Abstract
Cancer evolution at all stages is driven by both epigenetic abnormalities as well as genetic alterations. Dysregulation of epigenetic control events may lead to abnormal patterns of DNA methylation and chromatin configurations, both of which are critical contributors to the pathogenesis of cancer. These epigenetic abnormalities are set and maintained by multiple protein complexes and the interplay between their individual components including DNA methylation machinery, histone modifiers, particularly, polycomb (PcG) proteins, and chromatin remodeling proteins. Recent advances in genome-wide technology have revealed that the involvement of these dysregulated epigenetic components appears to be extensive. Moreover, there is a growing connection between epigenetic abnormalities in cancer and concepts concerning stem-like cell subpopulations as a driving force for cancer. Emerging data suggest that aspects of the epigenetic landscape inherent to normal embryonic and adult stem/progenitor cells may help foster, under the stress of chronic inflammation or accumulating reactive oxygen species, evolution of malignant subpopulations. Finally, understanding molecular mechanisms involved in initiation and maintenance of epigenetic abnormalities in all types of cancer has great potential for translational purposes. This is already evident for epigenetic biomarker development, and for pharmacological targeting aimed at reversing cancer-specific epigenetic alterations.
Keywords: cancer epigenetics, DNA methylation, polycomb proteins, cancer stem cells, biomarkers, epigenetic therapy
Introduction
Normal biological functions in a multicellular organism rely on intricate orchestration between the basic cellular features preset by genetic constitution, and a sophisticated network of cellular RNA expression patterns governed by epigenetic regulation. Epigenetics refers to the establishment of heritable changes in gene expression without alterations in primary DNA sequences. Such gene patterns play an essential role in various biological processes including embryonic developmental events, adult cell renewal, gene imprinting, and X-chromosome inactivation 1, 2. Regulation of these essential biological functions depends on the interplay between at least three major epigenetic mechanisms, discussed in great deal in other reviews in this issue (Gasser, Kouzarides, Crabtree, Zhu), including DNA methylation, histone modifications, and nucleosome remodeling. These controlling mechanisms for chromatin organization act coordinately to modulate expression of canonical coding and non-coding RNAs. If these processes are dysregulated, they may lead to many human diseases, including cancer, autoimmune diseases, and neurological disorders 3, 4, 5, 6.
In this review, we would like to provide an overview, based on current knowledge, of how epigenetic components are involved in the pathogenesis of cancer. In this context, we especially explore the relationships between epigenetic control events in normal development, and regulation of normal stem/progenitor cells, and that in oncogenesis. These areas are particularly relevant to the “cancer stem cell” hypothesis and its importance to tumorigenesis and to key aspects of cancer clinical management. The latter clinical applications of epigenetic mechanisms in cancer are explored and discussed, particularly in two major areas – biomarker development and therapies targeting epigenetic abnormalities.
Epigenetic alterations in cancer development
At multiple stages of tumor evolution, cells escape normal physiological regulation of proliferation, differentiation, and cell death, leading to uncontrolled cell growth 7. This progression course involves abnormal activation of oncogenes, inactivation of tumor suppressor genes, and altered expression of non-coding RNAs, which can also harbor these functions 8, 9, 10. The abnormal gene function states are driven by both genetic abnormalities, through mutations and genomic instability events, and epigenetic alterations involving the epigenetic machinery mentioned in this issue and which we review directly below.
DNA methylation
Of the epigenetic mechanisms dysregulated during oncogenesis, the best studied are those for DNA methylation. This chemical modification of DNA, as reviewed elsewhere in this issue (Zhu and colleagues), involves addition of a methyl group to the 5′ position of cytosines, predominantly in the context of “CpG” dinucleotides for the mammalian genome. The involved enzymatic step utilizes S-adenosyl-methionine as a methyl donor and is carried out by three separate DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) 11, 12.
In normal mammalian cells, the CpG dinucleotide is under-represented because it has been depleted over evolution via deamination of methylated cytosines and inaccurate repair of the deaminated product to thymines 13, 14. The majority of these remaining CpG dinucleotide sites are methylated and found in the non-coding repetitive elements and gene bodies. When distributed in CpG poor gene promoters, the sites are methylated in tissue-specific patterns, which often correlate with decreased transcriptional activity 2, 15. In contrast to the above, a small percentage of “CpG” dinucleotides are clustered in regions termed “CpG islands”, many of which surround gene promoters, the transcription start sites, and/or first exons 16. Some 85-90% of such islands remain constitutively free of DNA methylation with the remainder associated with transcriptional silencing involved in X-chromosome inactivation 2, 17, genomic imprinting 18, and transposable elements inhibition 2. Some promoter islands in other gene types can also be mosaically methylated in normal tissues, especially those with less CpG density (or what have been referred to as “intermediate density” islands) 1. Abnormal DNA methylation of promoter CpG islands is a fundamental abnormality of cancer, which is discussed extensively below in this review.
There are two major altered methylation patterns observed in cancer – global DNA hypomethylation and the above mentioned promoter DNA hypermethylation 4, 19, 20, 21. Multiple lines of evidence have indicated that global loss of methylcytosine is correlated with different stages of cancer progression and metastasis in various tumor types, including prostate, cervical, hepatocellular, and brain cancers 22, 23, 24, 25. Moreover, this change may appear at earlier stages such as in pre-invasive colon polyps 26. The potential cellular consequences of global DNA methylation loss are diverse, ranging from chromosome instability, genetic mutation, to reactivation of various cancer-related genes including cancer testis antigen MAGEA1, inserted viral oncogenes, or imprinted genes related to growth such as IGF2 25, 27, 28.
In virtually every type of cancer, in the same cancer cells that harbor genome-wide DNA hypomethylation, hundreds of genes simultaneously exhibit DNA hypermethylation of promoter CpG islands 29, 30. In point of fact, this epigenetic event affects more genes than do mutations 5, 31. The alteration is associated with very stable states of transcriptional silencing, and for many tumor suppressor genes such as the von Hippel-Lindau (VHL) gene 32, cyclin-dependent kinase inhibitors 2A (CDKN2A) 33, 34 and 2B (CDKN2B) 35, 36, 37, 38, and others, this change can serve as an alternative mechanism to mutation for tumor suppressor gene inactivation. For such genes, as embodied in the Knudson two-hit model, the above epigenetic silencing can provide the first hit to inactivate one, or both, alleles of a gene or can co-exist with a mutation in the opposite allele 39.
Epigenetic silencing of key genes can affect virtually all pathways, which participate in the development of cancer at different stages 4 – cancer initiation, progression, invasion, and metastasis. Examples include dysfunction of DNA repair genes, such as hMLH1 (DNA mismatch repair protein), which can be an early event in the development of endometrial and colon cancer 40, 41 and associated with a microsatellite instability phenotype 42. Another DNA repair gene, O6-methylguanine-DNA methyltransferase (MGMT), if inactivated by DNA hypermethylation, can predispose tumors to mutations of a specific type in critical genes including TP53 43 and K-Ras 44. Loss of cell cycle control can be involved with silencing of the aforementioned CDKN2A gene, and changes in cell migration and invasion can involve epigenetically mediated silencing of genes such as CDH1 45. Most recently, pathways modulated by microRNAs (miRNAs) have also been identified as involved with promoter DNA hypermethylation silencing of these non-coding transcription products 46, 47, 48, 49. Also, downregulation of miRNA expression has been linked to overexpression of DNMTs and, thus, facilitation of gene promoter DNA hypermethylation in cancer 50, 51.
Over the past 20 years, a growing number of proven and candidate tumor suppressor genes have been identified, and characterized, by virtue of their being DNA hypermethylated and silenced in cancer. One key aspect of this work is that many of these genes are seldom mutated, or have never been recognized as genetically altered in tumors 5. Thus, their loss of function in cancer appears to be due solely to epigenetic mechanisms. Examples include, tissue inhibitor of metalloproteinase-3 (TIMP3) 52, the secreted frizzled-related gene family, which acts normally to counteract Wnt pathway activity (SFRP1, SFRP2, SFRP4, and SFRP5) 53, Ras association gene (RASSF1A) 54, 55, and so on. Such gene identification has fueled the development, as will be discussed later in this review, of many new strategies to screen the cancer genome for DNA-hypermethylated genes, and to identify new genes later well proven to have potential to function as tumor suppressor genes 56, 57, 58, 59, 60.
How DNA hypermethylation silences genes in cancer cells is a critical aspect of research. Some aspects are known from understanding of basic molecular facets of the DNA methylation machinery and more detailed descriptions of these can be found in the review by Zhu and colleagues in this issue. Briefly, DNA methylation serves as a signal to recruit the methyl CpG-binding domain (MBD) family including MeCP2, MBD1, MBD2, MBD3, and MBD4 2, 61, 62. The MBD proteins can recruit histone deacetylases, which are key to many gene silencing protein complexes 2, 63, 64, 65, 66. Importantly, all three biologically active DNMTs also bind these enzymes 67, 68, 69. In addition, MBD proteins are participants in chromatin remodeling complexes, containing transcriptional corepressors such as Sin3A, which are recruited to methylated loci, thereby establishing repressive chromatin architecture leading to transcription repression. The NuRD complex (also known as Mi-2 complex) is one such multi-protein complex involved in methylation-mediated gene silencing, which contains MBD3, histone deacetylases, a chromatin remodeling ATPase, and others 70, 71. MBD protein-related transcriptional repression can be histone deacetylase-dependent 63, 64, 72 or -independent 73. The deacetylase-independent mechanisms in transcriptional repression by MBD proteins include steric inhibition of transcription complex assembly, and higher order chromatin structure changes associated with DNA methylation 74.
What is much less understood is how all of the above proteins and complexes and other molecular determinants participate in the initiation of the cancer-specific DNA hypermethylation, and do this in the setting where involved cells are also simultaneously losing normal regions of the same modification. This question is under intense investigation by multiple groups. All clues generated, to date, concern complex interactions of chromatin regulation inherent to developmental biology and cell renewal and interaction of the DNA methylation machinery with modifications of histones, and the proteins that guide these latter modifications. Some features of current hypotheses are discussed in later sections.
Links between abnormal patterns of DNA methylation and chromatin regulation in cancer cells
The active or repressed transcription states of genes are maintained, as reviewed elsewhere in this issue (Kouzarides, Crabtree, Workman) by communications between histone modifications and chromatin-modifying protein complexes. In this regard, polycomb and trithorax, two major groups of chromatin-modifying proteins, have been shown to direct cell fate determination and to preserve gene expression patterns through many rounds of cell division 75, 76, 77, 78, 79. This histone regulation is critical to normal development and adult cell renewal when properly orchestrated and serves in combination with patterns of nucleosome positioning as the key layer of control to establish gene expression patterns 80, 81. Elucidating these interactions and dynamics is proving critical to the understanding of epigenetic abnormalities in cancer.
In essence, mechanisms that may underlie aberrant gene silencing in cancer, particularly, are being linked to altered patterns of the above chromatin regulatory events as governed by the many different post-translational modifications on histone tails. As reviewed in this issue by Kouzarides and colleagues, there are various types of histone tail modifications, such as acetylation, methylation, phosphorylation, ubiquitination, among others, that combine to determine repressive versus active states of gene transcription. In turn, these modifications regulate gene expression through their interactions with chromatin-associated proteins, in marking regions of transcriptionally active euchromatin and regions of transcriptionally inactive heterochromatin 82. Key to our current review of cancer-related epigenetic abnormalties is that the balance between many of the above marks can be altered in cancer and these alone may cause dysregulated states of gene transcription. Moreover, these modifications are very interactive with DNA methylation and, thus, as we will discuss, can potentially be key to what triggers initiation and maintenance of cancer-specific abnormalities such as promoter CpG island DNA hypermethylation. For example, histone marks such as acetylated histone H3, and especially di- or trimethylated histone H3 lysine 4 (H3K4me2, H3K4me3), are antagonistic in experimental models to imposition of DNA methylation 83, 84, 85, 86. On the other hand, histone marks for repression of transcription, such as deacetylated histone H3, trimethylated histone H3 lysine 9 (H3K9me3), and trimethylated histone H3 lysine 27 (H3K27me3) are correlated with DNA methylation in normal and abnormal settings 87, 88.
Many of the above possible correlations between histone marks and transcription are evident in cancer. For instance, epigenetically silenced genes, including those marked by promoter DNA methylation, are marked simultaneously, to variable extent, by the repressive marks, H3K9me3 and H3K27me3. In this setting, the active marks of H3K4me2, H3K4me3, and H3K9 acetylation are reduced 87, 89, 90. Interestingly, when the DNA-hypermethylated genes are reactivated by 5-aza-2′-deoxycytidine, a DNA demethylating agent, or examined in a colon cancer cell line where the DNMT1 and 3b have been genetically disrupted and DNA-hypermethylated genes are demethylated and re-expressed, the repressive chromatin does not fully return to an active euchromatic state 89. Rather, the gene promoters are left in a “semi-heterochromatic” state, in which the gene promoters have restored levels, to a variable degree, of the active H3K4me mark but retained some levels of the repressive mark, H3K27me3 90. This chromatin pattern closely resembles one that Bernstein et al. 91 have termed bivalent chromatin. In normal embryonic stem cells (ESCs), this bivalent promoter pattern marks a set of CpG island-containing genes which are held in a poised, low transcription state to prevent premature lineage commitment 91, 92.
The studies of polycomb group (PcG) proteins and complexes are shedding important light on how genes may succumb to abnormal silencing in cancer 93, 94, 95. First, the link of PcG marking and bivalency has been advanced by the initial findings that some 50% of genes with promoter CpG island DNA hypermethylation in colon cancer, are among the ∼10% of PcG-marked genes, most of which have the CpG islands in a bivalent state, in ESC and embryonic progenitor cells 88, 96, 97. Second, several constituents of these PcG complexes have been shown to interact with DNMTs, and possibly to promote initiation and maintenance of cancer-specific silencing in adult cancer 98, 99. In this regard, EZH2, the PcG protein in the polycomb repressive complex 2/3 (PRC 2/3) that catalyzes the trimethylation of histone H3 lysine 27 (H3K27me3) may be a key player 100, 101. While EZH2-mediated gene silencing usually takes place in the absence of DNA methylation, a study by Vire et al. 99 suggests EZH2 can directly interact with DNMTs. Growing evidence suggests a strong link of EZH2 to oncogenesis and to cancer-specific gene silencing. Overexpression of EZH2 has been found in many cancers and the expression level is correlated with tumor progression and prognosis 102, 103, 104, 105, 106. Depletion of EZH2 leads to growth arrest of cancer cells 102, 107. Moreover, we have previously shown that EZH2 and H3K27me3 are retained to a variable degree at the promoter of genes, which are DNA hypermethylated and abnormally silenced in cancer 89. However, knockdown of EZH2 does not fully reactivate genes that are densely DNA hypermethylated and silenced in adult cancer 108, 109, suggesting EZH2 may not be absolutely required for maintenance of DNA methylation.
CBX7, another PcG protein, is a constituent of PRC1, and has also been shown to read the repressive histone marks, H3K9me3 and H3K27me3, and to participate in mediating gene silencing in the development of cancer 98, 110, 111, 112. Similar to EZH2, in an experimental setting, CBX7 is able to recruit DNA methylation machinery to gene promoters and facilitate repression of genes, which are frequently silenced in adult cancer 98. Given the close connection between chromatin-modifying complexes, gene silencing, and DNA methylation, PcG complexes and their constituents are actively being pursued for their link to abnormal gene silencing and how aberrant DNA methylation is initiated and maintained in cancer.
In addition to chromatin-modifying complexes outlined above, another major player, which participates in mammalian gene regulation is chromatin remodeling complexes. The SWI/SNF family is one such player, which mediates ATP-dependent chromatin remodeling processes and alters the position of nucleosomes along DNA. Malfunction of the SWI/SNF multisubunit complexes has been associated with cancer development 113, 114. For example, BAF47 (encoded by SMARCB1), a subunit of the SWI/SNF complexes, was found to be inactivated in various types of cancer, including rhabdoid tumors 115, central nervous system tumors 116, and chronic myeloid leukemia 117. Loss of heterozygosity of other components in the SWI/SNF complexes, BRM (encoded by SMARCA2) and BRG1 (encoded by SMARCA4), has been found in primary lung cancers 118, 119. Recently, another member of the SWI/SNF complex, BAF250A (encoded by ARID1A) was shown to be frequently mutated in ovarian clear cell carcinoma 120. Although the epigenetic changes caused by these mutations have yet to be defined, the frequent mutations, in many types of cancer, of genes encoding proteins involved in chromatin remodeling have indicated its important role in the pathogenesis of cancer.
Cancer epigenomics
Recent advances of high-throughput technology have enabled scientists to map the human cancer genome at single-nucleotide resolution 121, 122, 123, 124, 125, 126, 127. Similar to genomic studies, research in cancer epigenetics for gene discovery, has been steadily moving beyond candidate gene approaches to large-scale epigenomic designs for characterization of global epigenetic alterations in cancer, including global patterns of DNA methylation, histone modifications, and factor occupancy of their gene targets. The hope is that understanding the human cancer epigenome landscape will provide valuable insights into the molecular mechanisms involved in oncogenesis and offer implications in translational research 6, 128.
Indeed, the epigenetic alterations in cancer have been shown to be a global event. Several genome-wide methylation studies indicate that hundreds of genes can coordinately undergo CpG island promoter DNA hypermethylation and become silenced in adult cancers, including individual tumors from patients 31, 60, 129, 130. In some instances, genes can be silenced simultaneously throughout long stretches of chromosomes and the spreading of silencing seems to affect neighboring unmethylated genes through repressive chromatin 131. However, more often, DNA methylation appears to take place locally with no initial preference for nuclear position or chromatin architecture 132.
Epigenomic studies may provide an additional perspective to sites of cancer-specific abnormalities in DNA methylation-dependent gene regulation. A recent study by Irizarry et al. 133 suggests DNA methylation in the “shore” regions, hundreds of base pairs away from classic CpG islands and/or transcriptional start sites, can also contribute to gene regulation. The molecular mechanism of how this “shore” methylation regulates gene expression is not yet well understood. However, one possibility may be that “shore” methylation controls enhancer activities thereby modulating gene expression in cancer.
Characterization of genome-wide methylated cytosines has clinical implications as well. Almost any given type of cancer is a heterogeneous disease composed of distinct clinical and biological subtypes. Genomic and epigenomic profiling may help identify molecular signatures of existing and new subtypes, thereby helping to derive more accurate classifications to guide clinical management 57, 134. For example, as part of The Cancer Genome Atlas project, global analysis of promoter DNA methylation patterns in 272 glioblastomas identified a distinct subset of tumors with a glioma-CpG island methylator phenotype, which is closely associated with presence of somatic mutations of the IDH1 gene 57. Patients with these tumors are younger and have better clinical outcome. Similarly, biologically distinct subtypes in acute myeloid leukemia were discovered using global methylation profiles 135.
Global alterations of histone modifications are of great importance in basic cancer research as well. A study by Fraga et al. 136 showed losses of acetylated histone H4 lysine 16 and trimethylated histone H4 lysine 20 were associated with tumorigenesis in a mouse model of multistage skin carcinogenesis. Similar findings were also observed in breast and liver cancer development 137, 138. In addition to histone modifications, studies on various PcG proteins have provided a link between gene silencing and cancer development.
Epigenetics and cancer stem cell hypothesis
It has long been known that individual cancers harbor heterogeneous cell populations and recent work has emphasized their diverse tumorigenic potentials. Thus, the concept of the cancer stem cell hypothesis has arisen, which stresses that only certain subpopulation(s), known as cancer stem cells or cancer-initiating cells or tumor propagating cells, may sustain and perpetuate tumors 139, 140. In 1994, John Dick and colleagues demonstrated that CD34+CD38− leukemic stem cells possessed differentiative and proliferative capacities, and were capable of initiating human leukemia in NOD/SCID mice 141. This seminal study on leukemic stem cells provided a paradigm for later studies on cancer stem cells in solid tumors, such as glioblastomas 142, breast cancer 143, prostate cancer 144, hepatocellular carcinomas 145, 146, colon 147, 148, 149, pancreatic 150, and head and neck cancers 151, among others.
Over the past decade, a growing body of evidence has indicated a huge relevance of the cancer stem cell hypothesis to clinical cancer management. As demonstrated in multiple studies, the cancer-initiating cells are usually resistant to standard chemotherapy 152 or radiotherapy 153, leading to clinical recurrence and treatment failure. Furthermore, they may be capable of forming metastatic foci at distant sites. Thus, understanding the origins and molecular characteristics of these cells may pave the way for developing therapies that directly target and eliminate cancer-initiating cells, thereby helping to prevent tumor recurrence or distant metastasis.
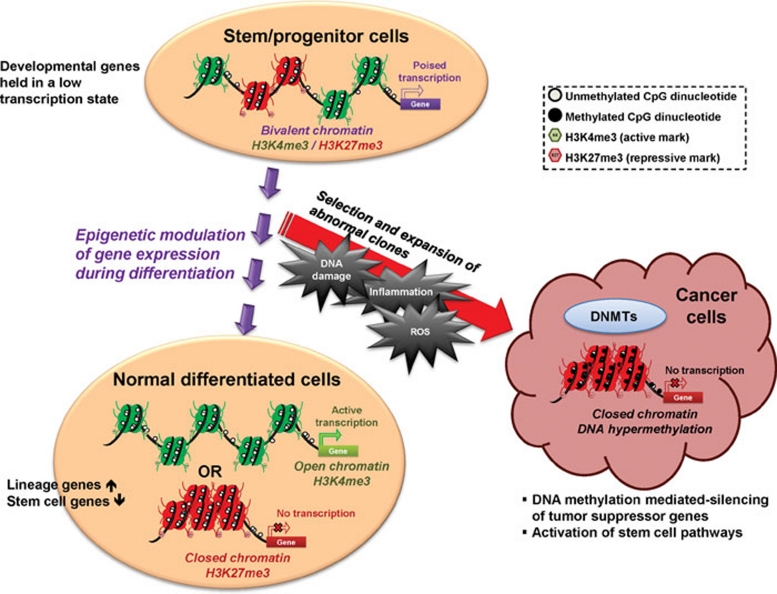
Despite the existence of the above discussed tumor cell subpopulations, the cancer stem cell hypothesis, including its clinical relevance and the precise origins of these cells, continue to generate many controversies. Therefore, much effort has focused on how cancer stem cells are derived and what role epigenetic events play during this process. A key hypothesis is that tumors are initiated through abnormal expansion of clonal stem/progenitor cells, which evolve in the setting of the chronic cell renewal events attendant to high-risk states for cancer, such as chronic inflammation. Here, in concert, genetic and epigenetic changes may help provide the survival advantage which allows these cell subpopulations to withstand the toxic environment of inflammation constituted by accumulating reactive oxygen species, and which then contributes to tumor initiation and progression 154, 155 (Figure 1).
Figure 1.
In normal stem/progenitor cells, the promoter regions of many CpG island-containing developmental genes are marked by both active (trimethylated histone H3 lysine 4; H3K4me3) and repressive marks (trimethylated histone H3 lysine 27; H3K27me3), termed “bivalent chromatin” by Bernstein et al. 91. This chromatin pattern holds these genes in a low, poised transcription state to prevent premature lineage commitment. When the stem/progenitor cells respond to environmental cues and start to differentiate, a shift of the balance between the active and repressive epigenetic marks takes place with corresponding changes in chromatin architecture, leading to the silencing of stemness genes and upregulation of lineage-specific genes. However, repeated environmental stress such as chronic inflammation or accumulating reactive oxygen species (ROS) may promote clonal expansion of cells with genetic or epigenetic abnormalities, which then contribute to tumor initiation and progression. During this course of oncogenesis, the repressive marks in the promoter regions of tumor suppressor genes may recruit DNA methylation machinery to impose abnormal CpG island methylation on these genes leading to permanent gene silencing. At the same time, these epigenetic abnormalities may also contribute to activation of stem cell pathways, such as the Wnt pathway, and bestow self-renewing properties on cancer cells.
Studies on epigenetic alterations both in stem cells and in cancer are providing important insights into the stem/progenitor cell origin of cancer 154. There is compelling evidence, as we outlined earlier, showing that the PRC, which we have discussed as linked to abnormal gene silencing in cancer, target similar sets of CpG island-containing genes in ESC as in cancer 88, 96, 97. A working hypothesis suggested vulnerability for these PcG-marked genes, which do not have promoter DNA methylation in ESC, to gain this change as stem/progenitor-like cells emerge during tumorigenesis 88, 97, 154 (Figure 1). This abnormal methylation may, then, help abnormally lock in activation of stem cell pathways and contribute to the self-renewing ability of cancer cell subpopulations. Many data suggest epigenetic-mediated silencing events, among others, may bestow such properties on cancer cells during oncogenesis 156. This is well demonstrated by abnormal activation of the Wnt signaling pathway at early stage of colon cancer development through epigenetic-mediated silencing of key genes, such as Wnt pathway antagonists, including the secreted frizzled-related gene family (SFRP1, SFRP2, SFRP4, and SFRP5) and SRY-box containing gene 17 (SOX17) 53, 157.
In parallel to the fact that different subpopulations within a tumor possess distinct biological phenotypes, epigenetic-mediated silencing may not be a universal phenomenon in every sub-population. In a glioblastoma cell line model, the methylation status of the promoter of one marker used to identify stem-like cells, CD133 (prominin-1), is heterogeneous between CD133+ and CD133− subpopulations. In most cases, the presence of methylation in the CD133− cells correlated with absence or decreased expression of this surface glycoprotein 158. This suggests aberrant DNA methylation in tumors can be dynamic and can be imposed during the transition between active and repressive state of gene transcription. Interestingly, however, promoter hypermethylation of other tumor suppressor genes, such as SFRP1 and SOX17, which is often considered to drive oncogenesis at early stages, is already present in the stem-like CD133+ subpopulation and preserved in the CD133− subpopulation, possibly consistent with the stem/progenitor cell origin of epigenetic abnormalities in cancer 158.
Clinical applications
The universal occurrence of epigenetic alterations in cancer has broad potential for important clinical applications. Similar to genetic changes, epigenetic alterations are heritable and stable. Therefore, their potentials as molecular markers in cancer patients are being extensively explored for cancer risk evaluation, early detection, prognosis stratification, and treatment response prediction 4, 159. On the other hand, unlike genetic mutations, epigenetic changes, including DNA methylation and histone modifications, are pharmacologically reversible, which makes them an attractive target in cancer therapeutics 160, 161.
Biomarker development
The use of monitoring sequences containing promoter CpG island DNA hypermethylation as a diagnostic tool in cancer is gaining widespread appreciation. The high prevalence and abundance of involved genes in cancer tissues, presence of the abnormality at early stages of oncogenesis, relative stability of the methylation marks, and ease of assaying the change in sites such as serum, sputum, stool, and so on with non- or minimally invasive procedures, make use of hypermethylated sequences an attractive biomarker approach 162.
The fact that CpG island promoter methylation of some genes may precede cancer development rationalizes its use to predict risks for cancer. A series of studies showed detection of a panel of DNA-hypermethylated genes in sputum can identify subjects with high risk for lung cancer development 163, 164. Moreover, methylation markers can be useful for early detection of cancer. For instance, presence of TFPI2 or GATA4 methylation in stool DNA has reasonably high predictive value of colorectal cancer and can be used as a non-invasive screening tool coupled with conventional screening methods 165, 166.
Similarly, accumulating data indicate gene-specific methylation can be a useful clinical marker for patient prognosis stratification. One example is RASSF1A, for which inactivation by promoter methylation is associated with poor prognosis in patients with different types of cancer 167, 168. Likewise, Brock et al. 169 showed that detection of p16 and CDH13 methylation simultaneously in DNA from tumors and mediastinal lymph nodes of patients with stage I non-small cell lung cancer who underwent curative resection is associated with early recurrence. This molecular re-staging strategy may, then, be powerful for predicting which patients with this disease may benefit from more than just surgery alone. These findings suggest that prognosis prediction markers may be used to guide clinical management. Sometimes, a panel of multiple genes may be required for such purposes. In a recent study by Shen et al. 170, a panel of 10 DNA hypermethylation genes was used to predict overall survival in patients with myelodysplastic syndrome. Notably, some attempts have been made to identify novel markers through genome-wide methylation profiling. With this approach, Figueroa et al. 135 were able to discover a panel of 15 genes predictive of overall survival in patients with acute myeloid leukemia.
In addition, DNA methylation patterns may be predictive of patients' response to chemotherapy and correlated with clinical outcome. One such example is for the gene encoding, O6-methylguanine-DNA methyltransferase (MGMT), a DNA repair protein, which reverses the addition of alkyl groups to the guanine base of DNA. Promoter methylation-mediated silencing of MGMT in gliomas is a useful predictor for response to alkylating agents, such as carmustine (BCNU) or temozolomide 171, 172, 173, 174. Similarly, methylation of a mismatch repair gene, hMLH1 in ovarian and colon cancer cell lines confers chemoresistance to many chemotherapeutic agents. Treatment with a DNA demethylating agent, 5-aza-2′-deoxycytidine, can reactivate hMLH1 and reverse the chemoresistance 175, 176. Likewise, epigenetic silencing of apoptotic peptidase activating factor 1 (APAF-1), a proapoptotic gene, confers chemoresistance to melanoma and leukemia cells through mediating resistance to cytochrome c-dependent apoptosis 177, 178. These findings demonstrate the potential for clinical use of DNA methylation markers in tailoring medical care to the need of individual patients.
Notably, assay of histone modifications may also provide a potential molecular strategy to monitor clinical outcome in cancer patients. Several studies have shown that lower global levels of dimethylated histone H3 lysine 4 (H3K4me2) and acetylated histone H3 lysine 18 predict clinical recurrence in prostate, lung, kidney, breast and pancreatic cancer patients 179, 180, 181, 182.
Epigenetic therapeutics in cancer
Targeting reversal of epigenetic alterations in cancer such as DNA methylation and histone modifications has emerged as an attractive strategy in cancer management owing to the reversible nature of these changes 160. Many compounds have been discovered to target proteins that control DNA methylation, histone acetylation, and histone methylation. Some of them are already being used clinically with encouraging effects, which highlights the potential of epigenetic therapy and facilitates the development of novel drugs to target epigenetic mechanisms in cancer. Two clinically used compounds with DNA demethylating activities, azacitidine (Vidaza; Celgene, Summit, NJ, USA) and decitabine (Dacogen; SuperGen, Dublin, CA, USA), have been approved by the FDA for their promising efficacy in hematological malignancies, especially in the pre-leukemic disorder, myelodysplastic syndrome 183, 184, 185, 186, 187. Both compounds are structurally similar to cytosine nucleosides and require incorporation into DNA to exert effects. They were synthesized in the 1960s as anti-metabolites and later found to have DNA demethylating activities through inhibition of DNMTs 188, 189. In earlier years, the high toxicities observed in cancer patients treated with the drugs at high doses limited their widespread uses, especially in solid tumors 190. Nevertheless, in the past decade, the drugs received renewed clinical interests and use of low dose regimens is yielding promising clinical efficacy with relatively mild side effects.
Many efforts have been made toward elucidating the actual mechanisms through which azacitidine and decitabine exert clinical efficacy. In addition to potential re-expression of tumor suppressor genes, which are silenced in association with DNA hypermethylation 4, 191, these drugs have multiple effects including cancer cell differentiation 192, 193, 194, 195, DNA damage 196, 197, formation of covalent adducts between DNMTs and azanucleoside-substituted DNA 198, 199, immune modulatory effects through reactivation of cancer/germ-line antigens 200, inhibition of NFκB anti-apoptotic pathway 201, and so on. Notably, as some data suggested, these drugs might regulate gene expression in a DNA methylation-independent manner through breaking up complex protein interactions by inhibiting and removal of DNMTs from the nucleus 67, 69, 202. It has also been speculated that global effects of the drugs, both DNA methylation-dependent and -independent, may reverse genome-wide epigenetic alterations in cancer through resetting multiple cellular pathways. Besides mechanisms of action, it would be equally important to study mechanisms of drug resistance for translational implications. Indeed, Qin et al. 203 found low deoxycytidine kinase, low nucleoside transporters (i.e., hENT1), and high cytosine deaminase are factors that confer resistance of cancer cell lines to decitabine. Moreover, in light of the cancer stem cell hypothesis and the epigenetic mechanisms involved, understanding whether the drugs have differential effects on different subpopulations may help guide future uses of these drugs in the clinic.
Another class of epigenetic-modifying agents used clinically is histone deacetylase (HDAC) inhibitors 204, 205, 206, 207, 208. In cancer cells, HDAC enzymes, among many other functions, can modulate chromatin configurations and mediate cancer-related gene silencing as components of repressive protein complexes containing DNMTs. Thus, inhibition of HDAC enzymes may reverse abnormal gene silencing in cancer. Many HDAC inhibitors have been shown to have potent anti-tumor effects and entered clinical trials. Two such inhibitors, vorinostat (also known as, suberoylanilide hydroxamic acid; SAHA) and romidepsin (also known as depsipeptide or FK228) have been approved by the FDA for treating cutaneous T-cell lymphoma 206, 207, 208. In addition to anti-tumor effects, other potential uses of HDAC inhibitors and other epigenetic-modifying agents in clinical oncology are being explored. A study by Sharma et al. 209 indicates drug resistance may derive via epigenetic mechanisms and can be reversed by various HDAC inhibitors. This suggests a novel use of epigenetic therapy in overcoming tolerance or resistance to standard chemotherapy in the clinical setting of cancer management, where drug resistance has always been a major concern.
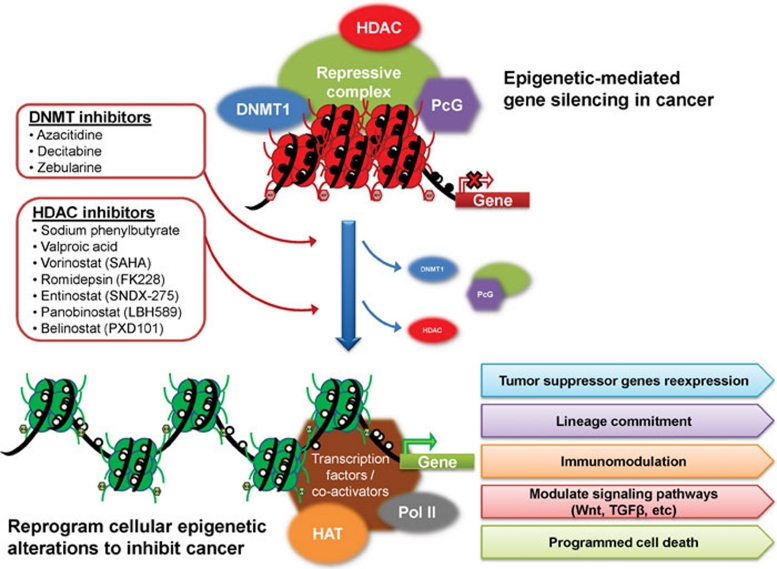
Given that DNA methylation-mediated aberrant gene silencing in cancer involves transcriptional repressive complexes containing both DNMTs and HDAC, targeting both enzymes with combination therapy of a DNMT inhibitor and an HDAC inhibitor appears to be an inviting approach in cancer management (Figure 2). Indeed, sequential application of an HDAC inhibitor following a DNA demethylating agent has shown synergistic effects in gene re-expression in vitro and enhanced anti-tumor effects clinically 210, 211. Moreover, epigenetic-modifying agents may couple with other standard chemotherapeutic agents to boost clinical efficacy with lower doses of either drug. Emerging data indicate azacitidine and decitabine may modify multiple cellular pathways through gene reactivation, and sensitize cancer cells to other drugs that target similar pathways.
Figure 2.
DNA methylation-mediated aberrant gene silencing in cancer involves transcriptional repressive complexes in the gene promoter region and interactions between DNA methylation machinery, chromatin modifiers (such as histone deacetylase, HDAC) and polycomb (PcG) proteins. Pharmacological inhibition of individual components in the repressive complex with DNMT inhibitors and HDAC inhibitors, either alone or in combination, may result in DNA demethylation and complex disintegration leading to reactivation of critical genes and reversal of genome-wide epigenetic alterations in cancer through resetting multiple cellular processes, including lineage commitment, immunomodulation, major cell signaling pathways, programmed cell death, and others. HAT: histone acetylase. Pol II: RNA polymerase II.
Despite the promising clinical efficacy of azacitidine or decitabine at low doses in hematological malignancies, when given alone 183, 184, 187, 212 or in combination with other drugs 211, 213, whether the drugs used at similar dosing schedules exert comparable anti-cancer activities on solid tumors is actively under investigation. In an ongoing lung cancer clinical trial at our institution, a low dose regimen for azacitidine and an HDAC inhibitor, entinostat (also known as SNDX-275 or MS-275) achieves robust and durable response in some patients with metastatic disease who failed several lines of previous chemotherapy. Clinical trials in other tumor types including breast and colon cancers are underway. Importantly, in addition to clinical efficacy, several areas warrant extensive research in the context of clinical trials to maximize patient benefits, such as optimization of dosing schedule and sequences, and searching for ways to identify those patients who would potentially benefit from epigenetic therapy.
Future directions
It is apparent that, over the past 20 years, our view of tumor biology has changed with a major addition being awareness that epigenetic abnormalities complement genetic alterations to drive all stages of cancer evolution. While the research in cancer epigenetics has already contributed to our understanding of fundamental steps in cancer formation, to our knowledge about control of normal and abnormal gene regulation by the chromatin landscape, and to the growing potential for use of information gained for translational purposes, major challenges remain. We are far from having full understanding of the molecular mechanisms that are responsible for the initiation and maintenance of the epigenetic abnormalities that help drive tumorigenesis. We must, then for example, pursue the possibilities for molecular progression of abnormal gene silencing during tumor progression as contributed by PcG mediation of transcriptional repression. What drives this initiation and progression and how do cancer risk states play a role? What are the targeting mechanisms for PcG and its inter-actors in this progression? How precisely do they tie together the concept of cancer stem-like cells to events for derivation and maintenance of stem and progenitor cells in normal developmental and adult cell renewal settings? In this regard, especially, the molecular ties between PcG and targeting of DNA methylation in normal and neoplastic settings need much further clarification. Most broadly speaking, the full epigenomes of all cancer types, and their subpopulations, need to be mapped and compared accurately with the normal cell compartments from which they arise. This effort must take into account the interplay between genetic abnormalities in cancer and how these depend upon the epigenetic landscape for their oncogenic potential. Also, the precise biological ramifications of the growing number of recognized cancer mutations in genes encoding for proteins involved in regulation of chromatin and DNA methylation must be delineated 113, 114, 120. Finally, we have much work ahead to exploit all of the above knowledge for translational purposes. We must continue the development of epigenetic biomarkers, which can enhance our capabilities to assess cancer risk, to make earlier cancer diagnoses, and to chart cancer prognosis and predict therapeutic responsiveness of different cancer subtypes. The potential for reversing epigenetic abnormalities for the purposes of cancer prevention and treatment is real but is probably in its very early stages in terms of delineating the best molecular targets, and developing or learning to use the drugs and agents that will be required. The future is a bright one and should hold bountiful rewards for both basic and translational cancer research.
References
- Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Chang SC, Tucker T, Thorogood NP, Brown CJ. Mechanisms of X-chromosome inactivation. Front Biosci. 2006;11:852–866. doi: 10.2741/1842. [DOI] [PubMed] [Google Scholar]
- Kacem S, Feil R. Chromatin mechanisms in genomic imprinting. Mamm Genome. 2009;20:544–556. doi: 10.1007/s00335-009-9223-4. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Kim YI, Giuliano A, Hatch KD, et al. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74:893–899. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Lin CH, Hsieh SY, Sheen IS, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001;61:4238–4243. [PubMed] [Google Scholar]
- Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- Cui H, Onyango P, Brandenburg S, et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- Zardo G, Tiirikainen MI, Hong C, et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet. 2002;32:453–458. doi: 10.1038/ng1007. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Merlo A, Mao L, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- Herman JG, Civin CI, Issa JP, et al. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- Uchida T, Kinoshita T, Hotta T, Murate T. High-risk myelodysplastic syndromes and hypermethylation of the p15Ink4B gene. Leuk Lymphoma. 1998;32:9–18. doi: 10.3109/10428199809059242. [DOI] [PubMed] [Google Scholar]
- Quesnel B, Fenaux P. P15INK4b gene methylation and myelodysplastic syndromes. Leuk Lymphoma. 1999;35:437–443. doi: 10.1080/10428199909169608. [DOI] [PubMed] [Google Scholar]
- Knudson AG. JrMutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Catasus L, Matias-Guiu X, et al. hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol. 1999;155:1767–1772. doi: 10.1016/S0002-9440(10)65492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Nuovo GJ, Zervos EE, et al. Age-related hypermethylation of the 5′ region of MLH1 in normal colonic mucosa is associated with microsatellite-unstable colorectal cancer development. Cancer Res. 2001;61:6991–6995. [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Stresemann C, Kuner R, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Bandres E, Agirre X, Bitarte N, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Tsang WP, Ng SS, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101:699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Herman JG, Corn PG, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Burbee DG, Forgacs E, Zochbauer-Muller S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP.Comprehensive high-throughput arrays for relative methylation (CHARM) Curr Protoc Hum GenetChapter 20:Unit 20.1.1–19. [DOI] [PMC free article] [PubMed]
- Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, et al. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Thiesen J, Stratling WH. Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 2000;28:2201–2206. doi: 10.1093/nar/28.10.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaludov NK, Wolffe AP. MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res. 2000;28:1921–1928. doi: 10.1093/nar/28.9.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Edwards JR, O'Donnell AH, Rollins RA, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ., 3rd, , Reinberg D. Histone H3 Lys 4 methylation: caught in a bind. Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, et al. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Van Neste L, Cope L, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Mohammad HP, Cai Y, McGarvey KM, et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69:6322–6330. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Weikert S, Christoph F, Kollermann J, et al. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int J Mol Med. 2005;16:349–353. [PubMed] [Google Scholar]
- Matsukawa Y, Semba S, Kato H, et al. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–6280. doi: 10.1038/sj.onc.1208771. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Martinez-Leal JF, Rizzo S, et al. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- Scott CL, Gil J, Hernando E, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci USA. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Biegel JA, Fogelgren B, Zhou JY, et al. Mutations of the INI1 rhabdoid tumor suppressor gene in medulloblastomas and primitive neuroectodermal tumors of the central nervous system. Clin Cancer Res. 2000;6:2759–2763. [PubMed] [Google Scholar]
- Grand F, Kulkarni S, Chase A, et al. Frequent deletion of hSNF5/INI1, a component of the SWI/SNF complex, in chronic myeloid leukemia. Cancer Res. 1999;59:3870–3874. [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih Ie M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Frigola J, Song J, Stirzaker C, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- Easwaran H, Van Neste L, Sen S, et al. Aberrant silencing of cancer related genes by CpG hypermethylation is independent of their spatial organization in the nucleus. Cancer Res. 2010;70:8015–8024. doi: 10.1158/0008-5472.CAN-10-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Kovalchuk O, Pogribny IP. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther. 2006;5:65–70. doi: 10.4161/cbt.5.1.2288. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Ross SA, Tryndyak VP, et al. Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4-20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis. 2006;27:1180–1186. doi: 10.1093/carcin/bgi364. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Suetsugu A, Nagaki M, Aoki H, et al. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;351:820–824. doi: 10.1016/j.bbrc.2006.10.128. [DOI] [PubMed] [Google Scholar]
- Yin S, Li J, Hu C, et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer. 2007;120:1444–1450. doi: 10.1002/ijc.22476. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui W, Wang Q, Barber JP, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68:190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation. Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction. Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Zhang W, Glockner SC, Guo M, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JM, Tsai HC, Glockner SC, et al. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res. 2008;68:8094–8103. doi: 10.1158/0008-5472.CAN-07-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol. 2008;68:1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- Glockner SC, Dhir M, Yi JM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellebrekers DM, Lentjes MH, van den Bosch SM, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res. 2009;15:3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- Tomizawa Y, Kohno T, Kondo H, et al. Clinicopathological significance of epigenetic inactivation of RASSF1A at 3p21.3 in stage I lung adenocarcinoma. Clin Cancer Res. 2002;8:2362–2368. [PubMed] [Google Scholar]
- Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Dis Markers. 2007;23:73–87. doi: 10.1155/2007/291538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- Shen L, Kantarjian H, Guo Y, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28:605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60:6039–6044. [PubMed] [Google Scholar]
- Strathdee G, MacKean MJ, Illand M, Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18:2335–2341. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Jia L, Srinivasula SM, Liu FT, et al. Apaf-1 protein deficiency confers resistance to cytochrome c-dependent apoptosis in human leukemic cells. Blood. 2001;98:414–421. doi: 10.1182/blood.v98.2.414. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuyakorn A, Paulus R, Farrell J, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsheikh SE, Green AR, Rakha EA, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- Issa JP, Garcia-Manero G, Giles FJ, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- Silverman LR, Demakos EP, Peterson BL, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- Kaminskas E, Farrell A, Abraham S, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- Cashen AF, Schiller GJ, O'Donnell MR, DiPersio JF. Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J Clin Oncol. 2010;28:556–561. doi: 10.1200/JCO.2009.23.9178. [DOI] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Hemimethylated duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic Acids Res. 1981;9:2933–2947. doi: 10.1093/nar/9.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio A, Weber JS. Review of the clinical experience with 5-azacytidine and 5-aza-2′-deoxycytidine in solid tumors. Curr Opin Investig Drugs. 2002;3:627–633. [PubMed] [Google Scholar]
- Oki Y, Aoki E, Issa JP. Decitabine--bedside to bench. Crit Rev Oncol Hematol. 2007;61:140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Biol Chem. 1982;257:2041–2048. [PubMed] [Google Scholar]
- Christman JK, Mendelsohn N, Herzog D, Schneiderman N. Effect of 5-azacytidine on differentiation and DNA methylation in human promyelocytic leukemia cells (HL-60) Cancer Res. 1983;43:763–769. [PubMed] [Google Scholar]
- Pinto A, Attadia V, Fusco A, et al. 5-Aza-2′-deoxycytidine induces terminal differentiation of leukemic blasts from patients with acute myeloid leukemias. Blood. 1984;64:922–929. [PubMed] [Google Scholar]
- Attadia V. Effects of 5-aza-2′-deoxycytidine on differentiation and oncogene expression in the human monoblastic leukemia cell line U-937. Leukemia. 1993;7 Suppl 1:9–16. [PubMed] [Google Scholar]
- Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhao Y, Li L, et al. An ATM- and Rad3-related (ATR) signaling pathway and a phosphorylation-acetylation cascade are involved in activation of p53/p21Waf1/Cip1 in response to 5-aza-2′-deoxycytidine treatment. J Biol Chem. 2008;283:2564–2574. doi: 10.1074/jbc.M702454200. [DOI] [PubMed] [Google Scholar]
- Ferguson AT, Vertino PM, Spitzner JR, et al. Role of estrogen receptor gene demethylation and DNA methyltransferase.DNA adduct formation in 5-aza-2′deoxycytidine-induced cytotoxicity in human breast cancer cells. J Biol Chem. 1997;272:32260–32266. doi: 10.1074/jbc.272.51.32260. [DOI] [PubMed] [Google Scholar]
- Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpf AR. A potential role for epigenetic modulatory drugs in the enhancement of cancer/germ-line antigen vaccine efficacy. Epigenetics. 2006;1:116–120. doi: 10.4161/epi.1.3.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre C, Grosjean J, Tailler M, et al. A novel effect of DNA methyltransferase and histone deacetylase inhibitors: NFkappaB inhibition in malignant myeloblasts. Cell Cycle. 2008;7:2139–2145. doi: 10.4161/cc.7.14.6268. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- Qin T, Jelinek J, Si J, Shu J, Issa JP. Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer cell lines. Blood. 2009;113:659–667. doi: 10.1182/blood-2008-02-140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore SD, Weng LJ, Zhai S, et al. Impact of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin Cancer Res. 2001;7:2330–2339. [PubMed] [Google Scholar]
- Stimson L, Wood V, Khan O, Fotheringham S, La Thangue NB. HDAC inhibitor-based therapies and haematological malignancy. Ann Oncol. 2009;20:1293–1302. doi: 10.1093/annonc/mdn792. [DOI] [PubMed] [Google Scholar]
- Whittaker SJ, Demierre MF, Kim EJ, et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol. 2010;28:4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- Piekarz RL, Frye R, Turner M, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvic M, Zhang C. Clinical and laboratory experience of vorinostat (suberoylanilide hydroxamic acid) in the treatment of cutaneous T-cell lymphoma. Br J Cancer. 2006;95:S13–S19. [Google Scholar]
- Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- Blum W, Klisovic RB, Hackanson B, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25:3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]