Abstract
Chromatin is not an inert structure, but rather an instructive DNA scaffold that can respond to external cues to regulate the many uses of DNA. A principle component of chromatin that plays a key role in this regulation is the modification of histones. There is an ever-growing list of these modifications and the complexity of their action is only just beginning to be understood. However, it is clear that histone modifications play fundamental roles in most biological processes that are involved in the manipulation and expression of DNA. Here, we describe the known histone modifications, define where they are found genomically and discuss some of their functional consequences, concentrating mostly on transcription where the majority of characterisation has taken place.
Keywords: histone, modifications, chromatin
Introduction
Ever since Vincent Allfrey's pioneering studies in the early 1960s, we have known that histones are post-translationally modified 1. We now know that there are a large number of different histone post-translational modifications (PTMs). An insight into how these modifications could affect chromatin structure came from solving the high-resolution X-ray structure of the nucleosome in 1997 2. The structure indicates that highly basic histone amino (N)-terminal tails can protrude from their own nucleosome and make contact with adjacent nucleosomes. It seemed likely at the time that modification of these tails would affect inter-nucleosomal interactions and thus affect the overall chromatin structure. We now know that this is indeed the case. Modifications not only regulate chromatin structure by merely being there, but they also recruit remodelling enzymes that utilize the energy derived from the hydrolysis of ATP to reposition nucleosomes. The recruitment of proteins and complexes with specific enzymatic activities is now an accepted dogma of how modifications mediate their function. As we will describe below, in this way modifications can influence transcription, but since chromatin is ubiquitous, modifications also affect many other DNA processes such as repair, replication and recombination.
Histone acetylation
Allfrey et al. 1 first reported histone acetylation in 1964. Since then, it has been shown that the acetylation of lysines is highly dynamic and regulated by the opposing action of two families of enzymes, histone acetyltransferases (HATs) and histone deacetylases (HDACs; for review, see reference 3). The HATs utilize acetyl CoA as cofactor and catalyse the transfer of an acetyl group to the ε-amino group of lysine side chains. In doing so, they neutralize the lysine's positive charge and this action has the potential to weaken the interactions between histones and DNA (see below). There are two major classes of HATs: type-A and type-B. The type-B HATs are predominantly cytoplasmic, acetylating free histones but not those already deposited into chromatin. This class of HATs is highly conserved and all type-B HATs share sequence homology with scHat1, the founding member of this type of HAT. Type-B HATs acetylate newly synthesized histone H4 at K5 and K12 (as well as certain sites within H3), and this pattern of acetylation is important for deposition of the histones, after which the marks are removed 4.
The type-A HATs are a more diverse family of enzymes than the type-Bs. Nevertheless, they can be classified into at least three separate groups depending on amino-acid sequence homology and conformational structure: GNAT, MYST and CBP/p300 families 5. Broadly speaking, each of these enzymes modifies multiple sites within the histone N-terminal tails. Indeed, their ability to neutralize positive charges, thereby disrupting the stabilizing influence of electrostatic interactions, correlates well with this class of enzyme functioning in numerous transcriptional coactivators 6. However, it is not just the histone tails that are involved in this regulation, but there are additional sites of acetylation present within the globular histone core, such as H3K56 that is acetylated in humans by hGCN5 7. The H3K56 side chain points towards the DNA major groove, suggesting that acetylation would affect histone/DNA interaction, a situation reminiscent of the proposed effects of acetylating the histone N-terminal tail lysines. Interestingly, knockdown of the p300 HAT has also been shown to be associated with the loss of H3K56ac 8, suggesting that p300 may also target this site. However, unlike GCN5 knockdown, p300 knockdown increases DNA damage, which may indirectly affect H3K56ac levels 7.
In common with many histone-modifying enzymes, the type-A HATs are often found associated in large multiprotein complexes 6. The component proteins within these complexes play important roles in controlling enzyme recruitment, activity and substrate specificity. For instance, purified scGCN5 acetylates free histones but not those present within a nucleosome. In contrast, when scGCN5 is present within the so-called SAGA complex, it efficiently acetylates nucleosomal histones 9.
HDAC enzymes oppose the effects of HATs and reverse lysine acetylation, an action that restores the positive charge of the lysine. This potentially stabilizes the local chromatin architecture and is consistent with HDACs being predominantly transcriptional repressors. There are four classes of HDAC 6: Classes I and II contain enzymes that are most closely related to yeast scRpd3 and scHda1, respectively, class IV has only a single member, HDAC11, while class III (referred to as sirtuins) are homologous to yeast scSir2. This latter class, in contrast to the other three classes, requires a specific cofactor for its activity, NAD+.
In general, HDACs have relatively low substrate specificity by themselves, a single enzyme being capable of deacetylating multiple sites within histones. The issue of enzyme recruitment and specificity is further complicated by the fact that the enzymes are typically present in multiple distinct complexes, often with other HDAC family members. For instance, HDAC1 is found together with HDAC2 within the NuRD, Sin3a and Co-REST complexes 10. Thus, it is difficult to determine which activity (specific HDAC and/or complex) is responsible for a specific effect. Nevertheless, in certain cases, it is possible to at least determine which enzyme is required for a given process. For example, it has been shown that HDAC1, but not HDAC2, controls embryonic stem cell differentiation 11.
Histone phosphorylation
Like histone acetylation, the phosphorylation of histones is highly dynamic. It takes place on serines, threonines and tyrosines, predominantly, but not exclusively, in the N-terminal histone tails 3. The levels of the modification are controlled by kinases and phosphatases that add and remove the modification, respectively 12.
All of the identified histone kinases transfer a phosphate group from ATP to the hydroxyl group of the target amino-acid side chain. In doing so, the modification adds significant negative charge to the histone that undoubtedly influences the chromatin structure. For the majority of kinases, however, it is unclear how the enzyme is accurately recruited to its site of action on chromatin. In a few cases, exemplified by the mammalian MAPK1 enzyme, the kinase possesses an intrinsic DNA-binding domain with which it is tethered to the DNA 13. This may be sufficient for specific recruitment, similar to bona fide DNA-binding transcription factors. Alternatively, its recruitment may require association with a chromatin-bound factor before it directly contacts DNA to stabilize the overall interaction.
The majority of histone phosphorylation sites lie within the N-terminal tails. However, sites within the core regions do exist. One such example is phosphorylation of H3Y41, which is deposited by the non-receptor tyrosine kinase JAK2 (see below) 14.
Less is known regarding the roles of histone phosphatases. Certainly, given the extremely rapid turnover of specific histone phosphorylations, there must be high phosphatase activity within the nucleus. We do know, e.g., that the PP1 phosphatase works antagonistically to Aurora B, the kinase that lays down genome-wide H3S10ph and H3S28ph at mitosis 15, 16.
Histone methylation
Histone methylation mainly occurs on the side chains of lysines and arginines. Unlike acetylation and phosphorylation, however, histone methylation does not alter the charge of the histone protein. Furthermore, there is an added level of complexity to bear in mind when considering this modification; lysines may be mono-, di- or tri-methylated, whereas arginines may be mono-, symmetrically or asymmetrically di-methylated (for reviews see references 3, 17, 18, 19).
Lysine methylation
The first histone lysine methyltransferase (HKMT) to be identified was SUV39H1 that targets H3K9 20. Numerous HKMTs have since been identified, the vast majority of which methylate lysines within the N-terminal tails. Strikingly, all of the HKMTs that methylate N-terminal lysines contain a so-called SET domain that harbours the enzymatic activity. However, an exception is the Dot1 enzyme that methylates H3K79 within the histone globular core and does not contain a SET domain. Why this enzyme is structurally different than all of the others is not clear, but perhaps this reflects the relative inaccessibility of its substrate H3K79. In any case, all HKMTs catalyse the transfer of a methyl group from S-adenosylmethionine (SAM) to a lysine's ε-amino group.
HKMTs tend to be relatively specific enzymes. For instance, Neurospora crassa DIM5 specifically methylates H3K9 whereas SET7/9 targets H3K4. Furthermore, HKMT enzymes also modify the appropriate lysine to a specific degree (i.e., mono-, di- and/or tri-methyl state). Maintaining the same examples, DIM5 can tri-methylate H3K9 21 but SET7/9 can only mono-methylate H3K4 22. These specific reaction products can be generated using only the purified enzymes; so the ability to discriminate between different histone lysines and between different methylated states is an intrinsic property of the enzyme. It turns out from X-ray crystallographic studies that there is a key residue within the enzyme's catalytic domain that determines whether the enzyme activity proceeds past the mono-methyl product. In DIM5, there is a phenylalanine (F281) within the enzyme's lysine-binding pocket that can accommodate all the methylated forms of the lysine, thereby allowing the enzyme to generate the tri-methylated product 23. In contrast, SET7/9 has a tyrosine (Y305) in the corresponding position such that it can only accommodate the mono-methyl product 22. Elegant mutagenesis studies have shown that mutagenesis of DIM5 F281 to Y converts the enzyme to a mono-methyltransferase, whereas the reciprocal mutation in SET7/9 (Y305 to F) creates an enzyme capable of tri-methylating its substrate 23. More generally speaking, it seems that the aromatic determinant (Y or F) is a mechanism widely employed by SET domain-containing HKMTs to control the degree of methylation 24, 25.
Arginine methylation
There are two classes of arginine methyltransferase, the type-I and type-II enzymes. The type-I enzymes generate Rme1 and Rme2as, whereas the type-II enzymes generate Rme1 and Rme2s. Together, the two types of arginine methyltransferases form a relatively large protein family (11 members), the members of which are referred to as PRMTs. All of these enzymes transfer a methyl group from SAM to the ω-guanidino group of arginine within a variety of substrates. With respect to histone arginine methylation, the most relevant enzymes are PRMT1, 4, 5 and 6 (reviewed in 18, 26).
Methyltransferases, for both arginine and lysine, have a distinctive extended catalytic active site that distinguishes this broad group of methyltransferases from other SAM-dependent enzymes 27. Interestingly, the SAM-binding pocket is on one face of the enzyme, whereas the peptidyl acceptor channel is on the opposite face. This indicates that a molecule of SAM and the histone substrate come together from opposing sides of the enzyme 27. Indeed, this way of entering the enzyme's active site may provide an opportunity to design selective drugs that are able to distinguish between histone arginine/lysine methyltransferases and other methyltransferases such as DNMTs.
Histone demethylases
For many years, histone methylation was considered a stable, static modification. Nevertheless, in 2002, a number of different reactions/pathways were suggested as potential demethylation mechanisms for both lysine and arginine 28, which were subsequently verified experimentally.
Initially, the conversion of arginine to citrulline via a deimination reaction was discovered as a way of reversing arginine methylation 29, 30. Although this pathway is not a direct reversal of methylation (see deimination below), this mechanism reversed the dogma that methylation was irreversible. More recently, a reaction has been reported which reverses arginine methylation. The jumonji protein JMJD6 was shown to be capable of performing a demethylation reaction on histones H3R2 and H4R3 31. However, these findings have yet to be recapitulated by other independent researchers.
In 2004, the first lysine demethylase was identified. It was found to utilize FAD as co-factor, and it was termed as lysine-specific demethylase 1 (LSD1) 32. The demethylation reaction requires a protonated nitrogen and it is therefore only compatible with mono- and di-methylated lysine substrates. In vitro, purified LSD1 catalyses the removal of methyl groups from H3K4me1/2, but it cannot demethylate the same site when presented within a nucleosomal context. However, when LSD1 is complexed with the Co-REST repressor complex, it can demethylate nucleosomal histones. Thus, complex members confer nucleosomal recognition to LSD1. Furthermore, the precise complex association determines which lysine is to be demethylated by LSD1. As already mentioned, LSD1 in the context of Co-REST demethylates H3K4me1/2, but when LSD1 is complexed with the androgen receptor, it demethylates H3K9. This has the effect of switching the activity of LSD1 from a repressor function to that of a coactivator (reviewed in 33; see below).
In 2006, another class of lysine demethylase was discovered 34. Importantly, certain enzymes in this class were capable of demethylating tri-methylated lysines 35. They employ a distinct catalytic mechanism from that used by LSD1, using Fe(II) and α-ketoglutarate as co-factors, and a radical attack mechanism. The first enzyme identified as a tri-methyl lysine demethylase was JMJD2 that demethylates H3K9me3 and H3K36me3 35. The enzymatic activity of JMJD2 resides within a JmjC jumonji domain. Many histone lysine demethylases are now known and, except for LSD1, they all possess a catalytic jumonji domain 36. As with the lysine methyltransferases, the demethylases possess a high level of substrate specificity with respect to their target lysine. They are also sensitive to the degree of lysine methylation; for instance, some of the enzymes are only capable of demethylating mono- and di-methyl substrates, whereas others can demethylate all three states of the methylated lysine.
Other modifications
Deimination
This reaction involves the conversion of an arginine to a citrulline. In mammalian cells, this reaction on histones is catalysed by the peptidyl deiminase PADI4, which converts peptidyl arginines to citrulline 29, 30. One obvious effect of this reaction is that it effectively neutralizes the positive charge of the arginine since citrulline is neutral. There is also evidence that PADI4 converts mono-methyl arginine to citrulline, thereby effectively functioning as an arginine demethylase 29, 30. However, unlike a 'true' demethylase, the PADI4 reaction does not regenerate an unmodified arginine.
β-N-acetylglucosamine
Many non-histone proteins are regulated via modification of their serine and threonine side chains with single β-N-acetylglucosamine (O-GlcNAc) sugar residues. Recently, histones were added to the long list of O-GlcNAc-modified proteins 37. Interestingly, in mammalian cells, there appears to be only a single enzyme, O-GlcNAc transferase, which catalyses the transfer of the sugar from the donor substrate, UDP-GlcNAc, to the target protein. Like most of the other histone PTMs, O-GlcNAc modification appears to be highly dynamic with high turnover rates and, as with the forward reaction, there appears to be only a single enzyme capable of removing the sugar, β-N-acetylglucosaminidase (O-GlcNAcase). So far, histones H2A, H2B and H4 have been shown to be modified by O-GlcNAc 37.
ADP ribosylation
Histones are known to be mono- and poly-ADP ribosylated on glutamate and arginine residues, but relatively little is known concerning the function of this modification 38. What we do know is that once again the modification is reversible. For example, poly-ADP-ribosylation of histones is performed by the poly-ADP-ribose polymerase (PARP) family of enzymes and reversed by the poly-ADP-ribose-glycohydrolase family of enzymes. These enzymes function together to control the levels of poly-ADP ribosylated histones that have been correlated with a relatively relaxed chromatin state 38. Presumably, this is a consequence, at least in part, of the negative charge that the modification confers to the histone. In addition, though, it has been reported that the activation of PARP-1 leads to elevated levels of core histone acetylation 39. Moreover, PARP-1-mediated ribosylation of the H3K4me3 demethylase KDM5B inhibits the demethylase and excludes it from chromatin, while simultaneously excluding H1, thereby making target promoters more accessible 40.
Histone mono-ADP-ribosylation is performed by the mono-ADP-ribosyltransferases and has been detected on all four core histones, as well as on the linker histone H1. Notably, these modifications significantly increase upon DNA damage implicating the pathway in the DNA damage response 38.
Ubiquitylation and sumoylation
All of the previously described histone modifications result in relatively small molecular changes to amino-acid side chains. In contrast, ubiquitylation results in a much larger covalent modification. Ubiquitin itself is a 76-amino acid polypeptide that is attached to histone lysines via the sequential action of three enzymes, E1-activating, E2-conjugating and E3-ligating enzymes 41. The enzyme complexes determine both substrate specificity (i.e., which lysine is targeted) as well as the degree of ubiquitylation (i.e., either mono- or poly-ubiquitylated). For histones, mono-ubiquitylation seems most relevant although the exact modification sites remain largely elusive. However, two well-characterised sites lie within H2A and H2B. H2AK119ub1 is involved in gene silencing 42, whereas H2BK123ub1 plays an important role in transcriptional initiation and elongation 43, 44 (see below). Even though ubiquitylation is such a large modification, it is still a highly dynamic one. The modification is removed via the action of isopeptidases called de-ubiquitin enzyme and this activity is important for both gene activity and silencing [3 and references therein].
Sumoylation is a modification related to ubiquitylation 45, and involves the covalent attachment of small ubiquitin-like modifier molecules to histone lysines via the action of E1, E2 and E3 enzymes. Sumoylation has been detected on all four core histones and seems to function by antagonizing acetylation and ubiquitylation that might otherwise occur on the same lysine side chain 46, 47. Consequently, it has mainly been associated with repressive functions, but more work is clearly needed to elucidate the molecular mechanism(s) through which sumoylation exerts its effect on chromatin.
Histone tail clipping
Perhaps the most radical way to remove histone modifications is to remove the histone N-terminal tail in which they reside, a process referred to as tail clipping. It was first identified in Tetrahymena in 1980 48, where the first six amino acids of H3 are removed. However, it is now apparent that this type of activity also exists in yeast and mammals (mouse) where the first 21 amino acids of H3 are removed 49, 50. In yeast, the proteolytic enzyme remains unknown, but the clipping process has been shown to be involved in regulating transcription 49. The mouse enzyme was identified as Cathepsin L, which cleaves the N-terminus of H3 during ES cell differentiation 50.
Histone proline isomerization
The dihedral angle of a peptidyl proline's peptide bond naturally interconverts between the cis and trans conformations, the states differing by 180°. Proline isomerases facilitate this interconversion, which has the potential to stably affect peptide configuration. One proline isomerase shown to act on histones is the yeast enzyme scFpr4, which isomerizes H3P38 51. This activity is linked to methylation of H3K36, presumably by scFpr4 affecting the recognition of K36 by the scSet2 methyltransferase and the scJMJD2 demethylase though the exact mechanism remains unclear 51, 52. Nevertheless, this example highlights the fact that proline isomerization is an important modification of the histone tail. It is, however, not a true covalent modification since the enzyme merely 'flips' the peptide bond by 180°, thereby generating chemical isomers rather than covalently modified products.
Mode of action of histone modifications
Histone modifications exert their effects via two main mechanisms. The first involves the modification(s) directly influencing the overall structure of chromatin, either over short or long distances. The second involves the modification regulating (either positively or negatively) the binding of effector molecules. Our review has a transcriptional focus, simply reflecting the fact that most studies involving histone modifications have also had this focus. However, histone modifications are just as relevant in the regulation of other DNA processes such as repair, replication and recombination. Indeed, the principles described below are pertinent to any biological process involving DNA transactions.
Direct structural perturbation
Histone acetylation and phosphorylation effectively reduce the positive charge of histones, and this has the potential to disrupt electrostatic interactions between histones and DNA. This presumably leads to a less compact chromatin structure, thereby facilitating DNA access by protein machineries such as those involved in transcription. Notably, acetylation occurs on numerous histone tail lysines, including H3K9, H3K14, H3K18, H4K5, H4K8 and H4K12 53. This high number of potential sites provides an indication that in hyper-acetylated regions of the genome, the charge on the histone tails can be effectively neutralized, which would have profound effects on the chromatin structure. Evidence for this can be found at the β-globin locus where the genes reside within a hyper-acetylated and transcriptionally competent chromatin environment that displays DNase sensitivity, and therefore general accessibility 54. Multiple histone acetylations are also enriched at enhancer elements and particularly in gene promoters, where they presumably again facilitate the transcription factor access 55. However, multiple histone acetylations are not an absolute pre-requisite for inducing structural change – histones specifically acetylated at H4K16 have a significant negative effect on the formation of the 30 nm fibre, at least in vitro56.
Histone phosphorylation tends to be very site-specific and there are far fewer sites compared with acetylated sites. As with H4K16ac, these single-site modifications can be associated with gross structural changes within chromatin. For instance, phosphorylation of H3S10 during mitosis occurs genome-wide and is associated with chromatin becoming more condensed 57. This seems somewhat counterintuitive since the phosphate group adds negative charge to the histone tail that is in close proximity to the negatively charged DNA backbone. But it may be that displacement of heterochromatin protein 1 (HP1) from heterochromatin during metaphase by uniformly high levels of H3S10ph 58, 59 (see below) is required to promote the detachment of chromosomes from the interphase scaffolding. This would facilitate chromosomal remodeling that is essential for its attachment to the mitotic spindle.
Ubiquitylation adds an extremely large molecule to a histone. It seems highly likely that this will induce a change in the overall conformation of the nucleosome, which in turn will affect intra-nucleosomal interactions and/or interactions with other chromatin-bound complexes. Histone tail clipping, which results in the loss of the first 21 amino acids of H3 will have similar effects. In contrast, neutral modifications such as histone methylation are unlikely to directly perturb chromatin structure since these modifications are small and do not alter the charge of histones.
Regulating the binding of chromatin factors
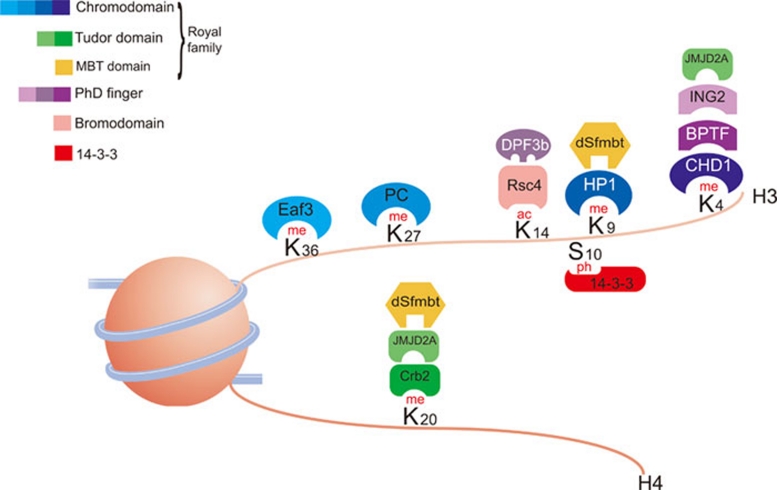
Numerous chromatin-associated factors have been shown to specifically interact with modified histones via many distinct domains (Figure 1) 3. There is an ever-increasing number of such proteins following the development and use of new proteomic approaches 60, 61. These large data sets show that there are multivalent proteins and complexes that have specific domains within them that allow the simultaneous recognition of several modifications and other nucleosomal features.
Figure 1.
Domains binding modified histones. Examples of proteins with domains that specifically bind to modified histones as shown (updated from reference 53).
Notably, there are more distinct domain types recognizing lysine methylation than any other modification, perhaps reflecting the modification's relative importance (Figure 1). These include PHD fingers and the so-called Tudor 'royal' family of domains, comprising chromodomains, Tudor, PWWP and MBT domains 62, 63, 64. Within this group of methyl-lysine binders, numerous domains can recognize the same modified histone lysine. For instance, H3K4me3 – a mark associated with active transcription – is recognized by a PHD finger within the ING family of proteins (ING1-5) (reviewed in 62). The ING proteins in turn recruit additional chromatin modifiers such as HATs and HDACs. For example, ING2 tethers the repressive mSin3a-HDAC1 HDAC complex to active proliferation-specific genes following DNA damage 65. Tri-methylated H3K4 is also bound by the tandem chromodomains within CHD1, an ATP-dependent remodelling enzyme capable of repositioning nucleosomes 66, and by the tandem Tudor domains within JMJD2A, a histone demethylase 67. In these cases, H3K4me3 directly recruits the chromatin-modifying enzyme.
A further example of specific methylated lysine binding is provided by the HP1 recognition of H3K9me3 – a mark associated with repressive heterochromatin. HP1 binds to H3K9me3 via its N-terminal chromodomain and this interaction is important for the overall structure of heterochromatin 68, 69. HP1 proteins dimerise via their C-terminal chromoshadow domains to form a bivalent chromatin binder. Interestingly, HP1 also binds to methylated H1.4K26 via its chromodomain 70. Since H1.4 is also involved in heterochromatin architecture, it is tempting to speculate that HP1 dimers integrate this positional information (H3K9me and H1.4K26me) in a manner that is important for chromatin compaction.
The L3MBTL1 protein is another factor that integrates positional information. Like HP1, L3MBTL1 dimerises thereby providing even more local contacts with the chromatin. It possesses three MBT domains, the first of which binds to H4K20me1/2 and H1bK26me1/2. In doing so, L3MBTL1 compacts nucleosomal arrays bearing the two histone modifications 71. Importantly, L3MBTL1 associates with HP1, and the L3MBTL1/HP1 complex, with its multivalent chromatin-binding potential, binds chromatin with a higher affinity than that of either of the two individual proteins alone.
Histone acetylated lysines are bound by bromodomains, which are often found in HATs and chromatin-remodelling complexes 72. For example, Swi2/Snf2 contains a bromodomain that targets it to acetylated histones. In turn, this recruits the SWI/SNF remodelling complex, which functions to 'open' the chromatin 73. Recently, it has also been shown that PHD fingers are capable of specifically recognizing acetylated histones. The DPF3b protein is a component of the BAF chromatin-remodelling complex and it contains tandem PHD fingers that are responsible for recruiting the BAF complex to acetylated histones 74.
Mitogen induction leads to a rapid activation of immediate early genes such as c-jun, which involves phosphorylation of H3S10 within the gene's promoter 75. This modification is recognized by the 14-3-3ζ protein, a member of the 14-3-3 protein family 76. Furthermore, studies in Drosophila melanogaster have indicated that this protein family is involved in recruiting components of the elongation complex to chromatin 77. Another example of a protein that specifically binds to phosphorylated histones is MDC1, which is involved in the DNA-repair process and is recruited to sites of double-strand DNA breaks (DSB). MDC1 contains tandem BRCT domains that bind to γH2AX, the DSB-induced phosphorylated H2A variant 78.
Histone modifications do not only function solely by providing dynamic binding platforms for various factors. They can also function to disrupt an interaction between the histone and a chromatin factor. For instance, H3K4me3 can prevent the NuRD complex from binding to the H3 N-terminal tail 79, 80. This simple mechanism seems to make sense because NuRD is a general transcriptional repressor and H3K4me3 is a mark of active transcription. H3K4 methylation also disrupts the binding of DNMT3L's PHD finger to the H3 tail 81. Indeed, this very N-terminal region of H3 seems to be important in regulating these types of interaction, though the regulation is not solely via modification of K4. For instance, phosphorylation of H3T3 prevents the INHAT transcriptional repressor complex from binding to the H3 tail 82.
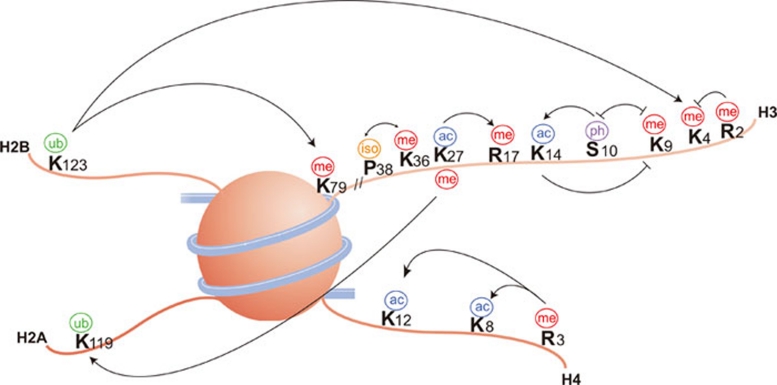
Histone modification cross-talk
The large number of possible histone modifications provides scope for the tight control of chromatin structure. Nevertheless, an extra level of complexity exists due to cross-talk between different modifications, which presumably helps to fine-tune the overall control (Figure 2). This cross-talk can occur via multiple mechanisms 53. (I) There may be competitive antagonism between modifications if more than one modification pathway is targeting the same site(s). This is particularly true for lysines that can be acetylated, methylated or ubiquitylated. (II) One modification may be dependent upon another. A good example of this trans-regulation comes from the work in Saccharomyces cerevisiae; methylation of H3K4 by scCOMPASS and of H3K79 by scDot1 is totally dependent upon the ubiquitylation of H2BK123 by scRad6/Bre1 43. Importantly, this mechanism is conserved in mammals, including humans 44. (III) The binding of a protein to a particular modification can be disrupted by an adjacent modification. For example, as discussed above, HP1 binds to H3K9me2/3, but during mitosis, the binding is disrupted due to phosphorylation of H3S10 59. This action has been described as a 'phospho switch'. In order to regulate binding in this way, the modified amino acids do not necessarily have to be directly adjacent to each other. For instance, in S. pombe, acetylation of H3K4 inhibits binding of spChp1 to H3K9me2/3 83. (IV) An enzyme's activity may be affected due to modification of its substrate. In yeast, the scFpr4 proline isomerase catalyses interconversion of the H3P38 peptide bond and this activity affects the ability of the scSet2 enzyme to methylate H3K36, which is linked to the effects on gene transcription 51. (V) There may be cooperation between modifications in order to efficiently recruit specific factors. For example, PHF8 specifically binds to H3K4me3 via its PHD finger, and this interaction is stronger when H3K9 and H3K14 are also acetylated on the same tail of H3 60. However, this stabilization of binding may be due to additional factors in a complex with PHF8 rather than a direct effect on PHF8 itself.
Figure 2.
Histone modification cross-talk. Histone modifications can positively or negatively affect other modifications. A positive effect is indicated by an arrowhead and a negative effect is indicated by a flat head (updated from reference 53).
There may also be cooperation between histone modifications and DNA methylation. For instance, the UHRF1 protein binds to nucleosomes bearing H3K9me3, but this binding is significantly enhanced when the nucleosomal DNA is CpG methylated 61. Conversely, DNA methylation can inhibit protein binding to specific histone modifications. A good example here is KDM2A, which only binds to nucleosomes bearing H3K9me3 when the DNA is not methylated 61.
Genomic localization of histone modifications
From a chromatin point of view, eukaryotic genomes can generally be divided into two geographically distinct environments 3. The first is a relatively relaxed environment, containing most of the active genes and undergoing cyclical changes during the cell cycle. These 'open' regions are referred to as euchromatin. In contrast, other genomic regions, such as centromeres and telomeres, are relatively compact structures containing mostly inactive genes and are refractive to cell-cycle cyclical changes. These more 'compact' regions are referred to as heterochromatin. This is clearly a simplistic view, as recent work in D. melanogaster has shown that there are five genomic domains of chromatin structure based on analysing the pattern of binding of many chromatin proteins 84. However, given that most is known about the two simple domains described above, references below will be defined to these two types of genomic domains.
Both heterochromatin and euchromatin are enriched, and indeed also depleted, of certain characteristic histone modifications. However, there appears to be no simple rules governing the localization of such modifications, and there is a high degree of overlap between different chromatin regions. Nevertheless, there are regions of demarcation between heterochromatin and euchromatin. These 'boundary elements' are bound by specific factors such as CTCF that play a role in maintaining the boundary between distinct chromatin 'types' 85. Without such factors, heterochromatin would encroach into and silence the euchromatic regions of the genome. Boundary elements are enriched for certain modifications such as H3K9me1 and are devoid of others such as histone acetylation 86. Furthermore, a specific histone variant, H2A.Z, is highly enriched at these sites 86. How all of these factors work together in order to maintain these boundaries is far from clear, but their importance is undeniable.
Heterochromatin
Although generally repressive and devoid of histone acetylations, over the last few years it has become evident that not all heterochromatin is the same. Indeed, in multicellular organisms, two distinct heterochromatic environments have been defined: (a) facultative and (b) constitutive heterochromatin.
(a) Facultative heterochromatin consists of genomic regions containing genes that are differentially expressed through development and/or differentiation and which then become silenced. A classic example of this type of heterochromatin is the inactive X-chromosome present within mammalian female cells, which is heavily marked by H3K27me3 and the Polycomb repressor complexes (PRCs) 87. This co-localization makes sense because the H3K27 methyltransferase EZH2 resides within the trimeric PRC2 complex. Indeed, recent elegant work has shed light on how H3K27me3 and PRC2 are involved in positionally maintaining facultative heterochromatin through DNA replication 88. Once established, it seems that H3K27me3 recruits PRC2 to sites of DNA replication, facilitating the maintenance of H3K27me3 via the action of EZH2. In this way, the histone mark is 'replicated' onto the newly deposited histones and the facultative heterochromatin is maintained.
(b) Constitutive heterochromatin contains permanently silenced genes in genomic regions such as the centromeres and telomeres. It is characterised by relatively high levels of H3K9me3 and HP1α/β 87. As discussed above, HP1 dimers bind to H3K9me2/3 via their chromodomains, but importantly they also interact with SUV39, a major H3K9 methyltransferase. As DNA replication proceeds, there is a redistribution of the existing modified histones (bearing H3K9me3), as well as the deposition of newly synthesized histones into the replicated chromatin. Since HP1 binds to SUV39, it is tempting to speculate that the proteins generate a feedback loop capable of maintaining heterochromatin positioning following DNA replication 68. In other words, during DNA replication, HP1 binds to nucleosomes bearing H3K9me2/3, thereby recruiting the SUV39 methyltransferase, which in turn methylates H3K9 in adjacent nucleosomes containing unmodified H3. Furthermore, this positive feedback mechanism helps to explain, at least in part, the highly dynamic nature of heterochromatin, not least its ability to encroach into euchromatic regions unless it is checked from doing so.
Euchromatin
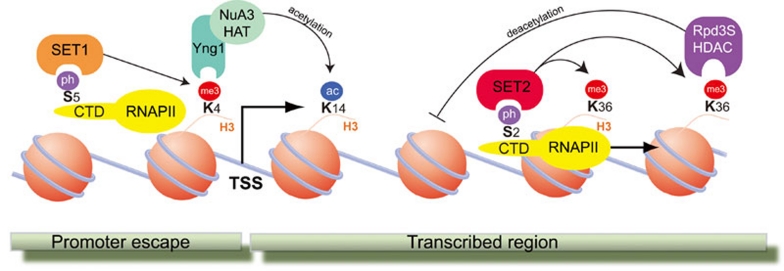
In stark contrast to heterochromatin, euchromatin is a far more relaxed environment containing active genes. However, as with heterochromatin, not all euchromatin is the same. Certain regions are enriched with certain histone modifications, whereas other regions seem relatively devoid of modifications. In general, modification-rich 'islands' exist, which tend to be the regions that regulate transcription or are the sites of active transcription 86. For instance, active transcriptional enhancers contain relatively high levels of H3K4me1, a reliable predictive feature 89. However, active genes themselves possess a high enrichment of H3K4me3, which marks the transcriptional start site (TSS) 86, 90. In addition, H3K36me3 is highly enriched throughout the entire transcribed region 91. The mechanisms by which H3K4me1 is laid down at enhancers is unknown, but work in yeast has provided mechanistic detail into how the H3K4 and H3K36 methyltransferases are recruited to genes, which in turn helps to explain the distinct distribution patterns of these two modifications (Figure 3). The scSet1 H3K4 methyltransferase binds to the serine 5 phosphorylated CTD of RNAPII, the initiating form of polymerase situated at the TSS 92. In contrast, the scSet2 H3K36 methyltransferase binds to the serine 2 phosphorylated CTD of RNAPII, the transcriptional elongating form of polymerase 93. Thus, the two enzymes are recruited to genes via interactions with distinct forms of RNAPII, and it is therefore the location of the different forms of RNAPII that defines where the modifications are laid down (reviewed in reference 3).
Figure 3.
Interplay of factors at an active gene in yeast (adapted from references 128 and 3).
Taken together, we are beginning to understand how some enzymes are recruited to specific locations, but our knowledge is far from complete. In addition, another question that needs to be considered relates to how different histone modifications integrate in order to regulate DNA processes such as transcription. Staying with H3K4me3 in budding yeast as an example, it has been shown that H3K4me3 recruits scYng1, which binds via its PHD finger 94. This in turn stabilizes the interaction of the scNuA3 HAT leading to hyperacetylation of its substrate, H3K14 (Figure 3). Thus, methylation at H3K4 is intricately linked to acetylation at H3K14. In a similar manner, and again in yeast, H3K36me3 has been shown to recruit the scRpd3S HDAC complex, which deacetylates histones behind the elongating RNAPII (Figure 3). This is important because it prevents cryptic initiation of transcription within coding regions 95, 96. Together, these examples show how the recruitment of two opposing enzyme activities (HATs and HDACs) is important at active genes in yeast. However, it is not clear whether these mechanisms are completely conserved in mammals. There is evidence for H3K4me3-dependent HAT recruitment, 55 but no evidence exists for H3K36me3-dependent HDAC recruitment.
In mammals, regulatory mechanisms governing the activity of certain genes can involve specific components more commonly associated with heterochromatic events. For example, repression of the cell-cycle-dependent cyclin E gene by the retinoblastoma gene product RB involves recruitment of HDACs, H3K9 methylating activity and HP1β 97, 98. Thus, the repressed cyclin E gene promoter appears to adopt a localized structure reminiscent of constitutive heterochromatin, i.e., presence of H3K9me2/3 and HP1. However, unlike true heterochromatin, this is a transitory structure that is lost as the cell progresses from G1- into S-phase when the cyclin E gene is activated. Thus, components of heterochromatin are utilized in a euchromatic environment to regulate gene activity.
Histone modifications and cancer
Crudely speaking, full-blown cancer may be described as having progressed through two stages, initiation and progression. As we discuss below, changes in 'epigenetic modifications' can be linked to both of these stages. However, before describing specific examples, we will consider the mechanisms by which aberrant histone modification profiles, or indeed the dysregulated activity of the associated enzymes, may actually give rise to cancer. Current evidence indicates that this can occur via at least two mechanisms; (i) by altering gene expression programmes, including the aberrant regulation of oncogenes and/or tumour suppressors, and (ii) on a more global level, histone modifications may affect genome integrity and/or chromosome segregation. Although it is beyond the scope of this review to fully discuss all of these possibilities, we will provide a few relevant examples highlighting these mechanisms.
Mouse models are invaluable tools for determining whether a particular factor is capable of inducing or initiating tumourigenesis. A good example is provided by the analysis of the MOZ-TIF2 fusion that is associated with acute myeloid leukaemia (AML) 99, 100. The MOZ protein is a HAT 101 and TIF2 is a nuclear receptor coactivator that binds another HAT, CBP 102. When the MOZ-TIF2 fusion was transduced into normal committed murine haematopoietic progenitor cells, which lack self-renewal capacity, the fusion conferred the ability to self-renew in vitro and resulted in AML in vivo103. Thus, the fusion protein induces properties typical of leukaemic stem cells. Interestingly, the intrinsic HAT activity of MOZ is required for neither self-renewal nor leukaemic transformation, but its nucleosome-binding motif is essential for both 103, 104. Importantly, the CBP interaction domain within TIF2 is also essential for both processes 103, 104. Thus, it seems that both self-renewal and leukaemic transformation involve aberrant recruitment of CBP to MOZ nucleosome-binding sites. Consequently, the transforming ability of MOZ-TIF2 most likely involves an erroneous histone acetylation profile at MOZ-binding sites. These findings provide a clear indication that the dysregulated function of histone modifying enzymes can be linked to the initiation stage of cancer development.
An activating mutation within the non-receptor tyrosine kinase JAK2 is believed to be a cancer-inducing event leading to the development of several different haematological malignancies, but there were few insights into how this could occur 105, 106. Recently however, JAK2 was identified as an H3 kinase, specifically phosphorylating H3Y41 in haematopoietic cells. JAK2-mediated phosphorylation of H3Y41 prevents HP1α from binding, via its chromoshadow domain, to this region of H3 and thereby relieves gene repression 14. This antagonistic mechanism was shown to operate at the lmo2 gene, a key haematopoietic oncogene 14, 107, 108.
In humans, extensive gene silencing caused by overexpression of EZH2 has been linked to the progression of multiple solid malignancies, including those of breast, bladder and prostate 109, 110, 111. This process almost certainly involves widespread elevated levels of H3K27me3, the mark laid down by EZH2. However, it has also recently been reported that EZH2 is inactivated in numerous myeloid malignancies, suggesting that EZH2 is a tumour suppressor protein 112, 113. This is clearly at odds to the situation in solid tumours where elevated EZH2 activity is consistent with an oncogenic function. One possible explanation for this apparent dichotomy is that the levels of H3K27me3 need to be carefully regulated in order to sustain cellular homeostasis. In other words, aberrant perturbation of the equilibrium controlling H3K27me3 (in either direction) may promote cancer development. In this regard, it is noteworthy that mutations in UTX (an H3K27me3 demethylase) have been identified in a variety of tumours 114, supporting the notion that H3K27me3 levels are a critical parameter for determining cellular identity.
Finally, changes in histone modifications have been linked to genome instability, chromosome segregation defects and cancer. For example, homozygous null mutant embryos for the gene PR-Set7 (an H4K20me1 HMT) display early lethality due to cell-cycle defects, massive DNA damage and improper mitotic chromosome condensation 115. Moreover, mice deficient for the SUV39 H3K9 methyltransferase demonstrate reduced levels of heterochromatic H3K9me2/3 and they have impaired genomic stability and show an increased risk of developing cancer 116.
It now seems clear that aberrant histone modification profiles are intimately linked to cancer. Crucially, however, unlike DNA mutations, changes in the epigenome associated with cancer are potentially reversible, which opens up the possibility that 'epigenetic drugs' may have a powerful impact within the treatment regimes of various cancers. Indeed, HDAC inhibitors have been found to be particularly effective in inhibiting tumour growth, promoting apoptosis and inducing differentiation (reviewed in 117), at least in part via the reactivation of certain tumour suppressor genes. Moreover, the Food and Drug Administration has recently approved them for therapeutic use against specific types of cancer, such as T-cell cutaneous lymphoma, and other compounds are presently in phase II and III clinical trials 118.
Other histone-modifying enzyme inhibitors, such as HMT inhibitors, are presently in the developmental phase. But before we plunge head-first into a full discovery programme for other inhibitors, we should consider a number of important issues relevant to the development of such initiatives (see 118 for full discussion). First, we do not fully understand how HDAC inhibitors achieve their efficacy. Do they for instance exert their effects via modulating the acetylation of histone or non-histone substrates? Second, the majority of HDAC inhibitors are not enzyme-specific, that is, they inhibit a broad range of different HDAC enzymes. It is not known whether this promotes their efficacy or whether it would be therapeutically advantageous to develop inhibitors capable of targeting specific HDACs. Thus, when developing new inhibitors such as those targeting HMTs, we need to consider whether we should aim for enzyme-specific inhibitors, enzyme subfamily specific inhibitors, or similarly to the HDAC inhibitors, pan-inhibitors. Nevertheless, the fact that these drugs are safe and the fact that they work at all, given the broad target specificity, are extremely encouraging. So the truth is that even though there is still a lot to learn about chromatin as a target, 'epigenetic' drugs clearly show great promise.
Future perspectives
We have identified many histone modifications, but their functions are just beginning to be uncovered. Certainly, there will be more modifications to discover and we will need to identify the many biological functions they regulate. Perhaps most importantly, there are three areas of sketchy knowledge that need to be embellished in the future.
The first is the delivery and control of histone modifications by RNA. There is an emerging model that short and long RNAs can regulate the precise positioning of modifications and they can do so by interacting with the enzyme complexes that lay down these marks 119, 120, 121, 122. Given the huge proportion of the genome that is converted into uncharacterised RNAs 123, 124, there is little doubt that this form of regulation is far more prevalent than is currently considered.
The second emerging area of interest follows the finding that kinases receiving signals from external cues in the cytoplasm can transverse into the nucleus and modify histones 14, 125. This direct communication between the extracellular environment and the regulation of gene function may well be more widespread. It could involve many of the kinases that are currently thought to regulate gene expression indirectly via signalling cascades. Such direct signalling to chromatin may change many of our assumptions about kinases, as drug targets and may rationalise even more the use of chromatin-modifying enzymes as targets.
The third and perhaps the most ill-defined process that will be of interest is that of epigenetic inheritance and the influence of the environment on this process. We know of many biological phenomena that are inherited from mother to daughter cell, but the precise mechanism of how this happens is unclear 126. Do histone modifications play an important role in this? The answer is yes, and as far as we know they are responsible for perpetuating these events. However, how does the epigenetic signal start off? Is the deposition of the modifications at the right place during replication enough to explain the process? Or is there a 'memory molecule', such an RNA, transmitted from mother to daughter cell 127, which can deliver histone modifications to the right place? These are fundamental questions at the heart of 'true' epigenetic research, and they will take us a while longer to answer.
References
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Dawson MA, Bannister AJ.Histone modificationsIn: Meyers R, ed. Encyclopedia of Molecular Cell Biology and Molecular Medicine. John Wiley and Sons; 2011. In Press. [Google Scholar]
- Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26:5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- Hodawadekar SC, Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–5540. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki M, Aihara H, Ito T. Role of histone phosphorylation in chromatin dynamics and its implications in diseases. Subcell Biochem. 2007;41:319–336. [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama K, Sugiura K, Hara T, et al. Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene. 2002;21:3103–3111. doi: 10.1038/sj.onc.1205432. [DOI] [PubMed] [Google Scholar]
- Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol Life Sci. 2009;66:407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Shi Y. Epigenetic regulation: methylation of histone and non-histone proteins. Sci China C Life Sci. 2009;52:311–322. doi: 10.1007/s11427-009-0054-z. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, et al. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson JR, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yang Z, Khan SI, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Tachibana M, Tamaru H, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SS. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci. 2009;66:2109–2121. doi: 10.1007/s00018-009-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static. Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW. {beta}-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going. Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Rozensal D, et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, et al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan D, Ingvarsdottir K, Sterner DE, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Kirmizis A, Nelson C, et al. Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol. 2009;16:17–22. doi: 10.1038/nsmb.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EM, Muratore-Schroeder TL, Cook RG, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zang J, Whetstine J, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kiefer CM, Hou C, Little JA, Dean A. Epigenetics of beta-globin gene regulation. Mutat Res. 2008;647:68–76. doi: 10.1016/j.mrfmmm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci USA. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne KS, Kutateladze TG. Structural insight into histone recognition by the ING PHD fingers. Curr Drug Targets. 2009;10:432–441. doi: 10.2174/138945009788185040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, III, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Karam CS, Kellner WA, Takenaka N, Clemmons AW, Corces VG. 14-3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS Genet. 2010;6:e1000975. doi: 10.1371/journal.pgen.1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- Adams-Cioaba MA, Min J. Structure and function of histone methylation binding proteins. Biochem Cell Biol. 2009;87:93–105. doi: 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Kouzarides T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24:647–652. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Insulators as mediators of intra- and inter-chromosomal interactions: a common evolutionary theme. J Biol. 2009;8:73. doi: 10.1186/jbiol165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature. Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Liang J, Prouty L, Williams BJ, Dayton MA, Blanchard KL. Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood. 1998;92:2118–2122. [PubMed] [Google Scholar]
- Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- Champagne N, Pelletier N, Yang XJ. The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene. 2001;20:404–409. doi: 10.1038/sj.onc.1204114. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Deguchi K, Ayton PM, Carapeti M, et al. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med. 2006;355:2452–2466. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- Levine RL, Pardanani A, Tefferi A, Gilliland DG. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer. 2007;7:673–683. doi: 10.1038/nrc2210. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci USA. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack MP, Rabbitts TH. Activation of the T-cell oncogene LMO2 after gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2004;350:913–922. doi: 10.1056/NEJMra032207. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikert S, Christoph F, Kollermann J, et al. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int J Mol Med. 2005;16:349–353. [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best JD, Carey N. Epigenetic opportunities and challenges in cancer. Drug Discov Today. 2010;15:65–70. doi: 10.1016/j.drudis.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Kouzarides T, Berger SL.Chromatin modifications and their mechanism of actionIn: Allis CD, Jenuwein T, Reinberg D, eds. Epigenetics, New York: Cold Spring Harbor Laboratory Press; 2007191–210. [Google Scholar]