Abstract
NF-κB was first discovered and characterized 25 years ago as a key regulator of inducible gene expression in the immune system. Thus, it is not surprising that the clearest biological role of NF-κB is in the development and function of the immune system. Both innate and adaptive immune responses as well as the development and maintenance of the cells and tissues that comprise the immune system are, at multiple steps, under the control of the NF-κB family of transcription factors. Although this is a well-studied area of NF-κB research, new and significant findings continue to accumulate. This review will focus on these areas of recent progress while also providing a broad overview of the roles of NF-κB in mammalian immunobiology.
Keywords: NF-κB, TLR, TCR, BCR, lymphoid organogenesis, inflammation, hematopoiesis
Introduction
As discussed in the introduction to the January special issue of Cell Research, the discovery and characterization of the NF-κB family of transcription factors resulted from studies in two major areas of research: immunology and cancer biology. Although the role of NF-κB in cancer biology is becoming progressively better established, historically much of our current knowledge of NF-κB resulted from efforts directed at understanding its role in the regulation and function of the immune response. This review will attempt to provide a comprehensive discussion of the diverse functions of NF-κB in immunobiology and update our previous efforts in reviewing the far-reaching role of NF-κB in this area of biology 1, 2, 3.
The mammalian immune response comprises various mechanisms by which the physiological and functional integrity of the host is maintained in the face of microbial insult. The bone marrow-derived cells of the immune system are chiefly, though by no means solely, responsible for performing these functions. Thus, we begin by reviewing the role of NF-κB in the development of these hematopoietic cells that mediate both innate and adaptive immune responses. With the relevant cell types and tissues in place, the immune response is triggered by host recognition of foreign pathogens. Our knowledge in this area of the immune response has expanded rapidly in the past decade. Pathogen recognition is followed by pathogen clearance, a highly variable response that may draw upon coordinated responses at the level of cell, tissue, and the organism. Finally, the immune response must be resolved, damage must be repaired and, whenever possible, the triggering insult remembered for future immunity.
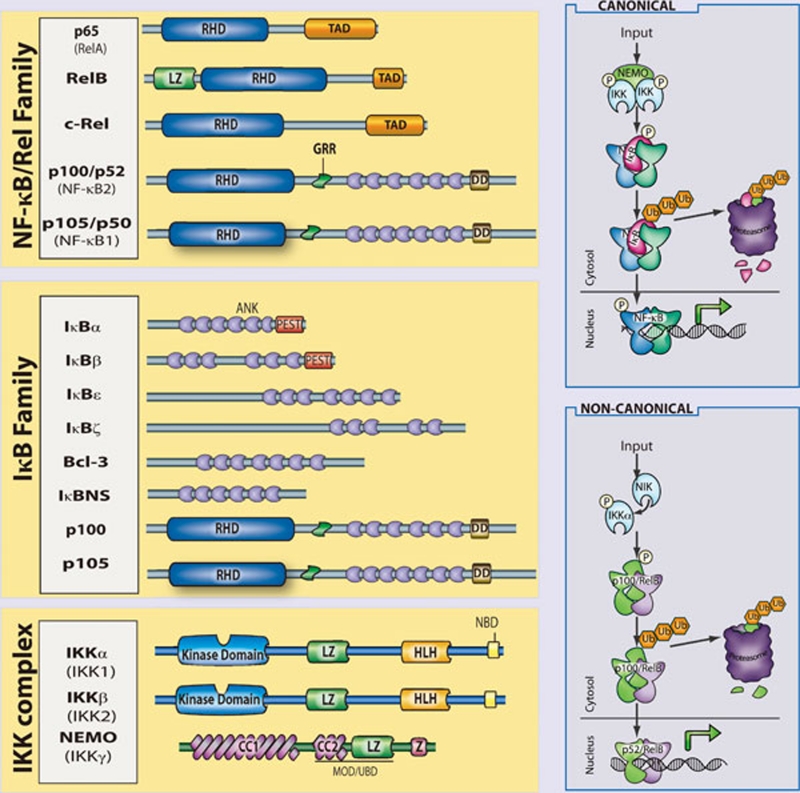
In mammals, the NF-κB family is composed of five related transcription factors: p50, p52, p65 (also RelA), c-Rel, and RelB (Figure 1). These transcription factors share an N-terminal DNA-binding/dimerization domain, known as the Rel homology domain, through which they can form homo- and heterodimers. NF-κB dimers can bind to a variety of related target DNA sequences called κB sites to modulate gene expression. RelB, c-Rel, and p65 contain C-terminal transcription activation domains (TADs) that enable co-activator recruitment and target gene expression. As they lack TADs, p50 and p52 can activate transcription by forming heterodimers with p65, c-Rel, or RelB, or by recruiting other TAD-containing proteins. However, as homodimers lacking the ability to drive transcription, they can repress transcription on binding to DNA.
Figure 1.
The mammalian NF-κB, IκB and IKK protein families. Relevant domains typifying each protein family are indicated and alternative nomenclatures are provided in parenthesis. The precursor proteins p100 and p105 function as both IκB and, when processed by the proteasome, NF-κB family members. Highly simplified schematics of canonical and non-canonical NF-κB pathways are shown. In the canonical pathway NEMO-containing IKK complexes are activated and induce phosphorylation and degradation of IκBα leading to the release of NF-κB dimers including p65:p50 dimers. In the non-canonical pathway, NEMO-independent activation of IKKα is mediated by the upstream kinase NIK. IKKα, with NIK, induces phosphorylation and processing of p100 to p52 resulting in the activation of predominantly p52:RelB complexes. Although they are illustrated separately here, as discussed in the text, signaling through these two pathways often is initiated by the same receptor – e.g., LTβR or CD40. (ANK, ankyrin-repeat domain; DD, death domain; RHD, REL homology domain; TAD, transactivation domain; LZ, leucine-zipper domain; GRR, glycine-rich region; HLH, helix-loop-helix domain; Z, zinc-finger domain; CC, coiled-coil domain; NBD, NEMO-binding domain; MOD/UBD, minimal oligomerization domain/ubiquitin-binding domain; PEST, proline, glutamic acid, serine, and threonine rich.)
In most cells, NF-κB complexes are inactive, residing predominantly in the cytoplasm in a complex with inhibitory IκB proteins (IκBα, IκBβ, IκBɛ, IκBζ, p100, p105, Bcl3, IκBns; Figure 1). When signaling pathways are activated, the IκB protein is degraded and NF-κB dimers enter the nucleus to modulate target gene expression. In almost all cases, the common step in this process is mediated by the IκB kinase (IKK) complex, which phosphorylates IκB and targets it for proteasomal degradation (see Liu and Chen, Cell Res 2011; 21:6–21). The IKK complex consists of two catalytically active kinases, IKKα (IKK1) and IKKβ (IKK2) and a regulatory scaffolding protein, NEMO (IKKγ Figure 1). These are the basic players in the NF-κB pathway; however, there is considerable complexity in how the individual proteins function and how they coordinate a nuanced NF-κB response (see Shih et al., Cell Res 2011; 21:86–102). The details of individual signaling pathways, the mechanism of action of upstream signaling intermediates, the role of various post-translational modifications, and cross-talk between NF-κB and other signaling pathways are covered in depth elsewhere in this special issue. Here, we focus on the function of NF-κB and the signaling pathways that regulate it in the mammalian immune response.
Development of the immune system
The mammalian immune system consists of a functionally linked group of anatomically disparate tissues and cell types. The dispersed cellular components of the immune system that arise from the bone marrow receive much of the attention in immunology and the study of NF-κB has likewise focused on these cells. However, lymphoid organs that facilitate coordination and dissemination of immune responses carried out by immune cells are also key sites of NF-κB function. Therefore, while this section is largely concerned with the role of NF-κB in hematopoiesis, the role of NF-κB in lymphoid organogenesis is also discussed briefly.
NF-κB and lymphoid organogenesis
NF-κB plays an important role in the development and function of primary (bone marrow, thymus) and secondary (lymph nodes, Peyer's patches, mucosal-associated lymphoid tissue, and the spleen) lymphoid tissues. There is clearly a role for NF-κB in the development and regulation of bone, and we refer the reader to the excellent review on the subject in this special issue (see Novack, Cell Res 2011; 21:169–182). The role of NF-κB in thymic architecture, originally thought to be limited to the important role of RelB in thymic architecture 4, has become clearer in terms of the development of medullary thymic epithelial cells 5, 6, 7, but still remains to be more fully elucidated 8. The secondary lymphoid tissues facilitate maintenance and activation of mature lymphocytes by providing an environment within which the interaction of lymphocytes and other leukocytes can be carefully orchestrated 9. The role of NF-κB in this process is now well appreciated. Multiple NF-κB knockouts exhibit defects in some aspect of secondary lymphoid organ development.
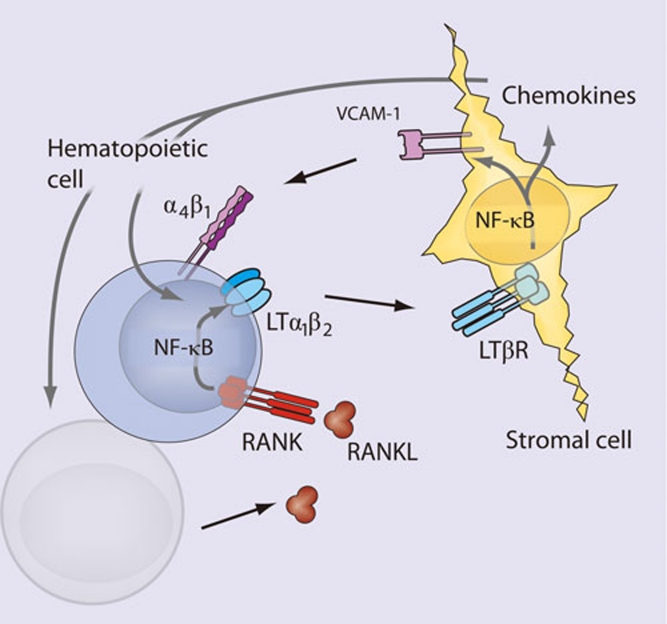
The initial events of lymphoid organogenesis involve the association of lymphotoxin (LT)α1β2-expressing hematopoietic cells and vascular cell adhesion molecule-1 (VCAM-1)-expressing stromal cells 10. This interaction initiates a positive feedback loop through NF-κB (Figure 2). LTα1β2, receptor activator of NF-κB ligand (RANKL), and TNFα are known to activate NF-κB, and figure prominently in lymphorganogenesis 10 (Figure 2). Also, mediators of lymphoid organogenesis and homeostasis, such as adhesion molecules (e.g., intercellular adhesion molecule, VCAM, peripheral node addressin, glycosylation-dependent cell adhesion molecule-1, and mucosal addressin cellular adhesion molecule (MadCAM)), cytokines (e.g., TNFα and LTα1β2) and organogenic chemokines (e.g., CXCL12 (GRO/MIP-2), CXCL13 (BLC), CCL19 (ELC), and CCL21 (SLC)), are regulated by NF-κB.
Figure 2.
NF-κB in lymphoid organogenesis. NF-κB regulates key components of a positive feedback loop between hematopoietic and stromal cells. LTα1β2-expressing hematopoietic cells induce production of VCAM-1 through the canonical NF-κB pathway and chemokines through the non-canonical pathway in LTβR-expressing stromal cells. Stromal expression of chemokines induces the upregulation of integrins (α4β1) on hematopoietic cells resulting in increased recruitment of LTα1β2-expressing cells, and increased signaling through stromal LTβR. RANKL signaling through RANK on the hematopoietic cell leads to activation of NF-κB, and further upregulation of LTα1β2.
Signaling through TNFR1, LTβR, and RANK activates p65-containing complexes and, hence, it is not surprising that rela−/−/tnfr1−/− double-knockout mice lack Peyer's patches and lymph nodes, and exhibit a disorganized spleen 11. The requirement for p65 in the development of these tissues is attributable to its function in stromal cells, and is likely because of a combination of effects: regulation of apoptosis (e.g., that induced by TNF); regulation of expression of organogenic factors, such as VCAM and LTα1β2; and enhancement of the non-canonical p52/RelB NF-κB pathway. The non-canonical NF-κB pathway has a well-established role in secondary lymphoid organ development. Deletion of components of the non-canonical NF-κB signaling pathway, including NIK, IKKα, LT, and RANK, all lead to severe defects in secondary lymphoid organogenesis 12, 13, 14, 15, 16, 17. The target of the non-canonical pathway, the p52/RelB dimer, is the primary transcriptional regulator of key organogenic factors including CXCL12, CXCL13, CCL19, CCL21, and MadCAM-1 18. Thus, the p52 single knockout lacks normal B-cell follicles, germinal centers, and Peyer's patches 19, 20, 21, while RelB knockouts lack Peyer's patches and exhibit compromised lymphnode development 18.
Splenic architecture is crucial for B-cell development as well as for the initiation and maturation of B-cell responses. The spleen is divided functionally and histologically into white and red pulp zones. Macrophages in the red pulp are responsible for destroying damaged erythrocytes, while the white pulp is populated by splenic lymphocytes organized into B-cell follicles and T-cell zones. Yet splenic architecture is also dynamic, as exemplified by the formation of germinal centers during the initiation and maturation of B-cell responses. As with other secondary lymphoid organs, NF-κB figures prominently in the development and maintenance of splenic architecture. Mice in which p65 has been deleted exhibit defects in both static and dynamic splenic architecture 11. Deletion of RelB, NIK, or IKKα leads to defects in splenic architecture, similar to those of ltbr−/− spleens 12, 13. Similar to p65/TNFR1 knockouts, mice with a defective non-canonical pathway fail to segregate B cell/T cell zones and they fail to form germinal centers following immunization. Marginal zone macrophages, which line the border between red and white pulp areas, are also absent or disorganized in RelB, p52, NIK, or IKKα knockouts 21, 22. Recent work has highlighted the role of LTα1β2 in the development and maintenance of the marginal sinus architecture, suggesting that the role of NF-κB in splenic architecture should perhaps be revisited with an eye toward an analysis of endothelial function. Some splenic defects are also attributable to effects on hematopoietic cells, most notably of the myeloid lineages.
In summary, both the canonical and non-canonical NF-κB pathways are required for the development of most secondary lymphoid organs (Figure 2). However, the non-canonical pathway, as assessed by examining mice deficient for IKKα, p52, NIK, or RelB, is especially important both during organogenesis and for maintenance of splenic architecture. Recent advances suggest that NF-κB signaling in endothelial cells, which are crucial regulators of cellular movement into and out of lymphoid organs, should be better characterized. The functional consequences of defects in these processes are severe and have direct ramifications for the ability of host to mount a robust immune response. As such, the function of NF-κB in the development and regulation of these organs represents an important facet of the role of NF-κB in immunobiology.
NF-κB and hematopoiesis
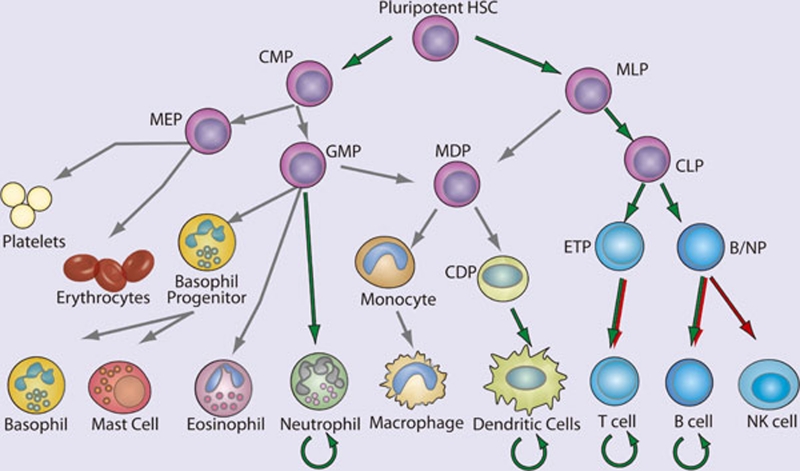
The immune system includes cells of the lymphoid and myeloid lineages: T cells, B cells, monocytes, macrophages, dendritic cells (DCs), natural killer (NK) cells, basophils, eosinophils, neutrophils, and mast cells (Figure 3). These bone marrow-derived cells are the core constituents of both the innate and adaptive immune responses. Proliferation, differentiation, and apoptosis are the defining characteristic of hematopoiesis, and NF-κB participates in the regulation of each of these processes. While the study of the role of NF-κB in development and homeostasis of hematopoetic cells has focused largely on B cell and T lymphocytes, it is likely that the NF-κB pathway is also important for the development of NK cells, DCs, monocytes, granulocytes, and other cellular components of the immune system. In general, an anti-apoptotic and pro-proliferative role for NF-κB is invoked in the hematopoietic system (Figure 3). Indeed, deletion of the kinase TAK1, an upstream component of both NF-κB and AP1 pathways, results in decreased antiapoptotic gene expression, hematopoietic stem cell apoptosis, and failure of hemtopoiesis 23. However, in actuality, the situation is much more complex. For example, ikba−/−ikbe−/− cells, which have elevated NF-κB activation are broadly defective in myelopoiesis and exhibit defects in NK cells and lymphoid lineages 24, 25. Thus, NF-κB function must be interrogated at each point of cellular differentiation in order to fully appreciate its contribution to hematopoiesis. Greater insight into the majority of events that lead to the development of immune cells, awaits both further characterization of the developmental processes and generation of better genetic tools to examine the consequences of perturbing NF-κB function.
Figure 3.
NF-κB in hematopoiesis. Red arrows indicate stages in which NF-κB activation is thought to contribute negatively and green arrows indicate a positive function in the development of the indicated lineages. Curved arrows indicate examples in which NF-κB contributes to the survival of cell population, either in the resting state or during immune responses. Gray arrows indicate developmental events for which NF-κB plays no role or for which the role of NF-κB has not been clearly demonstrated. (HSC, hematopoietic stem cell; CMP, common myeloid progenitor; MLP, myeloid/lymphoid progenitor; MEP, megakaryocyte erythrocyte progenitor; GMP, granulocyte monocyte progenitor; MDP, macrophage dendritic cell progenitor; CDP, common dendritic cell progenitor; CLP, common lymphoid progenitor; ETP, early thymic precursor; and B/NP, B-cell natural killer cell progenitor).
NF-κB in non-lymphocyte hematopoiesis
Although most, if not all cells of the body can exhibit some innate immune function, the ability to recognize microbes and initiate an antimicrobial response, we focus here only on the development of certain hematopoietic, non-lymphoid components of the innate immune system. In general, our understanding of the developmental origin of these innate cells lags far behind that of lymphocytes. Yet recent progress in characterizing the developmental process for several myeloid lineages should provide a better framework within which the contribution of NF-κB can be better assessed in the future.
Significant progress has been made recently in understanding DC development, including the characterization of a common DC progenitor 26, 27, 28, yet the contribution of NF-κB to early steps of DC lineage commitment remains poorly understood. The most notable contribution of NF-κB in this area is the requirement for RelB in development of the CD8α− DCs 4, 29, 30, 31. Although single knockouts of other NF-κB genes do not result in deficiencies in DC development, deletion of both p65 and p50 has a profound effect 32, 33. Again, however, the limitations of the experimental systems used to examine the contribution of NF-κB, particularly with regard to the need to either delete TNFR1 or utilize fetal liver transfer experiments, should be underscored. In the periphery, NF-κB is clearly required for DC maturation. Loss of IKKβ or inhibition of the IKK complex using a cell permeable peptide prevents DC maturation and antigen-presenting cell (APC) function 34, 35. In addition, peripheral differentiation and function of myeloid DCs, e.g., monocyte-derived DCs, is significantly impaired by NF-κB inhibition 36. DCs in the periphery survive for a short period of time following activation, but their survival can be prolonged by CD40L expression on T cells. CD40L activates both the canonical and non-canonical NF-κB pathways, and hence DCs deficient in both p50 and c-Rel, or DCs overexpressing a mutant super-repressor form of IκBα, demonstrate significantly decreased survival 32, 37. To date, the specific targets of NF-κB in DC development and, indeed, the role of NF-κB at individual stages of DC development, remain quite poorly characterized.
Genetic models of NF-κB deficiency and activation have revealed other roles for NF-κB in the myeloid lineage. Of the non-lymphocyte lineages, neutrophil development is perhaps best characterized in terms of the role of NF-κB. There remains, however, significant gaps in both our understanding of neutrophil development, and more so the role of NF-κB in this process. IκBα-knockout mice, which have elevated classical NF-κB activity, display robust granulocytosis 38. RelB-deficient mice exhibit an inflammatory phenotype with a marked increase in peripheral neutrophil numbers. Mice lacking c-Rel and p50, and heterozygous for p65, also exhibit neutrophilia 39. This increase in peripheral neutrophil numbers resulting from deletion of NF-κB appears to be related to both increased development and increased egress from the bone marrow 39. In mature neutrophils, the anti-apoptotic signal provided by NF-κB is crucial. In sharp contrast to lymphocytes, which are relatively long-lived in the absence of activation, neutrophils normally exhibit a very short lifespan but require protection from apoptosis during inflammation when they provide effector function. Neutrophils can respond by activating NF-κB in response to numerous pro-inflammatory stimuli, and the activated NF-κB can increase neutrophil survival 40. Although neutrophils are capable of activating NF-κB in response to many pro-inflammatory stimuli 41 they lack p52 and RelB 42, which are crucial for the maintenance of long-lived lymphocytes. Thus, in neutrophils the role of NF-κB is selective and cell type specific.
NF-κB in lymphopoiesis
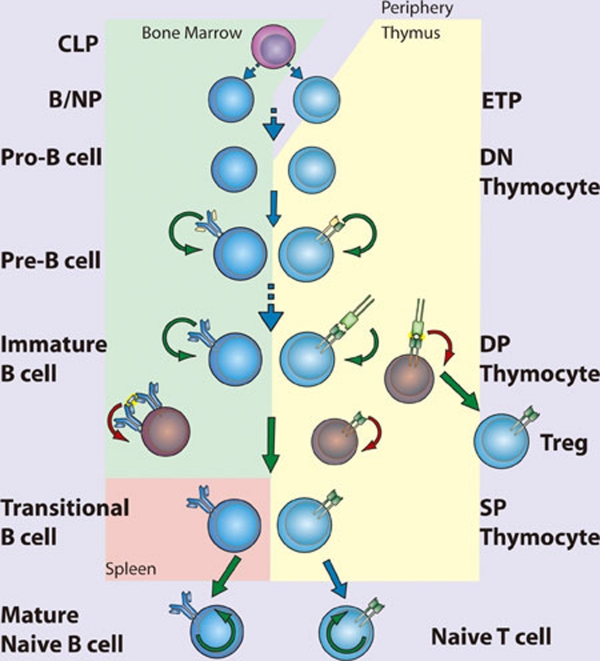
Development of T and B cells has, historically, been the subject of much greater scrutiny than the development of cells of the myeloid lineages. NF-κB has been examined in many aspects of lymphopoiesis and found to be vital for the development and function of adaptive immune cells 43 (Figure 4). Despite their potential longevity in the periphery, lymphocyte development is characterized by abundant apoptosis. As a consequence, the anti-apoptotic properties of NF-κB play a key role in lymphopoeisis. Indeed, in many instances the requirement for NF-κB can be overcome by transgenic expression of the anti-apoptotic factor Bcl-2 44. Indeed, the role of NF-κB in B cell development was recently re-examined 45. Although NF-κB was expendable in cells undergoing rearrangement of the eponymous Igκ loci, NF-κB was required to protect the resulting cells from apoptosis, and for these cells to progress to rearrangement of the Igλ locus. This deficit in NF-κB function could be circumvented through overexpression of Bcl-2 45. The necessity of NF-κB for lymphopoesis is strikingly illustrated in human genetic diseases in which the gene encoding NEMO is inactivated by mutation. Because the NEMO gene is located on the X chromosome, it is usually subject to random inactivation in individual cells in females. However, in female patients who are heterozygous for a mutant version of NEMO, all peripheral lymphocytes possess an intact NEMO gene, rather than the 50% predicted by random inactivation, suggesting that in the absence of NEMO-dependent NF-κB signaling, B and T cells fail to develop.
Figure 4.
NF-κB in lymphopoiesis. NF-κB plays a pro-survival role in common lymphoid precursor (CLP) cells which give rise to B- and T-cell lineages. B-cell development occurs in the bone marrow, where NF-κB protects pre-B cells from pro-apoptotic stimuli including TNFα. Signaling to NF-κB through the pre-B cell receptor mediates survival of Pre-B cells, which then undergo light chain recombination to produce a functional B cell receptor. NF-κB provides a necessary pro-survival signal during Igλ but not Igκ rearrangement. Expression of BCR leads to NF-κB-dependent differentiation into immature B cells. High levels of BCR signaling, i.e., through recognition of self-antigen, results in negative selection through the loss of NF-κB activity. Transitional B cells exit the bone marrow and migrate to the spleen, where they mature and differentiate, a process that also requires NF-κB. T-cell development occurs following migration of precursor cells into the thymus. Stimulation of NF-κB through pre-TCRα provides a pro-survival signal allowing recombination of the TCR α chain and maturation to the double-positive (DP) stage. Optimal signaling through the TCRα/β complex induces NF-κB-dependent survival pathways, while a failure to signal or high level signaling results in death by neglect or negative selection, respectively. Intermediate high NF-κB activation facilitates intrathymic regulatory T cell (Treg) development. NF-κB activity is required for the maintenance of long-lived B and T cells. (CLP, common lymphoid progenitor; ETP, early thymic progenitor; DN, double negative – CD4−CD8−; DP, double positive – CD4+CD8+; SP, single positive – either CD4+CD8− or CD4−CD8+)
The effects of NEMO inactivation in both mice and humans solidify the role of NF-κB in lymphopoiesis, although the details by which NF-κB functions in this process remain obscure. NF-κB plays diverse roles in lymphocyte development that can be grouped according to timing – that is, before, during, or after pre-antigen receptor signaling. Although no single NF-κB-subunit-knockout mouse has as severe a phenotype as NEMO knockouts with regard to the generation of mature lymphocytes, double knockouts of Rel proteins confirm the essential anti-apoptotic function of NF-κB. For example, loss of both p50 and p65, or both p65 and c-Rel, terminates lymphopoiesis before expression of the pre-antigen receptors 46, 47, suggesting that NF-κB regulates anti-apoptotic factors required for early lymphoid cell survival in response to pro-apoptotic stimuli (Figure 4). In fact, hematopoietic stem cells can activate NF-κB in response to TNFα, and in these cells NF-κB acts as a pro-survival factor 48. The role of NF-κB in early lymphocyte development seems clear – expression of either pre-T-cell receptor (pre-TCR) 49 or pre-BCR (pre-B-cell receptor) coincides with increasing NF-κB activity and induction of anti-apoptotic signals through NF-κB 50, 51. However, it remains unclear how NF-κB is activated downstream of the pre-AgR as the nature of the signaling pathway at play remains almost entirely uncharacterized. Whether NF-κB contributes to lineage choice made at these early stages or merely promotes survival is also unknown.
The extent of NF-κB activation serves as a rheostat in the selection of DP (double positive; CD4+CD8+) thymocytes (Figure 4). TCR-mediated NF-κB activation follows binding to peptide:major histocompatibility complex (MHC). A thymocyte that expresses a TCR that cannot bind MHC succumbs to “death by neglect”, whereas those that bind peptide:MHC are either positively or negatively selected depending on the strength of signaling. Thymocytes that bind self-peptide:MHC with very high affinity, are likely to be self-reactive, and hence are deleted through negative selection. Thus, only DP thymocytes that recognize self-peptide:MHC and signal within a defined range are positively selected and become single-positive (SP) T cells. During negative selection, NF-κB facilitates the induction of apoptosis following high affinity TCR ligation 52, perhaps by facilitating expression of pro-apoptotic genes and consequent sensitization to pro-apoptotic signals 53. The role of NF-κB in positive selection of thymocytes is more in keeping with the better-established role of NF-κB as an inducer of anti-apoptotic genes. Unlike in thymocytes, however, NF-κB functions as a pro-survival factor during negative selection of B cells. Immature B cells display constitutive NF-κB activity that is downregulated following BCR ligation 54 (Figure 4). Decreased NF-κB activity might then sensitize these cells to pro-apoptotic signals. Interestingly, some signaling components required for NF-κB activation in mature B and T cells can be genetically disrupted without affecting their development, suggesting that pathways leading to activation of NF-κB in developing B or T cells differ significantly from the pathways engaged following AgR ligation in mature lymphocytes.
Following positive and negative selection, DP thymocytes must make a lineage commitment as SP thymocytes (CD4+CD8− or CD4−CD8+) and thereafter emigrate from the thymus. This process requires NF-κB as deletion of NEMO in cd4-Cre mice results in loss of mature peripheral T cells 55. Equivalent deletion of the upstream kinase TAK1 has a similar outcome 56. The exact nature of NF-κB pathway requirement is somewhat unclear, as ikkb−/− chimeras, or cd4-Cre IKKβ conditional knockouts, are not defective in the production of naive T cells 55, 57. Further, the contribution of NF-κB to CD8 and CD4 lineages is not equivalent. CD8 SP cells have significantly higher levels of NF-κB activity than CD4 SP thymocytes 49, 58, yet the anti-apoptotic factor Bcl-2 is more highly expressed in CD4 than CD8 cells. Therefore, CD8 SP thymocytes are dependent on NF-κB for survival, while CD4 SP thymocytes are not 58. NF-κB is, however, clearly important in CD4 SP cell development, and forced activation of NF-κB in CD4 SP cells results in negative selection 58.
In addition to peripheral maturation and differentiation of T cells into various subsets, thymic differentiation into both TH17 cells and regulatory T cells (Tregs) is known to occur. The transcriptional control of TH17 cell differentiation and the role of thymic TH17 remain relatively obscure. One recent report has described an important role for IκBζ in TH17 differentiation 59. In this capacity, IκBζ acts with the nuclear orphan receptors RORγ and RORα to promote IL17A gene expression. Beyond this tangential role in TH17 development, a TH17-intrinsic role for NF-κB has been minimally interrogated. Instead the role of NF-κB in Treg development 60 is somewhat better understood. The differentiation of thymocytes into Foxp3/CD25-positive Tregs occurs in the thymus and depends on NF-κB. It is thought that NF-κB acts as a pioneer transcription factor enhancing accessibility of the foxp3 locus 61, 62, 63, 64.
Immature B cells complete development into either follicular or marginal zone B cells after exiting the bone marrow as transitional B cells. NF-κB-regulated expression of pro-survival factors is important to these final steps of B-cell development 65. Signaling through the TNFR superfamily member BAFFR is crucial for transitional B-cell survival through induction of anti-apoptotic Bcl-2 family members 66, 67, 68. Thus, BAFF-knockout mice exhibit a complete failure of transitional B-cell maturation, which mirrors that seen in Bcl-XL-knockout mice 66, 69, 70. BAFF activates both non-canonical and canonical NF-κB pathways, though the former is principally responsible for the anti-apoptotic function in transitional B cells. Nevertheless, deficiency in NEMO, IKKα, or IKKβ decreases the numbers of mature B cells 57, 71, 72. Likewise, p50/p52 or p65/c-Rel double-knockout progenitor cells are defective in their ability to mature beyond the transitional B-cell stage 47, 73. A requirement for both the canonical and non-canonical NF-κB pathways may explain why deletion of p50 and p52 produces a more complete block in B-cell development than loss of p65 and c-Rel. Thus, only those knockouts that target both the canonical and non-canonical NF-κB pathways have an effect that approximates the phenotype seen in BAFF or Bcl-XL deficiency. Constitutive hyperactivation of the canonical pathway can circumvent BAFF-R deletion 74, yet it remains unclear whether the canonical pathway merely supports the non-canonical pathway 75, or has an independent function.
NF-κB in innate immunity
Pattern recognition receptors
Recognizing pathogens is an absolute requirement for the initiation of effector functions. Mammals express several diverse classes of pattern recognition receptors (PRRs) that are dedicated to this purpose. These PPRs include transmembrane Toll-like receptors (TLRs), cytoplasmic Nod-like receptors (NLR) and RIG-I like receptors (RLR), scavenger receptors, C-type lectins, and the complement system. Although epithelial cells are frequently the first to encounter pathogens, they are also constantly exposed to non-pathogenic microbes. Therefore, while a variety of PRRs are differentially expressed in epidermis, gut, pulmonary, urinary, and reproductive epithelium, their expression and responsiveness is tightly controlled. Instead sentinel cells of the innate immune system, particularly tissue resident DCs and macrophages, express a more complete complement of PRRs, and exhibit a higher propensity to signal on exposure to pathogens.
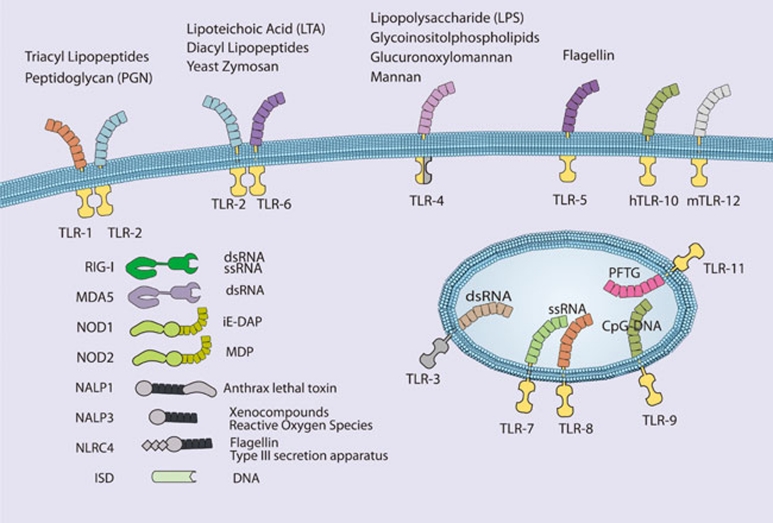
TLRs are evolutionarily conserved pattern recognition receptors that recognize unique, essential molecules characteristic of various classes of microbes 76. The 11 characterized mammalian TLRs have varied tissue distribution and serve as recognition receptors for pathogen-associated molecular patterns (PAMPs) present on bacteria, viruses, fungi, and parasites. Perhaps because of the modular nature of the TLR extracellular domain, which consists of multiple leucine-rich repeats (LRRs), these receptors are capable of recognizing more than one microbial molecule (Figure 5). Heterodimerization of TLRs further expands the repertoire of recognized molecules. In general, the ability of TLRs to distinguish between pathogen types is translated into appropriate innate and adaptive responses through the selective activation of NF-κB, IRFs, and other inducible transcription factors. TLR signaling pathways are complex and have been extensively reviewed elsewhere 2, 77, 78. In general, TLR signaling is often divided into MyD88-dependent and TRIF-dependent pathways: MyD88 signaling predominantly leads to the activation of NF-κB, while TRIF signaling leads to both IRF3 and, to a lesser extent, NF-κB activation.
Figure 5.
Pattern recognition receptors and their cognate ligands. TLRs 3, 7, 8, 9 and 11 have been reported to exhibit endosomal or intracellular localization while NOD1, NOD2, RIG-I, MDA-5, NALP1, NALP3, NLRC4, and the intracellular DNA sensor (ISD) function in the cytoplasm. Only a partial list of ligands or classes of ligands for each receptor is given.
In recent years, there has been significant activity in characterization of cytoplasmic PRRs, particularly those recognizing viral nucleic acids. These efforts followed the initial description of nucleotide oligomerization domain proteins (NOD), NOD1 and NOD2. NLRs, of which NOD1 and NOD2 are the best-known examples, are characterized by LRRs and NODs. NOD1, NOD2, and IPAF have CARD domains and can signal to NF-κB (see Blonska and Lin, Cell Res 2011; 21:55-70). NOD1 recognizes a peptidoglycan containing meso-diaminopimelic acid and induces NF-κB through a canonical pathway that includes activation of IKKβ. NOD2 recognizes muramyl dipeptide (MDP), a ubiquitous component of nearly all bacterial cell walls. The CARD-containing kinase RIP2 is required for NF-κB activation. The function of RIP2 tyrosine kinase activity is unclear, although it was recently shown that kinase inhibitors could prevent MDP-induced cytokine responses 79. Although RIP2 binds to NEMO, it is not thought that RIP2 tyrosine kinase activity mediates IKK phosphorylation directly. Instead it has been proposed that RIP2 directly mediates activation of the IKK complex through NEMO binding and induced proximity 80.
Retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) are DExD/H-box RNA helicase-containing cytoplasmic proteins that bind directly to dsRNA 81, 82, 83. The details of RNA binding to RIG-I and MDA5 continue to be the subject of intense research. Thus, it was thought that RIG-I recognizes dsRNA replicative intermediates of negative stranded single-stranded RNA (ssRNA) viruses, while MDA5 responds to long dsRNA replicative intermediates of plus strand ssRNA viruses 84, 85, 86, 87. Indeed, the structure of RIG-I bound to short dsRNA products has recently been solved 88, 89. RIG-I has also recently been implicated in the recognition of DNA viruses by binding short dsRNAs generated by RNA polIII 90. However, it has been shown that RIG-I can directly recognize ssRNA genomes containing 5′ triphosphates, and suggested that for negative stranded RNA viruses, this may be the relevant PAMP 91. On binding to either the viral genome, viral replication intermediate, or RNA product, RIG-I, and MDA5 induce the activation of IRF3 and NF-κB. The CARD-containing protein MAVS (also known as CARDIF, IPS1, and VISA) mediates signaling from RIG-I 92, 93, 94, 95. RIG-I and MDA5 differentially recognize various groups of RNA viruses and are independently required for robust antiviral responses 86. Therefore, together with TLRs, RLRs provide an additional mechanism of sensing viral infection and mediating type-I IFN (IFN-α/β) responses.
Pathogen recognition in innate immunity
Pathogens recognized by PRRs can be categorized as bacterial, viral, or eukaryotic, and each of these categories has several well-described PAMPs 96. Both in terms of accessibility and uniqueness to prokaryotes, the bacterial cell well is the most abundant source of PAMPs for TLRs and other PRRs. Lipoproteins, glycolipids, and protein components of bacterial cell wells have all been shown to function as PAMPs. The TLR4 ligand LPS, a glycolipid component of the outer membrane of Gram-negative bacteria, is the most thoroughly studied PAMP and the most potent TLR ligand known. Trace amounts of LPS activates the innate immune system via TLR4, leading to the production of numerous proinflammatory mediators, such as TNFα, IL-1, and IL-6. TLR4-mediated responses to LPS require CD14 and MD-2.
Conserved differences in bacterial nucleic acid structures can also be recognized by the innate immune system. TLR9 recognizes bacterial DNA containing unmethylated CpG motifs, and TLR9-deficient mice are not responsive to CpG DNA challenge 97. The low frequency and high rate of methylation of CpG motifs prevent recognition of mammalian DNA by TLR9 under physiological circumstances. A recent report indicated that the intracellular, endosomal restriction of TLR9 is critical for discriminating between self and non-self DNA, as host DNA, unlike microbial DNA, does not usually enter the endosomal compartment 98.
Nucleic acids are also key viral PAMPs, and are recognized by TLRs 3, 7, 8, and 9, as well as by RLRs. Signaling from viral PAMPs results in the activation of NF-κB and IRFs, which cooperatively mediate the production of IFN-α/β 99, 100. TLR3 recognizes dsRNA, a common viral replicative intermediate 101. TLR7 and TLR8, initially found to recognize synthetic antiviral compounds 102, 103, recognize guanosine- or uridine-rich ssRNA derived from RNA viruses 104, 105, 106. Interestingly, mammalian RNA is significantly less stimulatory than bacterial RNA, suggesting that nucleoside modifications facilitate discrimination between endogenous and pathogen RNA 107. In addition, subcellular localization of TLR7 and TLR8 to endocytic compartments, and limited constitutive cell type expression, may also facilitate self/non-self discrimination by minimizing exposure to host RNA. TLR9 recognizes viral CpG sequences and induces the induction of IFN-α 108, 109, 110. However, as membrane restriction prevents TLRs from sampling the cytosol where much of the viral lifecycle occurs, cytosolic PRRs provide comprehensive innate immune recognition. Thus, TLR3 and the adaptor TRIF are not required for viral induction of type I IFN in many tissues and cell types 111, although specialized innate antiviral cells, such as plasmacytoid DCs, rely more heavily on TLR mediated recognition of viral nucleic acids 112. Recognition of cytoplasmic dsDNA leading to NF-κB activation and type I interferon production has also been reported 113, 114. Although several candidates have been found, the relevant receptor has not yet been conclusively identified 115. This receptor(s) is predicted to be important for type I IFN production in response to viruses and intracellular pathogens, such as Listeria sp.
Although research on the recognition of PAMP/PRR pairs is an ongoing process of discovery, defective responses of MyD88-deficient cells indicate that many fungal and parasite species are capable of activating TLR pathways. TLR4 has been shown to recognize Aspergillus hyphae 116, and Cryptococcus neoformans capsular polysaccharide 117. TLR2 and TLR6 are required for recognition of yeast zymosan, while TLR4 is thought to recognize certain yeast mannans. The identification of parasite PAMPs has been more elusive, and their existence is somewhat controversial. Nevertheless, TLR2 heterodimers recognize various parasite GPI-anchored proteins and glycoinositolphospholipids from the parasitic protozoa Trypanosoma cruzi 118. TLR9 was initially reported to recognize the malarial pigment hemozoin, a byproduct of heme metabolism in infected erythrocytes 119, although subsequent studies suggest that DNA associated with hemozoin is the relevant TLR9 ligand 120. Instead, hemozoin may be recognized by the NLRP3 inflammasome 121. TLR11 recognizes a profilin-like protein that is conserved in apicomplexan parasites including Toxoplasma gondii 122. Recognition of helminths remains an area of controversy 123. In particular, it remains unclear how helminth PAMPs might preferentially induce TH2 responses that are characteristic of the immune responses to this class of pathogen. Direct skewing of adaptive responses by the mechanism of innate recognition is suggested to occur in other scenerios as well 124. For example, C-type lectins can directly affect TH activation and differentiantion 125, thus recognition of C. albicans α-mannans by dectin-2 induces a protective TH17-predominant response 126. Therefore, whether through cell type or PRR specificity, the recognition of specific PAMPS can shape the nature of the subsequent innate and adaptive immune responses.
Immediate anti-microbial responses
PAMP recognition initiates a complex series of events: the first is the mounting of immediate antimicrobial responses at the cellular level. This effective and evolutionarily conserved function of PRRs is dependent on activation of NF-κB gene programs. In particular, much of the early innate response has been demonstrated to depend on the canonical NF-κB pathway. Thus, rela−/−/ tnfr1−/− and ikbkb−/−/tnfr1−/− double-knockout mice have increased susceptibility to bacterial infection 57, 127, 128, 129. MEFs from Nemo-deficent mice do not exhibit NF-κB activation by LPS or IL-1 130. Therefore, activation of NF-κB responsive genes by the innate immune system depends on NEMO and likely progresses through the canonical NF-κB signaling pathway. Early anti-microbial effectors include NF-κB-dependent expression of defensins, which are cationic peptides that exert direct bactericidal activity by inducing membrane permeabilization. The classic mediators of defensin release are small intestinal Paneth cells, which secrete α-defensins into the intestinal lumen following exposure to bacterial PAMPs 131. The production of anti-microbial nitrogen and oxygen species, which are acutely toxic to a variety of microbes, augments the activity of anti-microbial peptides. Production of nitric oxide (NO) is mediated in part by inducible NO synthase (iNOS), which is coordinately regulated by NF-κB and STAT transcription factors 132.
Inflammation
Inflammation begins with epithelial or stromal cells of the infected tissue or tissue resident hematopoietic cells such as mast cells or DCs recognizing an inflammatory stimulus and propagating pro-inflammatory signals. These signals lead to the recruitment and activation of effector cells, initially neutrophils and later macrophages and other leukocytes, resulting in the tissue changes characteristic of inflammation – rubor, calor, dolor, and tumor (redness, heat, pain, and swelling, respectively). There is a vast amount of literature that correlates NF-κB activation with inflammation in a wide array of diseases and animal models. There are, likewise, numerous studies using gene targeting and inhibitors of NF-κB that have established the causative role of NF-κB in inflammatory processes.
NF-κB is responsible for the transcription of the genes encoding many pro-inflammatory cytokines and chemokines. The pathway from pathogen recognition to pro-inflammatory cytokine production demonstrates a particular reliance on NF-κB. The immediate targets of NF-κB-dependent pro-inflammatory cytokines, such as TNFα, tend to be receptors that, in turn, activate NF-κB. Therefore, NF-κB is crucial to the propagation and elaboration of cytokine responses. TNFα is particularly important for both local and systemic inflammation, and it is a potent and well-studied inducer of NF-κB. One important early target of these effectors is the vascular endothelium. Changes in vascular endothelial cells both recruit circulating leukocytes and provide them with a means of exiting the vasculature into the infected tissue. NF-κB regulates the expression of adhesion molecules, both on leukocytes and on endothelial cells, which allow the extravasation of leukocytes from the circulation to the site of infection 133. Thus, in the absence of p65, the recruitment of circulating leukocytes to sites of inflammation is impaired 127.
Recruited neutrophils are the key mediators of local inflammation and, as mentioned above, NF-κB is important for their survival and function under relatively toxic conditions 134. NF-κB is important for the production of the enzymes that generate prostaglandins and reactive oxygen species (e.g., iNOS and Cox, both NF-κB target genes) and may, furthermore, be involved in the signaling induced by prostaglandins 135, 136. NF-κB has also been implicated in the response to leukotrienes, which similar to prostaglandins are short-lived paracrine effectors. For example, leukotriene-induced IL-8 production appears to be dependent on rapid activation of NF-κB 137. Finally, matrix metalloporteinases are also crucial mediators of local inflammation and leukocyte chemotaxtis, and their expression is also regulated by NF-κB 138, 139, 140.
Resolution of inflammation and subsequent tissue repair is a crucial event, and its failure is a common source of pathology. Resolution of inflammation is an active process that is as complex as the inflammatory response itself, and involves numerous pathways that are not all directly relevant to NF-κB 141. Macrophages are key regulators of both inflammation and its resolution, and NF-κB plays a key role in the instruction of macrophage responses. Macrophage differentiation into pro-inflammatory (M1) or anti-inflammatory (M2) cells depends both on cell intrinsic NF-κB pathway activation as well as the local cytokine milieu, which is strongly influenced by NF-κB.
During acute inflammation, there are multiple negative feedback pathways that help to rein in inflammatory responses 3. It has long been known that cells, such as macrophages, become resistant to repeated pro-inflammatory stimuli 142. The IκB family protein Bcl-3 can function both as an inhibitor and mediator of NF-κB transcriptional programs 2. Following LPS stimulation, Bcl-3 is induced and binds p50 dimers, with which it can repress the expression of a subset of NF-κB target genes. In this manner, Bcl-3 mediates the selective inhibition of repeated LPS responses in macrophages 143. Although clearly not the only mechanism, induced transcriptional repression by Bcl-3/p50 likely contributes to LPS-tolerance. Furthermore by selectively affecting chromatin remodeling, Bcl-3 mediates repression of pro-inflammatory genes, and also facilitates the expression of the anti-inflammatory gene IL-10. Thus, it appears that Bcl-3 acts appropriately in the regulation of genes that are categorized as either “tolerizable or “non-tolerizable” 144. NF-κB p50 appears to be capable of mediating anti-inflammatory responses under several additional circumstances as well. For example, p50 negatively regulates IFNγ production by and proliferation of NK cells 145. Also, expression levels of p50 in M2-like tumor associated macrophages (TAMs) regulate the balance between anti- and pro-inflammatory functions. Thus, it was shown that TAMs overexpress p50 and that deletion of p50 renders these macrophages pro-inflammatory 146.
Inhibition of NF-κB during the resolution phase of inflammation can prolong the inflammatory process and prevent proper tissue repair 147. Furthermore, IKKα-deficient mice display increased inflammatory responses in models of local and systemic inflammation 148, and show increased production of pro-inflammatory chemokines and cytokines 148, 149. Rather unexpectedly, IKKα-deficient mice and embryonic-liver-derived macrophages from IKKα-deficient mice exhibit augmented inflammatory responses compared with wild-type mice 149. The mechanisms of IKKα-mediated repression of transcriptional responses seem to be through effects on the level of nuclear p65 and c-Rel 148. Although there are significant differences between these two reports with regard to the proposed mechanism of action, both, nevertheless, observe the same biological consequence of IKKα deficiency in macrophages. Furthermore, recent work has suggested that IKKβ also exhibits some anti-inflammatory capacity in macrophages. First, IKKβ suppresses the secretion of the potent pro-inflammatory cytokine IL-1β 150. Second, IKKβ is associated with the maintenance of an anti-inflammatory M2-like phenotype in TAMs 151. Third, IKKβ is thought to inhibit M1 macrophage function by interfering with the STAT pathway during infection 152.
Initiation of adaptive responses
The innate immune system invokes potent anti-microbial activities and maintains host protection under many settings. Nevertheless, initiating the adaptive immune response remains a crucial step for robust and durable pathogen clearance. The process of information transfer from the innate to adaptive immune system is mediated by activation and maturation of APCs. The nature of the PAMP and the signaling pathways activated by the cognate PRR, as well as the context within which pathogen recognition occurs, all influence the manner in which the APC instructs T and B cell responses.
DC maturation mediated by pathogen recognition is crucial for the initiation of the adaptive immune response. To activate naive T cells, DCs must alter their chemokine receptor expression to enable migration into lymphoid tissues. During activation DCs fine-tune their antigen processing machinery such that the presentation of pathogen epitopes is favored. Finally, the expression of co-stimulatory molecules including B7.1/B7.2 (or CD80/CD86) is upregulated. These co-stimulatory molecules ligate the co-stimulatory molecule CD28, thus providing the second signal necessary to induce full T-cell activation. The response to pathogens is tailored on the basis of the distribution of PRRs in different cell types, and the ability of different cell types to, in turn, interact with T cells in a biasing manner 153.
There continues to be debate surrounding how PRRs differentially induce TH1 versus TH2 responses. Maturation of DCs following viral infection depends on nucleic acid-binding PRRs 154, 155. Plasmacytoid DCs, which are robust inducers of interferon during viral infection, express viral PRRs including elevated expression of TLR7 and TLR9. Cytoplasmic nucleic acid-sensing PRRs, RLRs, are expressed more broadly, although they are further induced by type-I interferons, and they may, therefore, provide crucial amplification of antiviral responses following virus recognition by TLRs. While studies of anti-viral responses suggest that both cell type-specific distribution of PRRs, and selective activation of signaling pathways by certain PAMPs are key determinants of TH1 versus TH2 responses, the situation is less clear for other pathogens. It appears that multiple mechanisms converge in shaping how APCs interact with the adaptive immune system.
Role of NF-κB in the adaptive response
T-cell responses mediated by NF-κB
T-cell activation, differentiation, proliferation, and effector function are all influenced by gene programs regulated by NF-κB. The majority of effort directed at understanding the role of NF-κB in T cell activation has focused on CD4+ T cells (Figures 6 and 7). On the whole, the historical lack of CD8+ T-cell conditional knockouts and the selective loss of CD8 cells in the absence of functional NF-κB have impeded thorough characterization of the role of NF-κB in these cells. Nevertheless, there continues to be progress in this area and, in fact, there has been a resurgence of interest in recent years. CD4+ T cells have long been known to undergo differentiation into either TH1 or TH2 subsets depending on the cytokines present during activation 156. However, an appreciation of the ability of CD4+ helper T cells to differentiate into additional effector cell types, induced regulatory T cells, pro-inflammatory TH17 cells, TH22 cells TH9 cells, has prompted a fresh look at the contribution of NF-κB to T cell differentiation and effector function (Figure 7).
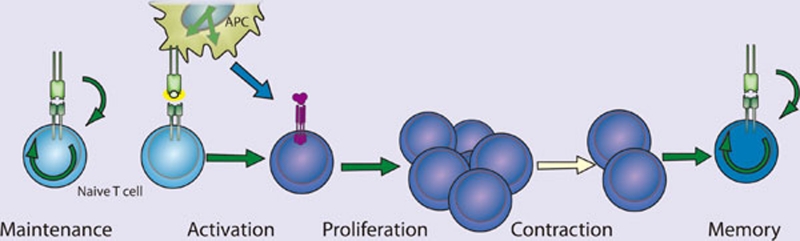
Figure 6.
NF-κB in T cell activation. NF-κB participates in the maintenance, activation, differentiation and proliferation of naive T cells. Tonic TCR stimulation promotes T cell maintenance, of both memory and naive cells, through NF-κB activation. Activation occurs when the naive T cell recognizes its cognate antigen presented by an activated APC expressing both peptide:MHC and B7 family co-stimulatory molecules. NF-κB-dependent proliferation and differentiation ensue and are influenced by the local cytokine milieu. NF-κB supports proliferation, differentiation and survival as indicated (green arrows).
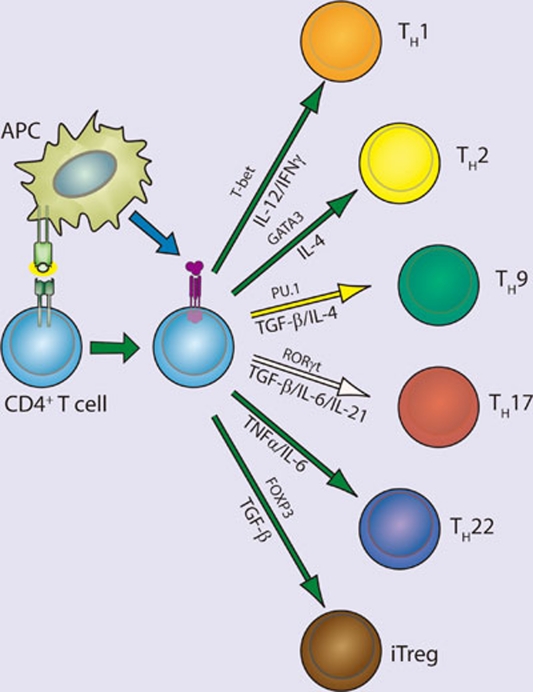
Figure 7.
TH cell differentiation. NF-κB participates in differentiation of several TH cell types following activation of naive CD4+ TH cells. Differentiation pathways in which NF-κB is implicated are indicated with a green arrow. Key transcription factors in each differentiation pathway are indicated above each arrow, while cytokine responsible for skewing TH cells toward a given pathway are indicated below each arrow. Additional cytokines and transcription factors implicated in several of the pathways are not depicted.
Activation of naive T cells requires antigen-specific and co-stimulatory signaling provided by activated APCs. Binding of the TCR to its cognate peptide presented in the binding cleft of MHC supplies the antigen-specific signal, while co-stimulatory signaling results from ligation of CD28 by B7 molecules expressed on activated APCs. Activated naive T-cells proliferate rapidly while simultaneously differentiating into effector cells (Figure 6). In the case of CD4+ T cells, proliferation leads to differentiation into immature effector cells, TH0, which subsequently differentiate into TH1, TH2, TH9, TH17, TH22, or inducible Treg (iTreg) cells depending on the cytokine milieu.
Stimulation of T cells through TCR and CD28 results, initially, in activation of p65-containing NF-κB complexes followed by delayed and sustained activation of c-Rel complexes 157, 158, 159, 160. Rapidly proliferating activated T cells rely on NF-κB activity for protection from apoptosis as well as for the production of cytokines, especially IL-2, supporting proliferation and differentiation (Figure 6). As expected, inhibition of NF-κB in activated T cells facilitates progression toward activation-induced cell death (AICD) or apoptosis 161, 162, and stimulation of p65-deficient naive T cells induces cell death 163. In contrast, T cells lacking c-Rel do not undergo apoptosis, but nevertheless, fail to proliferate in response to typical mitogenic stimuli 164. T cells, in which p105 cannot undergo inducible processing to p50 also fail to proliferate normally 165. In vitro, c-Rel is needed for production of IL-2 and therefore, for proliferation, but this need is not recapitulated in vivo 166. Interestingly, c-Rel-deficient T cells appear to have a defect in TH1 proliferation and production of IFNγ, indicating a selective role for NF-κB family members in TH1/TH2 differentiation, independent of that mediated by the innate response.
Multiple transcriptional activators and repressors regulate expression of IL-2. Amongst these, members of the NF-κB family play multiple roles. In naive T cells, which do not express IL-2, repressive p50 homodimers are found associated with the IL-2 promoter 167. Failure of T-cell proliferative responses in c-Rel-knockout mice is attributable to a failure to produce IL-2 164. In naive T cells, c-Rel is responsible for mediating chromatin remodeling across the IL-2 locus following CD3/CD28 co-stimulation 158. Thus, naive T cells can be primed by activation of c-Rel by pro-inflammatory cytokines, such as those elicited following stimulation with TLR ligands 157. Priming, which requires NF-κB activation, results in a more robust response to CD3/CD28 co-stimulation 168. Conversely, overexpression of p65 with c-Jun can overcome the requirement for co-stimulation in naive T cells 169.
TH0 differentiation into TH1, TH2, TH9, TH17, Treg effector cells depends on the induction of specific transcription factors, namely T-bet, GATA3, PU.1, RORγt, and Foxp3, respectively (Figure 7). There is increasing evidence that NF-κB family members are key arbitrators of the decision. For example, mice lacking p50 are unable to mount an asthma-like, airway TH2 response 170. Indeed, p50-deficient T cells fail to induce GATA3 expression during T-cell stimulation under TH2 differentiating conditions. These results suggest a transcriptional role for p50, and it was subsequently found that Bcl-3-deficient T cells also fail to undergo TH2 differentiation, suggesting that p50/Bcl-3 complexes are crucial for TH2 differentiation 171. Conversely, the same authors found that RelB-deficient T cells are deficient in TH1 differentiation due to decreased expression of T-bet. RelB may mediate upregulation of STAT4, which is responsible for T-bet induction downstream of IFNγ. Therefore, it appears that NF-κB activation during TCR stimulation may render cells competent for both proliferative and differentiating stimuli. As a corollary, NF-κB transactivation of the IL-2 gene is repressed as TH cells differentiate into TH1 or TH2 and become dependent on TH1 and TH2 cytokines (e.g., IFNγ and IL-4). It is thought that direct binding of T-bet to p65 that is associated with the IL-2 gene enhancer, may mediate the repression of IL-2 production in TH1 cells 172. In TH2 cells the lack of IL-2 transcription may be because of the decreased levels of p65 activation 173. As discussed above, several groups recently implicated NF-κB, and specifically c-Rel, in the regulation of Foxp3 expression and Treg development 61, 62, 63, 174. The relative contribution of c-Rel to iTreg development still requires further clarification. While a recent report demonstrated that IκBζ cooperates with ROR and is required for TH17 differentiation 59, the more general contribution of NF-κB in this process, either in the thymus or periphery, remains unclear. While the role of NF-κB in TH9 cells has yet to be interrogated, there are interesting clues suggesting an important role for NF-κB. Recently, PU.1 was reported to be the transcription factor responsible for TH9 differentiation 175. In unrelated recent work on acute myelogenous leukemia, it was noted that NF-κB regulates the transcription of PU.1 176. Furthermore, it has been suggested that IL-9 expression in T cells is regulated by NF-κB 177. Thus, it would seem reasonable to speculate that NF-κB may play an important role in TH9 differentiation (Figure 7). The transcription factors responsible for the differentiation of TH22 cells have not yet been identified. However, the possible role of TNFα as a TH22 differentiating cytokine is suggestive that NF-κB may be an important mediator of this differentiation process as well. In summary, NF-κB participates in both the initial T cell responses by supporting proliferation and regulating apoptosis, and is increasingly being shown to have an important role in the regulation of TH differentiation.
B-cell responses mediated by NF-κB
There are many parallels in the functions of NF-κB in T and B cells. NF-κB acts to support proliferation, regulate apoptosis, and control the processes of differentiation and maturation in B cells (Figure 8). B-cell responses are classified into two groups: thymus-dependent (TD) or thymus-independent (TI). In TD response follicular B cells require co-stimulatory signaling from TH cells expressing CD40L and cytokines, such as IL-4. These initial steps trigger germinal center formation, in which somatic hypermutation, isotype switching, and plasma cell differentiation occur. Thus, in individuals with a mutation in CD40L B cells fail to undergo class switch recombination in response to T-dependent antigens 178. Signaling through CD40 activates both canonical and non-canonical NF-κB pathways, although several lines of evidence suggest that the non-canonical pathway is not required in the response to TD antigens. Thus, B cells lacking p52 are fully capable of appropriate TD antigen responses when transferred 21, and B cells from relB−/− mice, although crippled in their proliferative response, undergo normal IgM secretion and class switching 179. In contrast to these results, B cells lacking either functional NIK or IKKα, exhibit defective responses 180, 181. It remains unclear whether the role of NIK and IKKα in this context is to induce p100 processing (non-canonical pathway) or to contribute to activation of the canonical pathway. Although both pathways are triggered by CD40 ligation, the current data suggest that canonical pathway activation is of great importance.
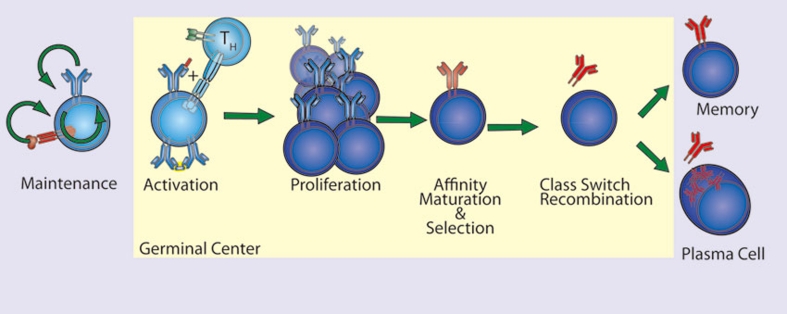
Figure 8.
NF-κB in B cell activation. NF-κB participates in the maintenance, activation, differentiation and proliferation of naive B cells. Tonic BCR and cytokine, BAFF, stimulation promotes naive B cell maintenance through NF-κB activation. Activation occurs when the naïve B cell recognizes its cognate antigen and receives co-stimulatory signaling (CD40L) from an activated TH cell within the germinal center. NF-κB-dependent proliferation and differentiation ensue and are coupled to BCR affinity maturation. NF-κB signaling in B cells expressing selected BCRs results in class switch recombination and differentiation into either memory B cells or plasma cells. NF-κB supports proliferation, differentiation and survival as indicated (green arrows).
Evidence also supports a role for the canonical pathway in class switch recombination. In contrast to B cells lacking RelB or p52, those from rela−/− mice exhibit markedly diminished class switching, despite a modest loss of lymphocyte proliferation following various stimuli 182. Likewise, c-Rel-deficient mice fail to generate protective humoral immune responses demonstrating the requirement for c-Rel in class switch recombination 164, 183, 184. B cells from nfbk1−/− mice exhibit decreased proliferation in response to mitogenic stimulation, and p50/p65 double-knockout B cells exhibit greater defects in proliferation and class switching 185, 186. Therefore, B cell proliferation and the TD B-cell maturation response are dependent on the canonical NF-κB pathway.
TI antigens are those that have an intrinsic ability to initiate B-cell responses in the absence of T-cell help. TI responses are carried out by marginal zone and CD5+ B cells. TI antigens function by acting as both antigen and B-cell mitogens, for example antigens that are PRR ligands or antigens with high avidity that are capable of crosslinking the BCR through repetitive structural features. In such cases it is expected that B-cell responses are more dependent on members of the canonical pathway that have well-documented roles in TLR and BCR signaling. For example, c-Rel-deficient B cells are highly sensitive to apoptosis following BCR cross-linking 187, 188, 189. As mentioned above, p50 and p50/RelA double-knockout B cells are deficient in responses to TI stimulation. Likewise, IKKβ-deficient B cells fail to mount TI or TD responses 190. These IKKβ-deficient B cells also exhibit increased spontaneous apoptosis, supporting that NF-κB is important in survival of B cells.
NF-κB and lymphocyte longevity
Lymphocyte homeostasis reflects balanced lymphocyte turnover and lymphopoiesis. Turnover of mature lymphocytes is complex as it depends upon both activation induced and homeostatic proliferation, and cell death. Lymphocyte survival is mediated through tonic stimulation provided by both the antigen receptor and certain cytokine receptors. As such, NF-κB can be assumed to play an important role in lymphocyte homeostasis and this assumption is supported by various genetic models.
T cells can be quite long-lived and naive T cells require continued contact with MHC:self-peptides for survival. These self-peptide complexes are most likely expressed on lymphoid DCs and generate a tonic low grade TCR signal. Survival of memory cells, on the other hand, is independent of continued contact with self-peptide:MHC complexes. B cells are formed at a far higher rate than T cells and therefore, B cells also undergo a significantly higher rate of turnover. Nonetheless, B cells too require maintenance signals to achieve peripheral homeostasis. The antigen receptor on B cells provides a basal level of signaling, albeit independent of the presence of antigen, which is required for maintenance of mature B cells. Thus, deletion of the BCR from mature B cells results in a complete loss of the peripheral B cell pool 191. The assumption is that BCR provides tonic activation of the NF-κB pathway resulting in the expression of anti-apoptotic proteins. Consistent with this assumption, loss of IKKβ, NEMO, or components of the CBM complex in mature B cells also results in complete loss of peripheral B cells 71, 190, 192. Furthermore, B cells from rela−/−, nfkb1−/−, nfkb2−/−, and c-rel−/− mice all have increased sensitivity to apoptosis and/or decreased survival ex vivo 67, 188, 193. In addition to contributing directly, it is thought that low level NF-κB activation in B cells contributes to survival through upregulation of p100, which is necessary for BAFFR-induced activation of the alternative pathway 67, 75, 166.
Several studies have implicated BAFFR in this aspect of the B lymphocyte survival 194, suggesting that the non-canonical NF-κB pathway is also relevant to B-cell survival. Consistent with this idea, constitutive activation of NIK or IKKα can circumvent the requirement for B cell BAFFR 74, 195. Conversely, B cells lacking IKKα exhibit defective survival 72, 196. The relevant targets of the non-canonical pathway downstream of BAFFR are thought to be Bcl-2 family members, e.g., the anti-apoptotic factor A1 187 or Bcl-XL, which are known to be regulated by p52/RelB-containing complexes, and are required for the maintenance of mature B cells. Indeed, Bcl-XL can complement B cells with a mutant BAFFR 197. Thus, tonic BCR stimulation, through upregulation of p100, may cooperate with BAFFR ligation and alternative pathway activation, to upregulate anti-apoptotic Bcl2 family members and mediate maintenance and survival of mature B cells.
Concluding remarks
The areas covered in the current review necessarily represent a subset of those in which NF-κB is a crucial regulator of immune responses. Recent work has highlighted the importance of NF-κB in the maintenance of epithelium barriers 198; as a regulator of inflammation associated with obesity and hyperlipidemia; and as a key factor in the development of inflammatory diseases and cancer. Our understanding of the immune system continues to improve, and coordinately our recognition of the many roles played by the NF-κB family of transcription factors also grows. As one example, the expanding universe of T-cell subsets opens up a new area for the analysis of the role of NF-κB in T-cell differentiation and effector function. It will be intriguing to see how NF-κB contributes to the development of TH9, TH17, TH22, and iTreg/TH3 cells. Deciphering the role of NF-κB in the effector function of these recently described cell types should also prove to be an area of great interest, particularly given the growing evidence for dysregulation of these cell types in autoimmune and inflammatory disease. There is much that remains unknown about the contributions of NF-κB in early and non-lymphoid hematopoiesis; in responses to danger signaling and allergens; in the coordination of pathogen-specific adaptive immune responses by the innate immune system; and in the differentiation and effector responses of both lymphoid and non-lymphoid innate immune cells. Thus moving forward, as in the past, much of the work done to understand the physiological and pathophysiological role of NF-κB will continue to focus on the role of this transcription factor family in Immunobiology.
Acknowledgments
Research in the authors' laboratory was supported by grants from the National Institutes of Health (to SG).
References
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- Burkly L, Hession C, Ogata L, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- Irla M, Hugues S, Gill J, et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity. 2008;29:451–463. doi: 10.1016/j.immuni.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Hikosaka Y, Nitta T, Ohigashi I, et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 2008;29:438–450. doi: 10.1016/j.immuni.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Shimo Y, Yanai H, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 2008;29:423–437. doi: 10.1016/j.immuni.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Anderson G, Jenkinson EJ, Rodewald HR. A roadmap for thymic epithelial cell development. Eur J Immunol. 2009;39:1694–1699. doi: 10.1002/eji.200939379. [DOI] [PubMed] [Google Scholar]
- Ruddle NH, Akirav EM. Secondary lymphoid organs: responding to genetic and environmental cues in ontogeny and the immune response. J Immunol. 2009;183:2205–2212. doi: 10.4049/jimmunol.0804324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol. 2010;10:664–674. doi: 10.1038/nri2832. [DOI] [PubMed] [Google Scholar]
- Alcamo E, Hacohen N, Schulte LC, Rennert PD, Hynes RO, Baltimore D. Requirement for the NF-kappaB family member RelA in the development of secondary lymphoid organs. J Exp Med. 2002;195:233–244. doi: 10.1084/jem.20011885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki S, Nakamura Y, Suzuka H, et al. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur J Immunol. 1994;24:429–434. doi: 10.1002/eji.1830240224. [DOI] [PubMed] [Google Scholar]
- Koike R, Nishimura T, Yasumizu R, et al. The splenic marginal zone is absent in alymphoplastic aly mutant mice. Eur J Immunol. 1996;26:669–675. doi: 10.1002/eji.1830260324. [DOI] [PubMed] [Google Scholar]
- Shinkura R, Kitada K, Matsuda F, et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat Genet. 1999;22:74–77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- Mebius RE. Organogenesis of lymphoid tissues. Nat Rev Immunol. 2003;3:292–303. doi: 10.1038/nri1054. [DOI] [PubMed] [Google Scholar]
- Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Dougall WC, Glaccum M, Charrier K, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz ZB, Weih DS, Sivakumar V, Weih F. RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxian S, Merkle H, Riemann M, et al. Abnormal organogenesis of Peyer's patches in mice deficient for NF-kappaB1, NF-kappaB2, and Bcl-3. Gastroenterology. 2002;122:1853–1868. doi: 10.1053/gast.2002.33651. [DOI] [PubMed] [Google Scholar]
- Caamano JH, Rizzo CA, Durham SK, et al. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J Exp Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G, Carlson L, Poljak L, et al. Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih DS, Yilmaz ZB, Weih F. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J Immunol. 2001;167:1909–1919. doi: 10.4049/jimmunol.167.4.1909. [DOI] [PubMed] [Google Scholar]
- Tang M, Wei X, Guo Y, et al. TAK1 is required for the survival of hematopoietic cells and hepatocytes in mice. J Exp Med. 2008;205:1611–1619. doi: 10.1084/jem.20080297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudeau B, Huetz F, Samson S, et al. IkappaBalpha/IkappaBepsilon deficiency reveals that a critical NF-kappaB dosage is required for lymphocyte survival. Proc Natl Acad Sci USA. 2003;100:15800–15805. doi: 10.1073/pnas.2535880100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson SI, Memet S, Vosshenrich CA, et al. Combined deficiency in IkappaBalpha and IkappaBepsilon reveals a critical window of NF-kappaB activity in natural killer cell differentiation. Blood. 2004;103:4573–4580. doi: 10.1182/blood-2003-08-2975. [DOI] [PubMed] [Google Scholar]
- Liu K, Victora GD, Schwickert TA, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Nussenzweig MC. Origin and development of dendritic cells. Immunol Rev. 2010;234:45–54. doi: 10.1111/j.0105-2896.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F, Carrasco D, Durham SK, et al. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- Wu L, D'Amico A, Winkel KD, Suter M, Lo D, Shortman K. RelB is essential for the development of myeloid-related CD8alpha− dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 1998;9:839–847. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- DiMolfetto L, Reilly C, Wei Q, Lo D. Dendritic-like cells from relB mutant mice. Adv Exp Med Biol. 1997;417:47–54. doi: 10.1007/978-1-4757-9966-8_8. [DOI] [PubMed] [Google Scholar]
- Ouaaz F, Arron J, Zheng Y, Choi Y, Beg AA. Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity. 2002;16:257–270. doi: 10.1016/s1074-7613(02)00272-8. [DOI] [PubMed] [Google Scholar]
- Abe K, Yarovinsky FO, Murakami T, et al. Distinct contributions of TNF and LT cytokines to the development of dendritic cells in vitro and their recruitment in vivo. Blood. 2003;101:1477–1483. doi: 10.1182/blood.V101.4.1477. [DOI] [PubMed] [Google Scholar]
- Andreakos E, Smith C, Monaco C, Brennan FM, Foxwell BM, Feldmann M. Ikappa B kinase 2 but not NF-kappa B-inducing kinase is essential for effective DC antigen presentation in the allogeneic mixed lymphocyte reaction. Blood. 2003;101:983–991. doi: 10.1182/blood-2002-06-1835. [DOI] [PubMed] [Google Scholar]
- Tas SW, de Jong EC, Hajji N, et al. Selective inhibition of NF-kappaB in dendritic cells by the NEMO-binding domain peptide blocks maturation and prevents T cell proliferation and polarization. Eur J Immunol. 2005;35:1164–1174. doi: 10.1002/eji.200425956. [DOI] [PubMed] [Google Scholar]
- van de Laar L, van den Bosch A, van der Kooij SW, et al. A nonredundant role for canonical NF-{kappa}B in human myeloid dendritic cell development and function. J Immunol. 2010. [DOI] [PubMed]
- Kriehuber E, Bauer W, Charbonnier AS, et al. Balance between NF-kappaB and JNK/AP-1 activity controls dendritic cell life and death. Blood. 2005;106:175–183. doi: 10.1182/blood-2004-08-3072. [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- von Vietinghoff S, Asagiri M, Azar D, Hoffmann A, Ley K. Defective regulation of CXCR2 facilitates neutrophil release from bone marrow causing spontaneous inflammation in severely NF-kappa B-deficient mice. J Immunol. 2010;185:670–678. doi: 10.4049/jimmunol.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois S, El Benna J, Dang PM, Pedruzzi E, Gougerot-Pocidalo MA, Elbim C. Inhibition of neutrophil apoptosis by TLR agonists in whole blood: involvement of the phosphoinositide 3-kinase/Akt and NF-kappaB signaling pathways, leading to increased levels of Mcl-1, A1, and phosphorylated Bad. J Immunol. 2005;174:3633–3642. doi: 10.4049/jimmunol.174.6.3633. [DOI] [PubMed] [Google Scholar]
- McDonald PP. Transcriptional regulation in neutrophils: teaching old cells new tricks. Adv Immunol. 2004;82:1–48. doi: 10.1016/S0065-2776(04)82001-7. [DOI] [PubMed] [Google Scholar]
- McDonald PP, Bald A, Cassatella MA. Activation of the NF-kappaB pathway by inflammatory stimuli in human neutrophils. Blood. 1997;89:3421–3433. [PubMed] [Google Scholar]
- Siebenlist U, Brown K, Claudio E. Control of lymphocyte development by nuclear factor-kappaB. Nat Rev Immunol. 2005;5:435–445. doi: 10.1038/nri1629. [DOI] [PubMed] [Google Scholar]
- Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, Korsmeyer SJ. bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell. 1991;67:879–888. doi: 10.1016/0092-8674(91)90361-2. [DOI] [PubMed] [Google Scholar]
- Derudder E, Cadera EJ, Vahl JC, et al. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat Immunol. 2009;10:647–654. doi: 10.1038/ni.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz BH, Scott ML, Cherry SR, Bronson RT, Baltimore D. Failure of lymphopoiesis after adoptive transfer of NF-kappaB-deficient fetal liver cells. Immunity. 1997;6:765–772. doi: 10.1016/s1074-7613(00)80451-3. [DOI] [PubMed] [Google Scholar]
- Grossmann M, Metcalf D, Merryfull J, Beg A, Baltimore D, Gerondakis S. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proc Natl Acad Sci USA. 1999;96:11848–11853. doi: 10.1073/pnas.96.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatt DW, Stillman WS, Yang Y, Gross S, Zheng JH, Irons RD. An essential role for NF-kappaB in human CD34(+) bone marrow cell survival. Blood. 1999;93:3302–3308. [PubMed] [Google Scholar]
- Voll RE, Jimi E, Phillips RJ, et al. NF-kappa B activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity. 2000;13:677–689. doi: 10.1016/s1074-7613(00)00067-4. [DOI] [PubMed] [Google Scholar]
- Jimi E, Phillips RJ, Rincon M, et al. Activation of NF-kappaB promotes the transition of large, CD43+ pre-B cells to small, CD43- pre-B cells. Int Immunol. 2005;17:815–825. doi: 10.1093/intimm/dxh263. [DOI] [PubMed] [Google Scholar]
- Feng B, Cheng S, Hsia CY, King LB, Monroe JG, Liou HC. NF-kappaB inducible genes BCL-X and cyclin E promote immature B-cell proliferation and survival. Cell Immunol. 2004;232:9–20. doi: 10.1016/j.cellimm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Hettmann T, DiDonato J, Karin M, Leiden JM. An essential role for nuclear factor kappaB in promoting double positive thymocyte apoptosis. J Exp Med. 1999;189:145–158. doi: 10.1084/jem.189.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto H, Surh CD, Sprent J. A role for Fas in negative selection of thymocytes in vivo. J Exp Med. 1998;187:1427–1438. doi: 10.1084/jem.187.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Lee H, Bellas RE, et al. Inhibition of NF-kappaB/Rel induces apoptosis of murine B cells. EMBO J. 1996;15:4682–4690. [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Tian J, Ji H, et al. I kappa B kinase 2 deficiency in T cells leads to defects in priming, B cell help, germinal center reactions, and homeostatic expansion. J Immunol. 2004;173:1612–1619. doi: 10.4049/jimmunol.173.3.1612. [DOI] [PubMed] [Google Scholar]
- Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci USA. 2006;103:11677–11682. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senftleben U, Li ZW, Baud V, Karin M. IKKbeta is essential for protecting T cells from TNFalpha-induced apoptosis. Immunity. 2001;14:217–230. doi: 10.1016/s1074-7613(01)00104-2. [DOI] [PubMed] [Google Scholar]
- Jimi E, Strickland I, Voll RE, Long M, Ghosh S. Differential role of the transcription factor NF-kappaB in selection and survival of CD4+ and CD8+ thymocytes. Immunity. 2008;29:523–537. doi: 10.1016/j.immuni.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Iwai Y, Oh-Hora M, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009;10:689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Kameswaran V, Tone Y, et al. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Isomura I, Palmer S, Grumont RJ, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med. 2009;206:3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CS. Kickstarting Foxp3 with c-Rel. Immunity. 2009;31:852–853. doi: 10.1016/j.immuni.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Grossmann M, O'Reilly LA, Gugasyan R, Strasser A, Adams JM, Gerondakis S. The anti-apoptotic activities of Rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. EMBO J. 2000;19:6351–6360. doi: 10.1093/emboj/19.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann B, Gommerman JL, Vora K, et al. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–2114. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Batten M, Groom J, Cachero TG, et al. BAFF mediates survival of peripheral immature B lymphocytes. J Exp Med. 2000;192:1453–1466. doi: 10.1084/jem.192.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JA, Dillon SR, Mudri S, et al. TACI-Ig neutralizes molecules critical for B cell development and autoimmune disease. impaired B cell maturation in mice lacking BLyS. Immunity. 2001;15:289–302. doi: 10.1016/s1074-7613(01)00183-2. [DOI] [PubMed] [Google Scholar]
- Motoyama N, Wang F, Roth KA, et al. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- Pasparakis M, Schmidt-Supprian M, Rajewsky K. IkappaB kinase signaling is essential for maintenance of mature B cells. J Exp Med. 2002;196:743–752. doi: 10.1084/jem.20020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisho T, Takeda K, Tsujimura T, et al. IkappaB kinase alpha is essential for mature B cell development and function. J Exp Med. 2001;193:417–426. doi: 10.1084/jem.193.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G, Carlson L, Xing L, et al. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Derudder E, Hobeika E, et al. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Stadanlick JE, Kaileh M, Karnell FG, et al. Tonic B cell antigen receptor signals supply an NF-kappaB substrate for prosurvival BLyS signaling. Nat Immunol. 2008;9:1379–1387. doi: 10.1038/ni.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Tigno-Aranjuez JT, Asara JM, Abbott DW. Inhibition of RIP2's tyrosine kinase activity limits NOD2-driven cytokine responses. Genes Dev. 2010;24:2666–2677. doi: 10.1101/gad.1964410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Lin J, et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Andrejeva J, Childs KS, Young DF, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Lu C, Ranjith-Kumar CT, Hao L, Kao CC, Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2010. [DOI] [PMC free article] [PubMed]
- Lu C, Xu H, Ranjith-Kumar CT, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54 Pt 1:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- Wang J, Basagoudanavar SH, Wang X, et al. NF-kappa B RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J Immunol. 2010;185:1720–1729. doi: 10.4049/jimmunol.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Kaisho T, Takeuchi O, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- Jurk M, Heil F, Vollmer J, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Krug A, French AR, Barchet W, et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F, Leifer CA, Gursel I, et al. Cutting edge: role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J Immunol. 2001;167:3555–3558. doi: 10.4049/jimmunol.167.7.3555. [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- Mambula SS, Sau K, Henneke P, Golenbock DT, Levitz SM. Toll-like receptor (TLR) signaling in response to Aspergillus fumigatus. J Biol Chem. 2002;277:39320–39326. doi: 10.1074/jbc.M201683200. [DOI] [PubMed] [Google Scholar]
- Shoham S, Huang C, Chen JM, Golenbock DT, Levitz SM. Toll-like receptor 4 mediates intracellular signaling without TNF-alpha release in response to Cryptococcus neoformans polysaccharide capsule. J Immunol. 2001;166:4620–4626. doi: 10.4049/jimmunol.166.7.4620. [DOI] [PubMed] [Google Scholar]
- Campos MA, Almeida IC, Takeuchi O, et al. Activation of Toll-like receptor-2 by glycosylphosphatidylinositol anchors from a protozoan parasite. J Immunol. 2001;167:416–423. doi: 10.4049/jimmunol.167.1.416. [DOI] [PubMed] [Google Scholar]
- Coban C, Ishii KJ, Kawai T, et al. Toll-like receptor 9 mediates innate immune activation by the malaria pigment hemozoin. J Exp Med. 2005;201:19–25. doi: 10.1084/jem.20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parroche P, Lauw FN, Goutagny N, et al. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proc Natl Acad Sci USA. 2007;104:1919–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shio MT, Eisenbarth SC, Savaria M, et al. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 2009;5:e1000559. doi: 10.1371/journal.ppat.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarovinsky F, Zhang D, Andersen JF, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- Perrigoue JG, Marshall FA, Artis D. On the hunt for helminths: innate immune cells in the recognition and response to helminth parasites. Cell Microbiol. 2008;10:1757–1764. doi: 10.1111/j.1462-5822.2008.01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Gringhuis SI, den Dunnen J, Litjens M, et al. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat Immunol. 2009;10:203–213. doi: 10.1038/ni.1692. [DOI] [PubMed] [Google Scholar]
- Saijo S, Ikeda S, Yamabe K, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Alcamo E, Mizgerd JP, Horwitz BH, et al. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J Immunol. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- Li ZW, Chu W, Hu Y, et al. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph D, Yeh WC, Wakeham A, et al. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- Farlik M, Reutterer B, Schindler C, et al. Nonconventional initiation complex assembly by STAT and NF-kappaB transcription factors regulates nitric oxide synthase expression. Immunity. 2010;33:25–34. doi: 10.1016/j.immuni.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck SL, Perkins ND, Carr DP, Nabel GJ. Inhibition of phorbol ester-induced cellular adhesion by competitive binding of NF-kappa B in vivo. Mol Cell Biol. 1993;13:6530–6536. doi: 10.1128/mcb.13.10.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C, Chilvers ER, Lawson MF, et al. NF-kappaB activation is a critical regulator of human granulocyte apoptosis in vitro. J Biol Chem. 1999;274:4309–4318. doi: 10.1074/jbc.274.7.4309. [DOI] [PubMed] [Google Scholar]
- Poligone B, Baldwin AS. Positive and negative regulation of NF-kappaB by COX-2: roles of different prostaglandins. J Biol Chem. 2001;276:38658–38664. doi: 10.1074/jbc.M106599200. [DOI] [PubMed] [Google Scholar]
- Catley MC, Chivers JE, Cambridge LM, et al. IL-1beta-dependent activation of NF-kappaB mediates PGE2 release via the expression of cyclooxygenase-2 and microsomal prostaglandin E synthase. FEBS Lett. 2003;547:75–79. doi: 10.1016/s0014-5793(03)00672-0. [DOI] [PubMed] [Google Scholar]
- Thompson C, Cloutier A, Bosse Y, et al. CysLT1 receptor engagement induces activator protein-1- and NF-kappaB-dependent IL-8 expression. Am J Respir Cell Mol Biol. 2006;35:697–704. doi: 10.1165/rcmb.2005-0407OC. [DOI] [PubMed] [Google Scholar]
- Lai WC, Zhou M, Shankavaram U, Peng G, Wahl LM. Differential regulation of lipopolysaccharide-induced monocyte matrix metalloproteinase (MMP)-1 and MMP-9 by p38 and extracellular signal-regulated kinase 1/2 mitogen-activated protein kinases. J Immunol. 2003;170:6244–6249. doi: 10.4049/jimmunol.170.12.6244. [DOI] [PubMed] [Google Scholar]
- Vincenti MP, Coon CI, Brinckerhoff CE. Nuclear factor kappaB/p50 activates an element in the distal matrix metalloproteinase 1 promoter in interleukin-1beta-stimulated synovial fibroblasts. Arthritis Rheum. 1998;41:1987–1994. doi: 10.1002/1529-0131(199811)41:11<1987::AID-ART14>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Vincenti MP, Brinckerhoff CE. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. 2002;4:157–164. doi: 10.1186/ar401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol. 2005;6:1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. 2009;130:7–15. doi: 10.1016/j.clim.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells J, Baer M, Young HA, et al. BCL-3 and NF-kappaB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J Biol Chem. 2004;279:49995–50003. doi: 10.1074/jbc.M404246200. [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Tato CM, Mason N, Artis D, et al. Opposing roles of NF-kappaB family members in the regulation of NK cell proliferation and production of IFN-gamma. Int Immunol. 2006;18:505–513. doi: 10.1093/intimm/dxh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani A, Schioppa T, Porta C, et al. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–11440. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- Lawrence T, Gilroy DW, Colville-Nash PR, Willoughby DA. Possible new role for NF-kappaB in the resolution of inflammation. Nat Med. 2001;7:1291–1297. doi: 10.1038/nm1201-1291. [DOI] [PubMed] [Google Scholar]
- Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Bottero V, et al. Enhanced NF-kappaB activation and cellular function in macrophages lacking IkappaB kinase 1 (IKK1) Proc Natl Acad Sci USA. 2005;102:12425–12430. doi: 10.1073/pnas.0505997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten FR, Arkan MC, Bollrath J, et al. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130:918–931. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann T, Lawrence T, McNeish I, et al. 'Re-educating' tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CH, Bebien M, Didierlaurent A, et al. An antiinflammatory role for IKKbeta through the inhibition of “classical” macrophage activation. J Exp Med. 2008;205:1269–1276. doi: 10.1084/jem.20080124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Koyama S, Ishii KJ, Coban C, Akira S. Innate immune response to viral infection. Cytokine. 2008;43:336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D, Liou HC, Sen R. c-Rel-dependent priming of naive T cells by inflammatory cytokines. Immunity. 2005;23:445–458. doi: 10.1016/j.immuni.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Rao S, Gerondakis S, Woltring D, Shannon MF. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J Immunol. 2003;170:3724–3731. doi: 10.4049/jimmunol.170.7.3724. [DOI] [PubMed] [Google Scholar]
- Grumont RJ, Gerondakis S. Murine c-rel transcription is rapidly induced in T-cells and fibroblasts by mitogenic agents and the phorbol ester 12-O-tetradecanoylphorbol-13-acetate. Cell Growth Differ. 1990;1:345–350. [PubMed] [Google Scholar]
- Ghosh P, Tan TH, Rice NR, Sica A, Young HA. The interleukin 2 CD28-responsive complex contains at least three members of the NF kappa B family: c-Rel, p50, and p65. Proc Natl Acad Sci USA. 1993;90:1696–1700. doi: 10.1073/pnas.90.5.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov VN, Nikolic-Zugic J. Transcription factor activation during signal-induced apoptosis of immature CD4(+)CD8(+) thymocytes. A protective role of c-Fos. J Biol Chem. 1997;272:8558–8566. [PubMed] [Google Scholar]
- Jeremias I, Kupatt C, Baumann B, Herr I, Wirth T, Debatin KM. Inhibition of nuclear factor kappaB activation attenuates apoptosis resistance in lymphoid cells. Blood. 1998;91:4624–4631. [PubMed] [Google Scholar]
- Wan YY, DeGregori J. The survival of antigen-stimulated T cells requires NFkappaB-mediated inhibition of p73 expression. Immunity. 2003;18:331–342. doi: 10.1016/s1074-7613(03)00053-0. [DOI] [PubMed] [Google Scholar]
- Kontgen F, Grumont RJ, Strasser A, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- Sriskantharajah S, Belich MP, Papoutsopoulou S, et al. Proteolysis of NF-kappaB1 p105 is essential for T cell antigen receptor-induced proliferation. Nat Immunol. 2009;10:38–47. doi: 10.1038/ni.1685. [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Siebenlist U. Roles of the NF-κB Pathway in Lymphocyte Development and Function. Cold Spring Harb Perspect Biol. 2010;2:a000182. doi: 10.1101/cshperspect.a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundstrom S, Anderson P, Scheipers P, Sundstedt A. Bcl-3 and NFkappaB p50-p50 homodimers act as transcriptional repressors in tolerant CD4+ T cells. J Biol Chem. 2004;279:8460–8468. doi: 10.1074/jbc.M312398200. [DOI] [PubMed] [Google Scholar]
- Mora A, Youn J, Keegan A, Boothby M. NF-kappa B/Rel participation in the lymphokine-dependent proliferation of T lymphoid cells. J Immunol. 2001;166:2218–2227. doi: 10.4049/jimmunol.166.4.2218. [DOI] [PubMed] [Google Scholar]
- Parra E, McGuire K, Hedlund G, Dohlsten M. Overexpression of p65 and c-Jun substitutes for B7-1 costimulation by targeting the CD28RE within the IL-2 promoter. J Immunol. 1998;160:5374–5381. [PubMed] [Google Scholar]
- Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J Immunol. 2005;175:2102–2110. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Hong JH, Glimcher LH. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J Exp Med. 2005;202:1289–1300. doi: 10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer JA, Liou JS, Todd MD, Glimcher LH, Lichtman AH. Regulation of cytokine gene expression in T helper cell subsets. J Immunol. 1994;152:77–86. [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Sehra S, Goswami R, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonadies N, Neururer C, Steege A, Vallabhapurapu S, Pabst T, Mueller BU. PU.1 is regulated by NF-kappaB through a novel binding site in a 17 kb upstream enhancer element. Oncogene. 2010;29:1062–1072. doi: 10.1038/onc.2009.371. [DOI] [PubMed] [Google Scholar]
- Early SB, Huyett P, Brown-Steinke K, Borish L, Steinke JW. Functional analysis of -351 interleukin-9 promoter polymorphism reveals an activator controlled by NF-kappaB. Genes Immun. 2009;10:341–349. doi: 10.1038/gene.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruffo A, Farrington M, Hollenbaugh D, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- Snapper CM, Rosas FR, Zelazowski P, et al. B cells lacking RelB are defective in proliferative responses, but undergo normal B cell maturation to Ig secretion and Ig class switching. J Exp Med. 1996;184:1537–1541. doi: 10.1084/jem.184.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills DM, Bonizzi G, Karin M, Rickert RC. Regulation of late B cell differentiation by intrinsic IKKalpha-dependent signals. Proc Natl Acad Sci USA. 2007;104:6359–6364. doi: 10.1073/pnas.0700296104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer U, Althage A, Odermatt B, Hengartner H, Zinkernagel RM. Immunodeficiency of alymphoplasia mice (aly/aly) in vivo: structural defect of secondary lymphoid organs and functional B cell defect. Eur J Immunol. 2000;30:2799–2807. doi: 10.1002/1521-4141(200010)30:10<2799::AID-IMMU2799>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco D, Cheng J, Lewin A, et al. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J Exp Med. 1998;187:973–984. doi: 10.1084/jem.187.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazowski P, Carrasco D, Rosas FR, Moorman MA, Bravo R, Snapper CM. B cells genetically deficient in the c-Rel transactivation domain have selective defects in germline CH transcription and Ig class switching. J Immunol. 1997;159:3133–3139. [PubMed] [Google Scholar]
- Horwitz BH, Zelazowski P, Shen Y, et al. The p65 subunit of NF-kappa B is redundant with p50 during B cell proliferative responses, and is required for germline CH transcription and class switching to IgG3. J Immunol. 1999;162:1941–1946. [PubMed] [Google Scholar]
- Snapper CM, Zelazowski P, Rosas FR, et al. B cells from p50/NF-kappa B knockout mice have selective defects in proliferation, differentiation, germ-line CH transcription, and Ig class switching. J Immunol. 1996;156:183–191. [PubMed] [Google Scholar]
- Grumont RJ, Rourke IJ, Gerondakis S. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont RJ, Rourke IJ, O'Reilly LA, et al. B lymphocytes differentially use the Rel and nuclear factor kappaB1 (NF-kappaB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J Exp Med. 1998;187:663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owyang AM, Tumang JR, Schram BR, et al. c-Rel is required for the protection of B cells from antigen receptor-mediated, but not Fas-mediated, apoptosis. J Immunol. 2001;167:4948–4956. doi: 10.4049/jimmunol.167.9.4948. [DOI] [PubMed] [Google Scholar]
- Li ZW, Omori SA, Labuda T, Karin M, Rickert RC. IKK beta is required for peripheral B cell survival and proliferation. J Immunol. 2003;170:4630–4637. doi: 10.4049/jimmunol.170.9.4630. [DOI] [PubMed] [Google Scholar]
- Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- Prendes M, Zheng Y, Beg AA. Regulation of developing B cell survival by RelA-containing NF-kappa B complexes. J Immunol. 2003;171:3963–3969. doi: 10.4049/jimmunol.171.8.3963. [DOI] [PubMed] [Google Scholar]
- Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: a tutorial on B cell survival. Annu Rev Immunol. 2003;21:231–264. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Calado DP, Derudder E, et al. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc Natl Acad Sci USA. 2008;105:10883–10888. doi: 10.1073/pnas.0805186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Amanna IJ, Dingwall JP, Hayes CE. Enforced bcl-xL gene expression restored splenic B lymphocyte development in BAFF-R mutant mice. J Immunol. 2003;170:4593–4600. doi: 10.4049/jimmunol.170.9.4593. [DOI] [PubMed] [Google Scholar]
- Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]