Abstract
Recent findings show that chromatin dynamics and nuclear organization are not only important for gene regulation and DNA replication, but also for the maintenance of genome stability. In yeast, nuclear pores play a role in the maintenance of genome stability by means of the evolutionarily conserved family of SUMO-targeted Ubiquitin ligases (STUbLs). The yeast Slx5/Slx8 STUbL associates with a class of DNA breaks that are shifted to nuclear pores. Functionally Slx5/Slx8 are needed for telomere maintenance by an unusual recombination-mediated pathway. The mammalian STUbL RNF4 associates with Promyelocytic leukaemia (PML) nuclear bodies and regulates PML/PML-fusion protein stability in response to arsenic-induced stress. A subclass of PML bodies support telomere maintenance by the ALT pathway in telomerase-deficient tumors. Perturbation of nuclear organization through either loss of pore subunits in yeast, or PML body perturbation in man, can lead to gene amplifications, deletions, translocations or end-to-end telomere fusion events, thus implicating SUMO and STUbLs in the subnuclear organization of select repair events.
Keywords: DNA damage, nuclear organization, SUMO, ubiquitin, STUbL, repair
Introduction
In eukaryotic cells, chromosomal activities, including transcriptional activation, repression and DNA replication, are tightly regulated at multiple levels, from that of protein-DNA interactions to higher-order nuclear organization. Recent studies suggest that the nucleus contains spatially and functionally distinct subcompartments, which in some cases depend on elements of nuclear architecture, such as the nuclear lamina or nuclear pores. Such structures presumably function as scaffolds for different chromosomal activities by recruiting either substrates, enzymes or both 1.
A well-documented example of a functional compartment organized by the nuclear envelope is the one created by telomere clustering in budding yeast 2, 3. These foci sequester the silencing factors that promote chromatin-mediated transcriptional repression at telomeres, which have redundant mechanisms for binding to the inner nuclear envelope 1. In differentiated mammalian cells, transcriptionally inactive and dense-staining heterochromatin is also found along the inner face of the nuclear envelope or around the nucleolus 1, and using a fusion of the perinuclear B-type lamin to a DNA methyltransferase (the so-called Dam-ID technique), Bas van Steensel and colleagues have recently identified specific sequences that associate with the nuclear envelope 4. Consistent with findings in yeast, these were shown to be enriched for transcriptionally inactive, late-replicating genes 4. It is proposed that in both yeast and man, perinuclear zones contribute to gene repression by recruiting chromatin modifiers, such as the yeast Silent information regulator (SIR) complex or, in mammals, other NAD-independent histone deacetylases 5, 6, 7, 8. The physiological relevance of this organization was tested in yeast by ablating the factors that sequester SIR proteins and telomeric repeats at the nuclear periphery 9. This led to the derepression of subtelomeric genes and the promiscuous silencing of other loci at dispersed sites of the genome 9.
Besides these perinuclear zones of repression, nuclear pores have emerged as distinct functional compartments of the nuclear periphery. In 1985, Gunter Blobel proposed the “Gene gating hypothesis” which suggested that nuclear pores would selectively facilitate transcription as well as co- and post-transcriptional processing of subsets of mRNAs 10. Although there is no evidence yet showing that specific pores bind specific sets of genes, recent studies implicate nuclear pores in a range of cellular processes, well beyond their function in macromolecular transport. Notably, in budding yeast and Drosophila certain highly transcribed, and in some cases stress-induced, genes are associated with nuclear pores 11, 12, 13, 14, 15, 16, 17, 18, 19. This association not only promotes efficient export of mRNAs through coupling of transcription with export 10, 11, 13, 14, but also helps modulate transcript levels of some loci. A few yeast genes, e.g. INO1 or HXK1, like X-linked genes in male flies, show a modest but reproducible increase in expression when allowed to associate with nuclear pores 12, 15, 19. On the other hand, there are other genes, notably a set of yeast ribosomal biogenesis genes, whose expression levels are lower when they are associated with nuclear pores 11, 18. This subset of promoters has a particular constellation of regulators and their localization at the nuclear pore depends on the actin-related protein Arp6 18. Reduced expression of the HSP26 gene also correlates with localization at pores 11. One way to reconcile these results would be to propose that pore association fine-tunes RNA polymerase II elongation by regulating mRNA proofreading, processing and export, thereby having different effects on different genes. What triggers the shift in nuclear localization of a gene to the pore remains unclear, although in some cases mRNA appears to be required.
Intriguingly, recent studies have implicated the SUMO (Small Ubiquitin-related Modifier) pathway as an important player in higher-order nuclear organization both at yeast nuclear pores and at promyelocytic leukaemia nuclear bodies in mammalian cells, (PML-NBs; reviewed in 20). Indeed, SUMO metabolism and SUMO-binding proteins are enriched at both nuclear pores and PML-NBs in man, and at nuclear pores in yeast. SUMO and PML-NBs appear to regulate telomerase-independent telomere length maintenance, particularly in relation to the alternative lengthening pathway for human telomeres that arises in tumors 21. Recently, sumoylation of Ku and the ssDNA-binding factor Cdc13 in yeast has been implicated in the regulation of telomerase-dependent telomere elongation (Ferreira H and SMG, personal communication; Hang LE and Zhao X, personal communication). Moreover, SUMO is implicated in the maintenance of rDNA stability by suppressing illegitimate recombination among the yeast rDNA repeats 22. Consistent with this widespread effect on genomic integrity, it was found that many other proteins involved in DNA repair are sumoylated, notably PCNA, Rad52, Rfa1, Rfa2, Sgs1 helicase, as well as its human homologues, BLM and WRN 23, 24, 25, 26, 27, 28. Many of these proteins are found either transiently or stably associated with PML-NBs in mammalian cells in response to DNA damage (reviewed in 20).
SUMO conjugation to substrates is catalyzed by three distinct enzymatic steps: SUMO is first activated by an E1 activating enzyme in an ATP dependent manner. The activated SUMO is then transferred to an E2 conjugating enzyme. Finally, E2 conjugase catalyzes the covalent attachment of SUMO to the substrate with the help of E3 ligase, which bridges E2 conjugase and the substrate, thus determining the substrate specificity of the reaction. Removal of SUMO requires desumoylating enzymes or SUMO proteases (Ulp1 and Ulp2 in yeast, and SENP proteins in mammals) 29. SUMO modification can elicit diverse biological consequences, altering protein-protein interactions, intracellular localization and enzymatic activity. Recent identification of SUMO-targeted ubiquitin ligases (STUbLs) provides another mode of action whereby SUMO modification serves as a signal for further modification by ubiquitin and substrate degradation 30.
Although many studies have suggested a role for nuclear architecture in gene regulation 1, 5, 6, 7, 8, relatively little is known about how higher order nuclear organization contributes to the maintenance of genome stability. It has been shown that DNA damage checkpoint and repair proteins relocalize from a diffuse distribution to distinct subnuclear foci in a temporally regulated manner upon DNA damage, in both yeast and mammalian cells 31, 32, 33, 34. However, it is not clear if double-strand breaks (DSBs) are recruited to scaffolds for repair, or if repair factors assemble at sites of damage. Indeed, sites of damage arising from a targeted endonuclease 31 or γ-irradiation 32 are relatively immobile in the mammalian nucleus, suggesting that most DSB repair by non-homologous end joining, does not involve relocalization of damage to sites like PML-NBs. It remains possible, however, that specific pathways of DNA repair require specific nuclear subcompartments, since the efficiency of certain types of repair varies with the chromosomal context in which the damage occurs 35.
In yeast, recent studies have addressed the question of how genome stability is coordinated with nuclear organization. The results implicate both SUMO metabolism and nuclear pores in the spatial regulation of DNA repair. We summarize here evidence showing that yeast nuclear organization plays an important role in the maintenance of rDNA stability, telomere length and in the recovery from DSBs or collapsed replication forks that lack templates for repair by classical homologous recombination (HR). We then draw parallels with PML bodies in mammalian cells, which like pores, harbor the STUbL ligase and a SENP protease, which target sumoylated substrates or SUMO itself for degradation.
DNA repair and SUMO in yeast
The genome is constantly assaulted by exogenous insults as well as endogenous stress arising from vital cellular activities. The resulting DNA lesions must be repaired to prevent loss or incorrect transmission of genetic information, since errors can result in developmental abnormalities or tumorigenesis. DNA double-strand breaks are the most deleterious type of DNA lesion, and they can be created either by exogenous stress (for example, ionizing radiation) or by aberrant recombination or collapse at a stalled DNA replication fork.
Protein sumoylation plays important roles in the maintenance of genome integrity from yeast to man (Table 1). Mutations in genes involved in the SUMO pathway, such as Ubc9, Mms21, Ulp1, Slx5 and Slx8, render cells sensitive to DNA damage, and many proteins involved in the maintenance of genome stability have been shown to become sumoylated during the damage response 23, 24, 25, 26, 27, 28. Specifically, it has been shown that the SUMO pathway plays an important role in homologous recombination and the resumption of stalled/collapsed replication forks 27, 36, 37, 38.
Table 1. Involvement of the SUMO pathways in DNA repair in yeast.
| Protein | SUMO pathway | Function in genome stability |
|---|---|---|
| Ubc9 | SUMO E2 conjugase | Sister chromatid exchange 37 Resolution of recombination intermediate 27 |
| Mms21 | SUMO E3 ligase | Sister chromatid exchange 37 Resolution of recombination intermediate 27 ALT maintenance of telomeres 21 |
| Siz1 | SUMO E3 ligase | PCNA sumoylation (recruit Srs2) 44, 45 |
| Ulp1 | SUMO protease | Suppression of recombination during replication 56 |
| Slx5/Slx8 | STUbL | Maintenance of replication fork stability 25, 53 Alternative lengthening of telomeres 59 |
In order to cope with DSBs and the task of resumption of replication at a collapsed fork, eukaryotic cells have evolved highly conserved mechanisms to detect and repair these deleterious lesions. In particular, homologous recombination (HR) promotes error-free repair of DSBs using genetic information that is retrieved from homologous sequences, such as sister chromatids or chromosomal homologues (Figure 1) 39. HR requires the RAD52 epistasis group proteins, including Rad51, Rad52 and Rad54, and the breast cancer susceptibility genes Brca1 and Brca2 in mammals. In mitotically dividing yeast cells, sister chromatid exchange is the predominant pathway for HR-mediated repair of DSBs 40. This is likely to be the major mode of recombinational repair in mammals as well, since homologous chromosomes are not closely juxtaposed in somatic cells.
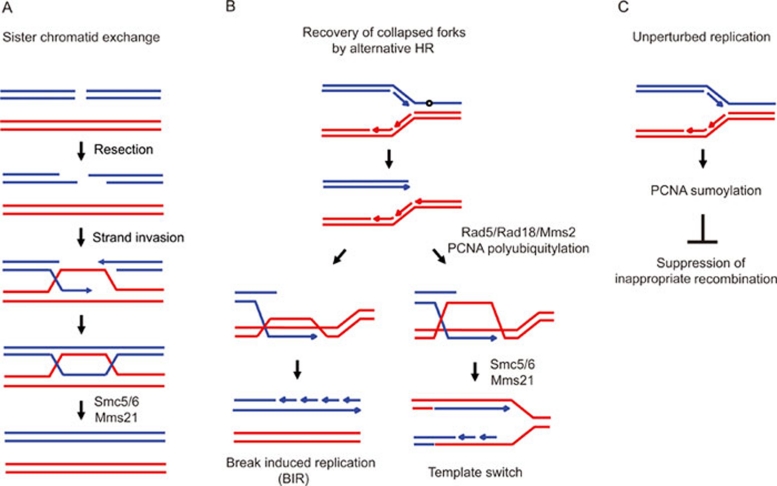
Figure 1.
DSB repair pathways and SUMO. (A) HR repairs DSBs using genetic information that is retrieved from sister chromatid 39. When a DSB occurs in one of two sister chromatids, the ends of the DSB are recognized by Mre11-Rad50-Xrs2 (MRX) complex in yeast (or Mre11-Rad50-Nbs1 (MRN) complex in mammals). To facilitate strand invasion into homologous sequences, the ends of the DSB are resected, which results in the formation of 3′ single stranded DNA overhangs. The 3′ ssDNA overhangs nucleate the formation of the Rad51 nucleoprotein filament, which has the ability to invade the homologous DNA duplex. The resolution of the joint molecule completes this error-free repair event. The Smc5/Smc6 complex, which binds a SUMO E3 ligase Mms21, is required for sister chromatid exchange in both yeast 37 and man 36. (B) When DNA replication forks encounter a damaged base (shown as a circle), it often leads to the collapse of replication forks, resulting in a one-end DSB. This collapsed replication fork can be repaired by template switch mediated by strand invasion. The template switch pathway is regulated by polyubiquitylation of sliding clamp PCNA at K164 by Rad5 (ubiquitin E2 conjugase) and Ubc13/Mms2 (E3 ligase) 38. Resolution of joint molecule requires Smc5/Sm6 and Mms21 SUMO E3 ligase 27. Alternatively, break induced replication (BIR) can be initiated by replication fork collapse or sudden shortening of telomeres. BIR involves the invasion of the broken end into the donor duplex, followed by DNA synthesis from the site of strand invasion to the end of the chromosome. (C) Sumoylation of PCNA mediated by E3 ligase Siz1 was observed during an unperturbed replication and was shown to recruit the anti-recombinogenic helicase Srs2 44, 45, thereby suppressing inapproprate recombination between newly synthesized sister chromatids.
In addition to the role of HR in DSB repair, alternative recombination pathways, such as break-induced replication (BIR) and template switch, play important roles in the resumption of stalled or collapsed replication forks, which can occur spontaneously during genome replication (Figure 1) 39. Whereas BIR often results in loss of heterozygosity, template switch is considered a pathway of error-free repair. In the case of template switch, it is regulated by polyubiquitylation of the homotrimeric sliding clamp PCNA, an essential platform for numerous proteins involved in replication, repair and chromatin assembly 41. Upon replication fork stalling/collapse, PCNA is polyubiquitylated at the highly conserved residue K164, by Rad5 (ubiquitin E2 conjugase) and Ubc13/Mms2 (E3 ligase), and this modification is thought to induce a switch of template 38, 41. Furthermore, Branzei et al. have shown that Rad18, together with Mms2–Ubc13, is required for the formation of X-shaped recombination intermediates at damaged replication forks, which potentially represent sister-chromatid junctions formed during a template switch process 38.
The Smc5/Smc6 complex, which binds the SUMO E3 ligase Mms21, has been shown to contribute to both sister chromatid exchange and HR-mediated replication fork resumption after insult by the alkylating agent MMS 37. This complex is specifically required for recombination between sister chromatids in both yeast 37 and man 36, and is recruited to DSBs in G2/M cells 37, 42. Moreover, it contributes to the maintenance of telomeric repeats by recombination in telomerase-deficient cells 21. Based on the structural homology of the Smc5/6 heterodimer with Smc1/Smc3 of cohesin, it can be assumed that the Smc5/Smc6 complex forms a ring structure that could encompass sister chromatids, although this has not been demonstrated on a molecular level.
By performing genome-wide mapping of Smc5 and Smc6 binding in yeast, Sjogren and colleagues have shown that the Smc5/Smc6 complex is recruited to collapsed replication forks, but not to forks that are reversibly stalled 42. This is consistent with its presence of or selective recruitment to DSBs. Consistent with a function in the rescue of collapsed forks, it has been shown that mutants in ubc9, which encodes the only yeast SUMO E2 conjugase, and in mms21, accumulate X-shaped recombination intermediates during replication of damaged templates 27. Similarly, the smc6 mutant in fission yeast accumulates aberrant recombination intermediates at rDNA upon fork collapse 43. In mammals SMC5/SMC6 and MMS21 are localized in PML-NBs and are essential for HR-mediated telomere maintenance 21. Taken together the results suggest that both the Smc5/Smc6 complex and its sumoylation activity have a role in resolving joint molecules formed either at collapsed replication forks or when strand invasion into an adjacent chromosome is necessary for repair (Figure 1A, 1B).
While the recombination-based fork restart requires the HR machinery, recombination at replication forks in general must be tightly controlled, normally being repressed to limit inappropriate recombination between newly synthesized strands. Asymmetric recombination generates chromosomal truncations or deletions, particularly in repetitive domains. SUMO again plays a role here: in yeast, sumoylation of PCNA mediated by E3 ligase Siz1 was observed during an unperturbed S phase and was shown to recruit the anti-recombinogenic helicase Srs2 (Figure 1) 44, 45, thereby suppressing unwanted recombination events. Interestingly, sumoylated PCNA was also required for the template switch pathway 38, suggesting that combinations of ubiquitylation and sumoylation channel damage to distinct modes of repair.
STUbL family members: targeting sumoylated proteins for degradation
A recently identified family of SUMO-targeted E3 ubiquitin ligases (STUbL), has been shown to play an important role in the maintenance of genome integrity from yeast to man (reviewed in 30). In the budding yeast, SLX5 and SLX8 genes, like the anti-recombination helicase gene SRS2, are required for viability in mutants lacking the Sgs1 DNA helicase 46. Slx5 and Slx8 form a heterodimeric Ub ligase that specifically recognizes sumoylated substrate through SUMO interaction motifs (SIMs). Both Slx5 and Slx8 harbor such SIMs, while Slx8 bears a RING finger domain with Ub ligase activity 25, 46, 47, 48, 49, 50, 51. The Slx5 protein physically interacts with components of the proteasome 52, and deletion of either SLX5 or SLX8 leads to the accumulation of sumoylated proteins 47, 51.
Mutants lacking Slx5 or Slx8 are hypersensitive to the continuous exposure to hydroxyurea (HU) 46, 48, 49 and accumulate spontaneous DNA damage during S phase as monitored by the accumulation of repair foci containing the recombination factor Rad52 or the Mec1/Ddc2 checkpoint kinase 25, 53. This suggests that these proteins have an important role during DNA replication. Consistently, Slx8 has been shown to form distinct foci during S phase and some of the foci colocalize with replication centers visualized by PCNA-CFP 25. Finally, slx5Δ and slx8Δ mutants show increased rates of spontaneous gross chromosomal rearrangements such as translocations and deletions 49, 53.
Importantly, results obtained in fission yeast argue that an accumulation of SUMO conjugates is responsible for the HU sensitivity of the slx8 mutant, since both phenotypes can be suppressed by deleting the major fission yeast SUMO E3 ligase Pli1 48. Similarly, overexpression of the desumoylating enzyme Ulp2 suppresses the DNA damage sensitivity of cells lacking both Rfp1 and Rfp2, the fission yeast functional homologues of Slx5 54. Collectively, these results implicate the Slx5/Slx8 STUbL in the maintenance of genome integrity during DNA replication, presumably through the ubiquitylation and subsequent degradation of a SUMO-modified target. Although it is still unclear which repair pathway is controlled by this degradation event, one possibility is that Slx5/Slx8 regulates a recombination event that is normally “inappropriate”, but which becomes necessary for replication restart upon fork collapse.
Nuclear envelope and genome stability: SUMO connections
Although protein sumoylation is clearly implicated in the maintenance of genome stability, exactly how SUMO exerts its function in repair is unclear. It may affect protein-protein interactions, or protein targeting; it may regulate enzymatic activity or else contribute to genomic stability by regulating the spatial organization of damage within the nucleus.
Evidence in support of this last option again comes largely from budding yeast. The yeast desumoylating enzyme Ulp1 is associated with nuclear pores through components of the inner pore basket, Mlp1 and Mlp2 58. Ulp1 is an essential enzyme, and its mutation or even mislocalization away from pores (e.g., in mlp1/mlp2 mutants) impairs both transport through nuclear pores and genome stability 55, 56. Notably, in addition to causing impaired nuclear transport, the temperature-sensitive mutant of Ulp1 accumulates single-stranded DNA during genome replication and has a hyper-recombination phenotype 56. Furthermore, both the conditional ulp1 mutant and deletions of mlp1 or mlp2 are synthetically lethal with the loss or mutation of genes involved in HR 52, 56, 57. The genomic instability observed in mutants of Mlp1/Mlp2 or Nup84 complex components correlates with a drop in Ulp1 protein levels; consistently, overexpression of Ulp1 partially suppresses the genomic instability observed in these mutants 57, 58. These studies suggested that nuclear pores may play an important role in the maintenance of genome stability by facilitating or coordinating SUMO metabolism. However, since pores and pore-associated factors affect many cellular processes, including macromolecular transport, mRNA quality control and gene regulation, it was initially unclear whether the reported genomic instability phenotypes were direct or indirect effects of nuclear pore function.
This has been clarified by the recent discovery that certain types of damaged DNA, such as persistent DSBs and collapsed replication forks, are physically recruited to nuclear pores 53 (Figure 2A, 2B). By tagging sites of an induced DSB or collapsed replication fork with lacO repeats and LacI-GFP, it was shown in budding yeast that damage itself is recruited to pores with the same kinetics as the Slx5/Slx8 STUbL complex binding to sites of damage 30, 53. The authors proposed that this repair pathway represents an alternative repair/recombination pathway, since the recruitment of DSBs was not observed if a donor for repair was present. Indeed, canonical HR events have been shown to take place in intranuclear foci, and not at nuclear pores (V Dion, SMG, personal communication). It is proposed that a sumoylated protein accumulates at collapsed forks or resected breaks, that requires Slx5/Slx8-mediated ubiquitylation and proteasomal degradation to promote appropriate repair. Proteosome subunits are also bound to and required for repair of certain types of damage. The exact relationship of the Slx5/Slx8-mediated pathway to the pore-associated SUMO protease Ulp1 may be complex, however, since Ulp1 is necessary both for priming sumoylation and for desumoylation events.
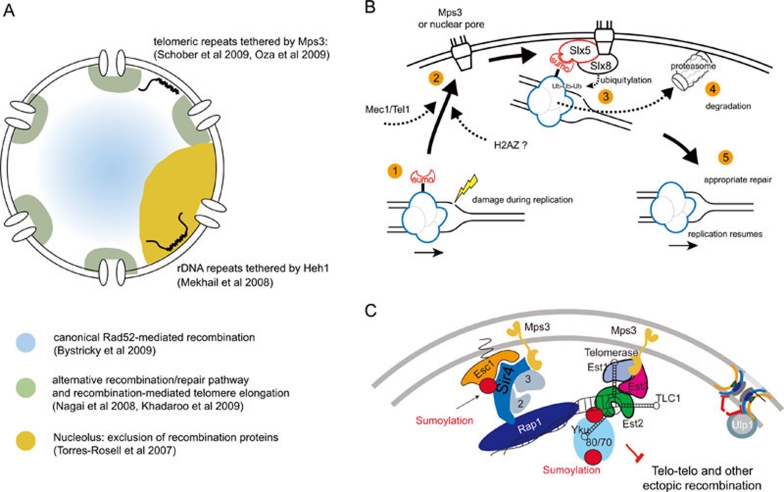
Figure 2.
DNA repair compartments in yeast nucleus. (A) Model of subnuclear compartmentalization of DNA repair pathways in budding yeast. Canonical recombination, including sister chromatid exchange, appears to occur predominantly in the nuclear interior based on the frequency and distribution of both spontaneous and induced Rad52 foci (blue) 67. In contrast, alternative recombination and repair pathways, which act on collapsed replication forks, may be facilitated by processing events at nuclear pores through action of the Slx5/Slx8 complex (green) 53, 59. Peripheral tethering of telomeres by an inner nuclear membrane protein Mps3 contributes to the suppression of recombination 65, 68. In the nucleolus (gold) the rDNA repeats are bound to the inner nuclear membrane by the yeast protein Heh1 69. Peripheral tethering may limit the access of sites to the recombination machinery that mediates sister chromatid exchange. (B) Model of Mec1-dependent relocation of damage to Nup84/Slx5/Slx8 complexes at the nuclear envelope 53. A sumoylated protein may accumulate at collapsed forks or resected breaks, requiring Slx5/Slx8 ubiquitylation and proteasomal degradation to enable appropriate repair. The proposed order of events are: (1) A sumoylated protein accumulates at collapsed fork or irreparable DSB. (2) The damage is recruited to pores in a Mec1-dependent manner. The shift correlates with resection and the binding of RPA and Rad51 and may require Htz1 sumoylation 66. (3) A sumoylated protein is ubiquitylated by Slx5/Slx8. (4) This leads to proteasomal degradation of the modified protein. (5) This degradation is proposed to facilitate appropriate repair (modified with permission from 53). (C) The SUN domain protein Mps3 tethers telomeres to the nuclear periphery, thereby contributing to the suppression of recombination between telomeric repeats 65, 68. Peripheral tethering possibly limits the access of the telomeric repeats to the recombinational machinery mediating sister chromatid exchange, which was shown to be enriched in the nuclear interior 67.
In further studies it was shown that critically short telomeres are also recruited to nuclear pores for processing and repair 59 (Figure 2A, 2B), along with factors involved in checkpoint kinase recruitment and recombination, notably Cdc13, RPA and Rad52 59, 60. Importantly, Slx5/Slx8 mutants are impaired or delayed for the generation of type II survivors 61, in which telomeric TG repeats are maintained by a Rad51-independent recombination pathway in the absence of telomerase 62. From these data we propose a model which suggests that a critically short telomere, generated by the loss of telomerase activity or collapse of replication forks, can be recognized as damage 60, 63 and be transferred to nuclear pores, where Slx5/Slx8-mediated ubiquitylation and degradation of a target protein facilitates alternative telomere elongation. What this target is remains unknown.
This particular pathway of recovery, provoked by the presence of critically short telomeres in the absence of telomerase, is reminiscent of the situation in mammalian cells, in which the alternative lengthening of telomeres involves the relocalization of telomeres to PML bodies, along with factors involved in recombination, such as RPA, Rad51, Rad52 and Smc5/Smc6 21, 64. Considering the observation that sumoylation of the telomere-binding protein, TRF1, is required for the localization of mammalian telomeres to PML bodies 21, we speculate that sumoylation of yeast telomere proteins might be similarly involved in the relocalization process. Indeed, both Yku70 and Yku80, as well as Cdc13, are SUMO targets (Ferreira H and SMG, personal communication; Hang LE and Zhao X, personal communication). Loss of sumoylation appears to stimulate telomerase activity, suggesting that telomerase is normally antagonized by a sumoylated target. On the other hand, in cells that lack telomerase altogether, sumoylation, pore-association, and Slx5/Slx8-mediated degradation of a sumoylated target, may trigger a crucial pathway of telomere repair.
Sites along the inner nuclear envelope that are distinct from nuclear pores appear to constitute a distinct compartment that helps regulate the DNA damage response. Two independent studies have shown that a persistent DSB can also bind the inner nuclear membrane by contacting the SUN-domain protein, Mps3 65, 66 (Figure 2A, 2C), and Peterson and colleagues have proposed that the tethering of a persistent DSB to the inner nuclear membrane by Mps3 helps suppress ectopic homologous recombination 65. Consistently, spontaneous Rad52 foci are enriched in the nuclear interior 67, and the loss of Mps3-telomere interaction results in hyper-recombination between subtelomeric elements in a tel1Δ background 68 (Figure 2A, 2C). Thus non-pore sites along the nuclear envelope may suppress recombination. A further manifestation of this comes from a gain-of-function experiment in which rDNA repeats were tethered to an inner nuclear membrane protein, Heh1 69. This was shown to reduce the genomic instability caused by unequal sister chromatid exchange within the repetitive rDNA cluster in yeast (Figure 2A, 2C). We speculate that peripheral tethering limits the access of rDNA and of telomeric or subtelomeric repeats to the machinery of sister chromatid exchange.
Ironically, Kalocsay et al. showed that the recruitment of an irreparable DSB to the nuclear periphery requires Rad51, as well as the histone H2A variant, H2AZ 66. The mutant lacking H2AZ is impaired in DSB resection, which is consistent with data from the Gasser laboratory showing that DSB resection and the binding of Mec1/Ddc2 are essential for the observed relocation 53. The requirement for Rad51, which binds ssDNA to form an invasion competent filament, suggests that relocalization occurs after a thwarted or inappropriate strand invasion event. Interestingly, it appears that the sumoylation of H2AZ is among the first modifications triggering the shift of an irreparable break to the nuclear periphery 66. One speculative scenario is that SUMO-H2AZ becomes targeted for degradation by Slx5/Slx8-mediated ubiquitylation, since H2AZ is deposited to an irreparable DSB at an early time point (30 min after DSB induction) and is subsequently evicted from the break 66. Inconsistent with this proposal, however, are the kinetics of association of the DSB with the nuclear pore (which requires ∼2 h) and the fact that H2A.Z probably gets evicted during the processing of the DSB to a 3′ overhang, necessary for HR repair 53. Moreover, given that mutations in the repair machinery mediating HR are synthetic lethal with deletion of SLX5, SLX8 or the nuclear pore component, NUP84, it is likely that these act on distinct pathways 53. Further work is needed to identify the triggers for damage relocalization, as well as the relevant targets of Slx5/Slx8 or the proteosome, and forms of repair that they regulate.
In conclusion, two peripheral compartments in yeast appear to represent distinct compartments; sumoylation, desumolyation and ubiquitination at pores appear to facilitate an alternative type of recombinational repair 53, 59, while the second compartment, which correlates with Mps3 sites along the inner nuclear membrane, appears to suppress recombination 65, 68 (Figure 2). It is of course possible that the two compartments are mechanistically linked, and that peripheral relocalization of damage favours alternative repair and recombination pathways, while suppressing canonical HR. The presence of SUMO-metabolizing enzymes at nuclear pores, and the repair defects of mutant in either nuclear pore or SUMO-modifying proteins, argue that the SUMO pathway plays a role in the relocalization of certain types of DNA damage to perinuclear subcompartments. It appears that the processing of sumoylated targets then helps coordinate possible repair pathways of certain types of damage. How does SUMO modification facilitate relocalization? The most likely scenario is that proteins harbouring SIMs 30 are found at pores or other sites along the nuclear envelope, serving as direct binding sites for the sumoylated proteins bound at persistent DSBs, collapsed forks or shortened telomeres. Identification of these molecules will be needed to extend our understanding of these processes.
PML nuclear bodies and SUMO pathway
In mammalian cells SUMO modification is also found at nuclear pores, and sumoylated proteins involved in DNA damage repair accumulate in specific subnuclear domains called promyelocytic leukemia nuclear bodies (PML-NBs) 20, 70. Remarkably, SUMO not only localizes to PML-NBs, but is also required for their assembly and formation. PML-NBs are a very prominent feature of mammalian nuclei, numbering between 5 and 30 per cell, with many functions that are still incompletely understood 20. The structures are formed from the constituent proteins PML and Sp100, and harbor factors involved in repair like MRE11 and the damage-associated RecQ helicase, BLM 20, 71.
The PML protein itself carries three SUMO-1 modification sites 72, 73 and a SIM 70. The SIM on PML mediates binding of PML-SUMO to itself and to other sumoylated proteins, forming large complexes. This SUMO-mediated crosslinking is the basis of PML-NB formation: following self-aggregation of PML-SUMO, the PML-NB recruits other sumoylated or SIM-containing proteins, such as Sp100 and DAXX 70, 74, 75, 76, 77, 78. Importantly, the PML protein that remains diffuse within the nucleoplasm lacks SUMO-1 modification 75, as does a precursor aggregate of PML, called a primary PML body 76, 77, 78, 79.
The PML protein itself was initially identified as a PML-RARα (Retinoic Acid Receptor alpha) fusion protein, which arises from a chromosomal translocation in patients with acute promyelocytic leukemia (APL). The PML-RARα fusion protein heterodimerizes with wild-type PML and disrupts PML-NB formation 80. Consistently, the typical PML-NB pattern is lost in APL patients. Treatment with all-trans retinoic acid (ATRA), which partially alleviates the disease, leads to the reformation of PML-NBs 81, 82.
The behavior of PML-NBs in APL and their response to ATRA or to arsenic trioxide (ATO, see below), implicate PML-NBs in human oncogenesis. Yet this is clearly not the only function of PML-NBs; they have been associated with a wide-range of nuclear activities (reviewed in 20, 83), including telomere maintenance by the ALT recombination pathway 21, 64, 84, apoptosis 85, DNA replication and repair 86, gene regulation and tissue-specific transcription 87, and viral infection response 88.
Relevant to the question of repair, PML-NBs were shown to co-localize with and recruit single-stranded DNA (ssDNA) molecules in response to exogenously induced DNA damage 89, 90. It is unclear if ssDNA foci are recruited into PML-NBs, or if damaged chromosomal DNA is processed into ssDNA once it is within a PML-NB. Importantly, the telomeric DNA recruited to PML-NBs for ALT are also linear, presumably containing stretches of ss TG-DNA rather than t-loops 91. The association and disassociation of repair and checkpoint signaling proteins with PML-NBs both prior to and after irradiation is complex 20, 90, 91, 92, 93, 94, 95. Nonetheless two points are noteworthy. The DNA damage response marker, γ-H2AX (histone H2AX phosphorylated on serine 139), which results from activation of ATM kinase, extends over large chromatin domains at radiation-induced breaks and colocalizes with PML-NBs hours after exposure to ionizing radiation 90, 92. Secondly, also at late time points radiation-induced MRE11 foci (IRIF), as well as p53, colocalize with PML-NBs. Given that the association of damage and foci occurs long after damage is induced, it is possible that PML-NBs recruit damage that could not be repaired by conventional NHEJ, requiring, for example, repair by recombination. This is supported by the further association of RAD51, BLM and MRE11 with PML, and the localization of recombination-mediated telomere maintenance at PML-NBs 21, 64. Consistently, pml−/− cells have a high rate of sister chromatid exchange 94, which also arises in cells defective for certain steps of HR. Consistent with the observation that PML-NB behavior depends strongly on the type of genotoxic stress incurred (e.g. severe alkylating DNA damage causes them to disperse 95), we propose that PML-NBs, like pores in yeast, cater to a specific subclass of recombination-mediated repair events.
PML: protein degraded by SUMO-dependent ubiquitination pathway
The link between the proteosome and Slx5/Slx8-mediated ubiquitination in yeast 52, appears to be conserved with PML and the human STUbL, RNF4. Indeed, PML was the first protein shown to be degraded by a SUMO-triggered, ubiquitin-mediated pathway, a pathway elucidated thanks to the exquisite sensitivity of APL to treatment by ATO 96. ATO enhances the conjugation of SUMO onto both PML and the PML-RARα fusion protein. Although different types of stress trigger increases in global SUMO conjugation levels, arsenic very specifically triggers sumoylation of PML (or the PML-RARα fusion). Recent work confirmed that the modified PML then becomes an in vivo target of the STUbL RNF4 96 97, 98, 99. Specifically, ATO-induced PML sumoylation correlates with its subsequent K48-linked polyubiquitination and degradation by the proteasome 100. This is thought to account for the chemotherapeutic efficacy of arsenic for APL.
Given that PML sumoylation recruits not only RNF4, ubiquitin and proteasomes to PML-NBs, but also other sumoylated proteins, these sites could physically integrate damage-dependent sumoylation, with ubiquitination and degradation, much like the proposed function of nuclear pores in yeast. Whether RNF4 activity leads to a particular pathway of repair, such as the strand-invasion required for ALT, is unclear for human cells. However, in yeast Slx5/Slx8 are indeed required for formation of the so-called “type II survivors” in telomerase-deficient cells, which reflects a Rad51-independent recombination pathway 61, 62. The exact relationship of these telomeric events with the role of Slx5/Slx8, sumoylation and the proteosome at collapsed replication forks or DSBs in yeast 53 is unclear, although they share the involvement of sumoylated proteins, STUbLs, and the nuclear pore. Similarly, ALT, SUMO, a STUbL and the proteosome all are found associated with PML-NBs. Although a fission yeast mutant lacking STUbL activity, can be complemented by the human orthologue, RNF4 48, it remains to be seen whether RNF4 itself regulates a recombinational pathway of repair.
Perspectives
The impact of nuclear organization in processes such as DNA repair is only beginning to be understood. We have summarized here how the spatial regulation of DSBs and DNA repair factors influences the maintenance of genome stability in yeast. In particular, SUMO pathway appears to play important roles in linking DNA repair and higher order nuclear organization, in both yeast and man. The yeast nuclear envelope — including pores and Mps3-determined sites — functions as a scaffold for various different DNA repair events, such as Slx5/Slx8-mediated pathways, the suppression of recombination among telomeric and rDNA repeats, replication fork restart, and telomerase-independent telomere maintenance. Perturbation of the scaffold structure, as well as the SUMO-modifying enzymes involved, leads to hazardous outcomes, including translocations, deletions and end-to-end telomere fusion event.
The pathways may have parallels in mammalian cells, although the best documented dependence on PML sumoylation and degradation in mammals arises from oxidative stress 20. The human STUbL, RNF4, is necessary for arsenic-induced PML degradation and the therapeutic response of acute promyelocytic leukaemia to this treatment. Whereas PML-NBs are also implicated in the response to DNA damage, the role played by RNF4 in repair remains to be determined. SUMO itself has long been suspected to play a role in recruitment of proteins to specific subnuclear compartments implicated in DNA repair, thus the profound defects in sumoylation-pathway homeostasis observed in STUbL mutants, suggest that subnuclear positioning and DNA repair by SUMO/ubiquitin-controlled pathways are linked. Further characterization of STUbL family members should give us a mechanistic understanding of the ways in which ubiquitination and sumoylation, DNA repair and subnuclear localization contribute to genome integrity. The notion that the three-dimensional organization of the nucleus and chromatin serves as a regulatory element for controlling homologous recombination or mechanisms that preserve genomic integrity, is a concept ripe for exploration.
Acknowledgments
The Gasser laboratory thanks the Novartis Research Foundation for continued support, as well as the Swiss National Science Foundation, NCCR programme “Frontiers in Genetics”. ND thanks the EU for a Marie Curie postdoctoral fellowship to the Gasser laboratory.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, et al. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, et al. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Nagai S, et al. The functional importance of telomere clustering: Global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast. The nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Shimada K, Oma Y, et al. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 2010;6:e1000910. doi: 10.1371/journal.pgen.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Kawabe Y, Seki M, Seki T, et al. Covalent modification of the Werner's syndrome gene product with the ubiquitin-related protein, SUMO-1. J Biol Chem. 2000;275:20963–20966. doi: 10.1074/jbc.C000273200. [DOI] [PubMed] [Google Scholar]
- Eladad S, Ye TZ, Hu P, et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol. 2006;8:1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, et al. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Melchior F. SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, van Veelen L, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Cortes-Ledesma F, Ira G, et al. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol. 2006;8:1032–1034. doi: 10.1038/ncb1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Lindroos HB, Strom L, Itoh T, et al. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Ampatzidou E, Irmisch A, O'Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, et al. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Roberts TM, Yang J, Desai R, Brown GW. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Yang L, Mullen JR, Brill SJ. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 2006;34:5541–5551. doi: 10.1093/nar/gkl685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jones GM, Prelich G. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics. 2006;172:1499–1509. doi: 10.1534/genetics.105.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy A, Calonge TM, Outwin EA, O'Connell MJ. Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem. 2007;282:20388–20394. doi: 10.1074/jbc.M702652200. [DOI] [PubMed] [Google Scholar]
- Stade K, Vogel F, Schwienhorst I, et al. A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J Biol Chem. 2002;277:49554–49561. doi: 10.1074/jbc.M207991200. [DOI] [PubMed] [Google Scholar]
- Soustelle C, Vernis L, Freon K, et al. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol Cell Biol. 2004;24:5130–5143. doi: 10.1128/MCB.24.12.5130-5143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Liu X, Garcia-Rubio M, et al. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell. 2007;18:2912–2923. doi: 10.1091/mbc.E07-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wu CY, Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biol. 2004;167:605–611. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadaroo B, Teixeira MT, Luciano P, et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol. 2009;11:980–987. doi: 10.1038/ncb1910. [DOI] [PubMed] [Google Scholar]
- Abdallah P, Luciano P, Runge KW, et al. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Lee JY, Abraham V, et al. Evidence that the S.cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res. 2006;34:506–516. doi: 10.1093/nar/gkj452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, et al. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Bystricky K, Van Attikum H, Montiel MD, et al. Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol Cell Biol. 2009;29:835–848. doi: 10.1128/MCB.01009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty E, Jensen K, Freemont P. PML nuclear bodies and their spatial relationships in the mammalian cell nucleus. Front Biosci. 2009;14:1182–1196. doi: 10.2741/3302. [DOI] [PubMed] [Google Scholar]
- Duprez E, Saurin AJ, Desterro JM, et al. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 1999;112 (Pt 3):381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, et al. Identification of three major sentrinization sites in PML. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Zhu J, Puvion F, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMorte VJ, Dyck JA, Ochs RL, Evans RM. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, et al. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- Kakizuka A, Miller WH, Jr, Umesono K, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- Koken MH, Puvion-Dutilleul F, Guillemin MC, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, et al. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Borden KL. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol. 2002;22:5259–5269. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Lallemand-Breitenbach V, Zhu J, de Thé H. PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Boe SO, Haave M, Jul-Larsen A, et al. Promyelocytic leukemia nuclear bodies are predetermined processing sites for damaged DNA. J Cell Sci. 2006;119(Pt 16):3284–3295. doi: 10.1242/jcs.03068. [DOI] [PubMed] [Google Scholar]
- Carbone R, Pearson M, Minucci S, Pelicci PG. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 2002;21:1633–1640. doi: 10.1038/sj.onc.1205227. [DOI] [PubMed] [Google Scholar]
- Fasching CL, Neumann AA, Muntoni A, Yeager TR, Reddel RR. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 2007;67:7072–7077. doi: 10.1158/0008-5472.CAN-07-1556. [DOI] [PubMed] [Google Scholar]
- Varadaraj A, Dovey CL, Laredj L, et al. Evidence for the receipt of DNA damage stimuli by PML nuclear domains. J Pathol. 2007;211:471–480. doi: 10.1002/path.2126. [DOI] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Ahmed K, et al. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol. 2006;175:55–66. doi: 10.1083/jcb.200604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- Conlan LA, McNees CJ, Heierhorst J. Proteasome-dependent dispersal of PML nuclear bodies in response to alkylating DNA damage. Oncogene. 2004;23:307–310. doi: 10.1038/sj.onc.1207119. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen Z, Lallemand-Breitenbach V, de Thé H. How acute promyelocytic leukaemia revived arsenic. Nat Rev Cancer. 2002;2:705–713. doi: 10.1038/nrc887. [DOI] [PubMed] [Google Scholar]
- Weisshaar SR, Keusekotten K, Krause A, et al. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 2008;582:3174–3178. doi: 10.1016/j.febslet.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Geoffroy MC, Jaffray EG, Walker KJ, Hay RT. Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol Biol Cell. 2010;21:4227–4239. doi: 10.1091/mbc.E10-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]