additional defect in neurulation. Genes Dev. 2000;14:1729–1733. [PMC free article] [PubMed] [Google Scholar]
- Makris C, Godfrey VL, Krahn-Senftleben G, et al. Female mice heterozygous for IKKg/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol Cell. 2000;5:969–979. doi: 10.1016/s1097-2765(00)80262-2. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Bloch W, Courtois G, et al. NEMO/IKKg-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- Rudolph D, Yeh WC, Wakeham A, et al. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKg-deficient mice. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D. Circuitry of nuclear factor κB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Weih F, Caamano J. Regulation of secondary lymphoid organ development by the nuclear factor-κB signal transduction pathway. Immunol Rev. 2003;195:91–105. doi: 10.1034/j.1600-065x.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- Stein B, Rahmsdorf HJ, Steffen A, Litfin M, Herrlich P. UV-induced DNA damage is an intermediate step in UV-induced expression of human immunodeficiency virus type 1, collagenase, c-fos, and metallothionein. Mol Cell Biol. 1989;9:5169–5181. doi: 10.1128/mcb.9.11.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebel A, Rahmsdorf HJ, Ullrich A, Herrlich P. Dephosphorylation of receptor tyrosine kinases as target of regulation by radiation, oxidants or alkylating agents. EMBO J. 1996;15:5314–5325. [PMC free article] [PubMed] [Google Scholar]
- Kokura S, Yoshida N, Sakamoto N, et al. The radical scavenger edaravone enhances the anti-tumor effects of CPT-11 in murine colon cancer by increasing apoptosis via inhibition of NF-κB. Cancer Lett. 2005;229:223–233. doi: 10.1016/j.canlet.2005.06.039. [DOI] [PubMed] [Google Scholar]
- van Maanen JM, Retel J, de Vries J, Pinedo HM. Mechanism of action of antitumor drug etoposide: a review. J Natl Cancer Inst. 1988;80:1526–1533. doi: 10.1093/jnci/80.19.1526. [DOI] [PubMed] [Google Scholar]
- van Maanen JM, Verkerk UH, Broersen J, et al. Semi-quinone formation from the catechol and ortho-quinone metabolites of the antitumor agent VP-16–213. Free Radic Res Commun. 1988;4:371–384. doi: 10.3109/10715768809066905. [DOI] [PubMed] [Google Scholar]
- Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22:5734–5754. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- Valerie K, Yacoub A, Hagan MP, et al. Radiation-induced cell signaling: inside-out and outside-in. Mol Cancer Ther. 2007;6:789–801. doi: 10.1158/1535-7163.MCT-06-0596. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Asano T, Nakayama K, et al. Nuclear IKKb is an adaptor protein for IκBa ubiquitination and degradation in UV-induced NF-κB activation. Mol Cell. 2010;39:570–582. doi: 10.1016/j.molcel.2010.07.030. [DOI] [PubMed] [Google Scholar]
- Devary Y, Rosette C, DiDonato JA, Karin M. NF-κB activation by ultraviolet light not dependent on a nuclear signal. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- Bender K, Gottlicher M, Whiteside S, Rahmsdorf HJ, Herrlich P. Sequential DNA damage-independent and -dependent activation of NF-κB by UV. EMBO J. 1998;17:5170–5181. doi: 10.1093/emboj/17.17.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Banin S, Ouyang H, et al. ATM is required for IκB kinase (IKKk) activation in response to DNA double strand breaks. J Biol Chem. 2001;276:8898–8903. doi: 10.1074/jbc.M009809200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Dimtchev A, Lavin MF, Dritschilo A, Jung M. A novel ionizing radiation-induced signaling pathway that activates the transcription factor NF-κB. Oncogene. 1998;17:1821–1826. doi: 10.1038/sj.onc.1202088. [DOI] [PubMed] [Google Scholar]
- Piret B, Schoonbroodt S, Piette J. The ATM protein is required for sustained activation of NF-κB following DNA damage. Oncogene. 1999;18:2261–2271. doi: 10.1038/sj.onc.1202541. [DOI] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKg by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- Panta GR, Kaur S, Cavin LG, et al. ATM and the catalytic subunit of DNA-dependent protein kinase activate NF-κB through a common MEK/extracellular signal-regulated kinase/p90(rsk) signaling pathway in response to distinct forms of DNA damage. Mol Cell Biol. 2004;24:1823–1835. doi: 10.1128/MCB.24.5.1823-1835.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Seufzer BJ, et al. NF-κB activation by camptothecin. A linkage between nuclear DNA damage and cytoplasmic signaling events. J Biol Chem. 2000;275:9501–9509. doi: 10.1074/jbc.275.13.9501. [DOI] [PubMed] [Google Scholar]
- Zhang H, Barcelo JM, Lee B, et al. Human mitochondrial topoisomerase I. Proc Natl Acad Sci USA. 2001;98:10608–10613. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerzberger-Davis SM, Nakamura Y, Seufzer BJ, Miyamoto S. NF-κB activation by combinations of NEMO SUMOylation and ATM activation stresses in the absence of DNA damage. Oncogene. 2007;26:641–651. doi: 10.1038/sj.onc.1209815. [DOI] [PubMed] [Google Scholar]
- Rashi-Elkeles S, Elkon R, Weizman N, et al. Parallel induction of ATM-dependent pro- and antiapoptotic signals in response to ionizing radiation in murine lymphoid tissue. Oncogene. 2006;25:1584–1592. doi: 10.1038/sj.onc.1209189. [DOI] [PubMed] [Google Scholar]
- Ashburner BP, Shackelford RE, Baldwin AS Jr, Paules RS. Lack of involvement of ataxia telangiectasia mutated (ATM) in regulation of nuclear factor-κB (NF-κB) in human diploid fibroblasts. Cancer Res. 1999;59:5456–5460. [PubMed] [Google Scholar]
- Mabb AM, Wuerzberger-Davis SM, Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-κB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Hur GM, Lewis J, Yang Q, et al. The death domain kinase RIP has an essential role in DNA damage-induced NF-κB activation. Genes Dev. 2003;17:873–882. doi: 10.1101/gad.1062403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, et al. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilmann M, Hinz M, Arslan SC, et al. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IκB kinase activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret B, Piette J. Topoisomerase poisons activate the transcription factor NF-κB in ACH-2 and CEM cells. Nucleic Acids Res. 1996;24:4242–4248. doi: 10.1093/nar/24.21.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-κB activation requires PARP-1 function to confer radioresistance. Oncogene. 2009;28:832–842. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HS, Lee DH, Kim DH, et al. cIAP1, cIAP2, and XIAP act cooperatively via nonredundant pathways to regulate genotoxic stress-induced nuclear factor-κB activation. Cancer Res. 2009;69:1782–1791. doi: 10.1158/0008-5472.CAN-08-2256. [DOI] [PubMed] [Google Scholar]
- Ducut Sigala JL, Bottero V, Young DB, et al. Activation of transcription factor NF-κB requires ELKS, an IκB kinase regulatory subunit. Science. 2004;304:1963–1967. doi: 10.1126/science.1098387. [DOI] [PubMed] [Google Scholar]
- Hinz M, Stilmann M, Arslan SC, et al. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation. Mol Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold CM, Wu ZH, Huang TT, Miyamoto S. Calcium-dependent regulation of NEMO nuclear export in response to genotoxic stimuli. Mol Cell Biol. 2007;27:497–509. doi: 10.1128/MCB.01772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer AL, Helmink BA, Innes CL, et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MH, Kuraishy AI, Deshpande C, et al. AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Mol Cell. 2010;39:873–885. doi: 10.1016/j.molcel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean-Raillard J, Tailler M, Ades L, et al. ATM mediates constitutive NF-κB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene. 2009;28:1099–1109. doi: 10.1038/onc.2008.457. [DOI] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, et al. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Rosato RR, Kolla SS, Hock SK, et al. Histone deacetylase inhibitors activate NF-κB in human leukemia cells through an ATM/NEMO-related pathway. J Biol Chem. 2010;285:10064–10077. doi: 10.1074/jbc.M109.095208. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- Lo YC, Lin SC, Rospigliosi CC, et al. Structural basis for recognition of diubiquitins by NEMO. Mol Cell. 2009;33:602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Bagneris C, Ageichik AV, Cronin N, et al. Crystal structure of a vFlip-IKKg complex: insights into viral activation of the IKK signalosome. Mol Cell. 2008;30:620–631. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Rushe M, Silvian L, Bixler S, et al. Structure of a NEMO/IKK-associating domain reveals architecture of the interaction site. Structure. 2008;16:798–808. doi: 10.1016/j.str.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A, Sato Y, Yamashita M, et al. Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett. 2009;583:3317–3322. doi: 10.1016/j.febslet.2009.09.028. [DOI] [PubMed] [Google Scholar]
- Grubisha O, Kaminska M, Duquerroy S, et al. DARPin-assisted crystallography of the CC2-LZ domain of NEMO reveals a coupling between dimerization and ubiquitin binding. J Mol Biol. 2010;395:89–104. doi: 10.1016/j.jmb.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Agou F, Ye F, Goffinont S, et al. NEMO trimerizes through its coiled-coil C-terminal domain. J Biol Chem. 2002;277:17464–17475. doi: 10.1074/jbc.M201964200. [DOI] [PubMed] [Google Scholar]
- Fontan E, Traincard F, Levy SG, et al. NEMO oligomerization in the dynamic assembly of the IkappaB kinase core complex. FEBS J. 2007;274:2540–2551. doi: 10.1111/j.1742-4658.2007.05788.x. [DOI] [PubMed] [Google Scholar]
- Ivins FJ, Montgomery MG, Smith SJ, et al. NEMO oligomerization and its ubiquitin-binding properties. Biochem J. 2009;421:243–251. doi: 10.1042/BJ20090427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill T, Dwyer AJ, Ziv Y, et al. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- Palkowitsch L, Leidner J, Ghosh S, Marienfeld RB. Phosphorylation of serine 68 in the IκB kinase (IKK)-binding domain of NEMO interferes with the structure of the IKK complex and tumor necrosis factor-a-induced NF-κB activity. J Biol Chem. 2008;283:76–86. doi: 10.1074/jbc.M708856200. [DOI] [PubMed] [Google Scholar]
- Cordier F, Grubisha O, Traincard F, et al. The zinc finger of NEMO is a functional ubiquitin-binding domain. J Biol Chem. 2009;284:2902–2907. doi: 10.1074/jbc.M806655200. [DOI] [PubMed] [Google Scholar]
- Laplantine E, Fontan E, Chiaravalli J, et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- Lessard L, Begin LR, Gleave ME, Mes-Masson AM, Saad F. Nuclear localisation of nuclear factor-κB transcription factors in prostate cancer: an immunohistochemical study. Br J Cancer. 2005;93:1019–1023. doi: 10.1038/sj.bjc.6602796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Fang F, St Clair DK, et al. SN52, a novel nuclear factor-κB inhibitor, blocks nuclear import of RelB:p52 dimer and sensitizes prostate cancer cells to ionizing radiation. Mol Cancer Ther. 2008;7:2367–2376. doi: 10.1158/1535-7163.MCT-08-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Fang F, Sun Y, St Clair DK, St Clair WH. RelB-dependent differential radiosensitization effect of STI571 on prostate cancer cells. Mol Cancer Ther. 2010;9:803–812. doi: 10.1158/1535-7163.MCT-09-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre B, Perkins ND. The Skp2 promoter integrates signaling through the NF-κB, p53, and Akt/GSK3b pathways to regulate autophagy and apoptosis. Mol Cell. 2010;38:524–538. doi: 10.1016/j.molcel.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Anest V, Hanson JL, Cogswell PC, et al. A nucleosomal function for IκB kinase-alpha in NF-κB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-a is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- Liao G, Sun SC. Regulation of NF-κB2/p100 processing by its nuclear shuttling. Oncogene. 2003;22:4868–4874. doi: 10.1038/sj.onc.1206761. [DOI] [PubMed] [Google Scholar]
- Vatsyayan J, Qing G, Xiao G, Hu J. SUMO1 modification of NF-κB2/p100 is essential for stimuli-induced p100 phosphorylation and processing. EMBO Rep. 2008;9:885–890. doi: 10.1038/embor.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner F, Moreno R, Schmitz ML. SUMOylation-dependent localization of IKKe in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol Cell. 2010;37:503–515. doi: 10.1016/j.molcel.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Zhou D, Brown SA, Yu T, et al. A high dose of ionizing radiation induces tissue-specific activation of nuclear factor-κB in vivo. Radiat Res. 1999;151:703–709. [PubMed] [Google Scholar]
- Wang Y, Meng A, Lang H, et al. Activation of nuclear factor κB in vivo selectively protects the murine small intestine against ionizing radiation-induced damage. Cancer Res. 2004;64:6240–6246. doi: 10.1158/0008-5472.CAN-04-0591. [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr. How regulated protein translocation can produce switch-like responses. Trends Biochem Sci. 1998;23:461–465. doi: 10.1016/s0968-0004(98)01316-4. [DOI] [PubMed] [Google Scholar]
- Sorensen FB, Jacobsen F. Stereological estimates of nuclear volume in the prognostic evaluation of primary flat carcinoma in situ of the urinary bladder. Histopathology. 1991;18:531–539. doi: 10.1111/j.1365-2559.1991.tb01480.x. [DOI] [PubMed] [Google Scholar]
- Mogensen O, Sorensen FB, Bichel P, Jakobsen A. Nuclear volume and prognosis in ovarian cancer. Int J Gynecol Cancer. 1992;2:141–146. doi: 10.1046/j.1525-1438.1992.02030141.x. [DOI] [PubMed] [Google Scholar]
- Mogensen O, Sorensen FB, Bichel P, Jakobsen A. Mean nuclear volume: a supplementary prognostic factor in endometrial cancer. Int J Gynecol Cancer. 1999;9:72–79. doi: 10.1046/j.1525-1438.1999.09829.x. [DOI] [PubMed] [Google Scholar]
- Kanamaru H, Sasaki M, Miwa Y, Akino H, Okada K. Prognostic value of sarcomatoid histology and volume-weighted mean nuclear volume in renal cell carcinoma. BJU Int. 1999;83:222–226. doi: 10.1046/j.1464-410x.1999.00912.x. [DOI] [PubMed] [Google Scholar]
- Arai Y, Okubo K, Terada N, et al. Volume-weighted mean nuclear volume predicts tumor biology of clinically organ-confined prostate cancer. Prostate. 2001;46:134–141. doi: 10.1002/1097-0045(20010201)46:2<134::aid-pros1017>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Dey P, Powari M. Mean nuclear volume in cervical intraepithelial neoplasia and carcinoma. Anal Quant Cytol Histol. 2002;24:70–72. [PubMed] [Google Scholar]
- Bonnelykke-Behrndtz ML, Sorensen FB, Damsgaard TE. Stereological quantification of tumor volume, mean nuclear volume and total number of melanoma cells correlated with morbidity and mortality. APMIS. 2008;116:903–911. doi: 10.1111/j.1600-0463.2008.01100.x. [DOI] [PubMed] [Google Scholar]
- Ozer E, Canda T, Kuyucuodlu F. p53 mutations in bilateral breast carcinoma. Corrleation with Ki-67 expression and the mean nuclear volume. Cancer Lett. 1998;122:101–106. doi: 10.1016/s0304-3835(97)00374-1. [DOI] [PubMed] [Google Scholar]