Abstract
Pluripotency, the ability of a cell to differentiate and give rise to all embryonic lineages, defines a small number of mammalian cell types such as embryonic stem (ES) cells. While it has been generally held that pluripotency is the product of a transcriptional regulatory network that activates and maintains the expression of key stem cell genes, accumulating evidence is pointing to a critical role for epigenetic processes in establishing and safeguarding the pluripotency of ES cells, as well as maintaining the identity of differentiated cell types. In order to better understand the role of epigenetic mechanisms in pluripotency, we have examined the dynamics of chromatin modifications genome-wide in human ES cells (hESCs) undergoing differentiation into a mesendodermal lineage. We found that chromatin modifications at promoters remain largely invariant during differentiation, except at a small number of promoters where a dynamic switch between acetylation and methylation at H3K27 marks the transition between activation and silencing of gene expression, suggesting a hierarchy in cell fate commitment over most differentially expressed genes. We also mapped over 50 000 potential enhancers, and observed much greater dynamics in chromatin modifications, especially H3K4me1 and H3K27ac, which correlate with expression of their potential target genes. Further analysis of these enhancers revealed potentially key transcriptional regulators of pluripotency and a chromatin signature indicative of a poised state that may confer developmental competence in hESCs. Our results provide new evidence supporting the role of chromatin modifications in defining enhancers and pluripotency.
Keywords: hESCs, epigenomics, histone modifications, enhancers
Introduction
Human embryonic stem cells (hESCs) are derived from the inner cell mass of the blastocyst 1. Due to their ability to self-renew while retaining the potential to differentiate into most other cell types in the body, there has been growing interest to explore hESCs in regenerative medicine and as a model system to study early human development.
A prevailing theory holds that pluripotency is established and maintained by a network of transcription regulatory proteins 2, 3. A core transcriptional regulatory network consisting of transcription factors OCT4, SOX2, NANOG and their regulatory target genes has been discovered to control the gene expression program to maintain self-renewal and pluripotency in human and mouse ESCs 3, 4, 5. Oct4, Sox2 and Nanog bind to key cis-regulatory sequences, including enhancers and promoters, recruiting co-activator complexes or transcriptional machinery to allow transcription and production of proteins 4, 5, 6. Importantly, they regulate each other's transcription, through feed forward loops, and produce two bistable states: in the presence of appropriate levels of all three proteins, the network is able to maintain the stable expression of each gene; on the other hand, if any of these proteins is absent or inappropriately expressed, the cells would then exit from this state and take on another stable state where some or all the three proteins are repressed. Besides regulating themselves, Oct4, Sox2 and Nanog also control the expression of a large number of other pluripotency genes, many of which encode for additional transcription factors that work together with the three to further stabilize the pluripotent state.
While it is clear that the transcriptional regulatory network plays a critical role in establishing and maintaining pluripotency of ES cells, other factors have also been shown to be important. Among them are epigenetic factors, such as DNA methylation and histone modifications, cis-regulating molecular marks that can exert long-lasting influences on gene activities in the cells 7, 8, 9. Without DNA methylation, differentiation to certain lineages is severely blocked 10, 11. Alterations that lead to abnormal chromatin modifications are also known to cause defects in either maintenance of pluripotency or cell fate specification 12, 13, 14. Therefore, there has been considerable interest in the examination of the DNA methylation profiles and chromatin landscape in pluripotent stem cells and lineage-committed cell types.
Promoter bivalency, characterized by trimethylation of both histone H3 lysine 4 (H3K4me3) and lysine 27 (H3K27me3) at gene promoters, has been proposed as an epigenetic mechanism for regulating development and proliferation 15, 16, 17, 18, 19, 20. These promoters are considered poised for activation (or repression) at a later stage, since they contain both active and repressive modifications. These genes are not transcribed in ES cells, but could either become activated during differentiation with concurrent loss of the methylation mark on H3K27 or silenced as indicated by loss of H3K4me3 15, 21. Given that not all repressed genes are targeted by H3K27me3, identifying which genes are influenced by this modification will be critical to understanding cell fate changes.
Regulation of chromatin state at specific enhancers may represent another epigenetic mechanism involved in pluripotency. We and others have demonstrated that transcriptional enhancers are characterized by the histone modification H3K4me1, and the presence of this mark is strongly correlated with tissue- and cell-type specific gene expression, suggesting a role for chromatin state in regulating enhancer activities 22, 23, 24. Recent studies have further shown that transcription factor binding and function at enhancers are strongly influenced by preexisting chromatin structure 25, 26, 27. More recently, Rada-Iglesias et al. examined the chromatin state in hESCs differentiating along a neural epithelium lineage, and uncovered two classes of enhancers distinguished by the presence or absence of lysine 27 acetylation on histone H3 (H3K27ac). The presence of this mark is indicative of active state of enhancers and correlated with activation of stem cell genes in the hESCs, while its absence coupled with presence of H3K4me1 would suggest a poised state that could become activated upon differentiation 28. Similar findings were also reported in mouse ES cells that undergo differentiation to neural progenitor cells 29.
To further characterize the role of chromatin modifications in self-renewal, pluripotency and differentiation, we have employed a new model of ES cell differentiation, which involves the treatment of hESCs grown in defined medium by BMP4, inducing them to exit the pluripotent state and enter a mixture state of mesendodermal and trophectodermal cells. We mapped epigenetic differences in undifferentiated ES cell (hESC) and differentiated ES cell (denoted as DFC) genomes. We have identified potential enhancers using chromatin signature patterns, and examined the dynamics of chromatin state at both promoters and enhancers during the differentiation of these cells. We found remarkable differences of chromatin dynamics at human promoters and enhancers. The chromatin state at promoters is generally stable during differentiation, with a small fraction undergoing changes that primarily involve a switch between active acetylation and repressive methylation at H3K27, which further allowed us to define a set of genes that appears to be important for maintenance of ES cell pluripotency, and another set that is involved in differentiation. The use of epigenetic changes distinguishes them from the larger group of differentially expressed genes. Our experiments identified over 50 000 potential enhancers in the undifferentiated ES cell and differentiated ES cell genomes, with a majority of the enhancers displaying marked changes in chromatin states in a manner that correlates with differential expression of their predicted target genes. These enhancers, and the factors predicted to bind them based on motif analysis, now provide a basis for future investigation into the regulatory networks of hESCs and differentiation. In addition, we also identified a set of poised enhancers marked by a distinct chromatin signature near developmentally important genes induced early in differentiation, underscoring the importance of enhancer elements in regulating differentiation. More importantly, this unique set of enhancer elements likely provides one means by which stem cells could respond to stimuli and differentiate to various cell types, in part becoming a key characteristic of pluripotency.
Results and Discussion
Genome-wide maps of chromatin state in hESC before and after differentiation
Low-passage (20-50) hESCs (H1) were grown in feeder cell-free medium TeSR1 as described 30. These hESCs express several known markers of stem cells including, OCT4, SOX2, NANOG, GDF3, DPPA4, DNMT3B, GABRB3, TDGF1, LEFTB, IFITM1, NODAL, GRB7, PODXL and CD9 31. To differentiate the hESCs, the cells were treated with BMP4 for 4-6 days (denoted as DFCs from here on), generating a heterogeneous cell population that is a mixture of mesendoderm (lineage markers: GATA4, GATA6, SOX17, FOXF1, GATA5 and CXCR4) 32, 33, 34, 35, 36, and trophectoderm (CDX2, GATA2 and GATA3; Supplementary information, Table S1) 37, 38, 39. We utilized chromatin immunoprecipitation coupled with genome-wide tiling microarrays (ChIP-chip) to map chromatin modifications in the genomes of both hESCs and DFCs 40. We focused on four modifications – H3K4me1, H3K4me3, H3K27ac and H3K27me3. Our previous studies demonstrated that the patterns of H3K4me1 and H3K4me3 profiles along the genome allow for identification of potential enhancers and promoters in particular cells 24. Additionally, the methylation at H3K27 has been demonstrated to play a critical role in silencing of gene expression in ES cells 41, 42. Our recent study suggested that H3K27 may also be acetylated at active gene promoters, as well as enhancers 23. By comparing the genome-wide maps of these four chromatin modifications from hESCs to those in DFCs, we hypothesized that we would be able to identify promoters and enhancers contributing to hESC and DFC identity.
Dynamic switch between acetylation and methylation at H3K27 during hESC differentiation
Promoters are key transcriptional regulatory sequences that integrate extracellular and intracellular inputs to control transcriptional initiation of genes. Previous studies have identified methylation of H3K4 and H3K27 at promoters to be important for the poised state of some key developmental regulator genes 15, 16, 17, 18, 19. To find whether additional promoters display dynamic changes in chromatin modification during ES cell differentiation, we examined modifications on H3K4 and H3K27 in both hESCs and DFCs. We found that the presence of H3K4me3 reveals little information in terms of gene activation: on a global scale, enrichment of this mark appears cell-type invariant during differentiation (Figure 1A and Supplementary information, Figure S1). This observation is in agreement with several recent studies finding this modification to be present at 70-80% of known transcription start sites (TSSs) 18, 19, 23, 43, 44, 45, 46. Interestingly, when we examined modifications to H3K27, we found a number of promoters displaying a switch between acetylation and methylation (Supplementary information, Figure S1). Trimethylation of this residue (H3K27me3) is a known marker of repressed promoters 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, in contrast to acetylation (H3K27ac), which is generally a hallmark of active chromatin 48, 49. These modifications were shown to be mutually exclusive in fly and human cells, with reciprocal changes in H3K27 acetylation and methylation mediated by changes in Drosophila CBP expression 50. Our results are in agreement, indicating that on a genome-wide scale these two modifications residing on the same residue are mutually exclusive: H3K27me3-marked promoters show no enrichment for H3K27ac, while those marked by H3K27ac are not enriched for H3K27me3.
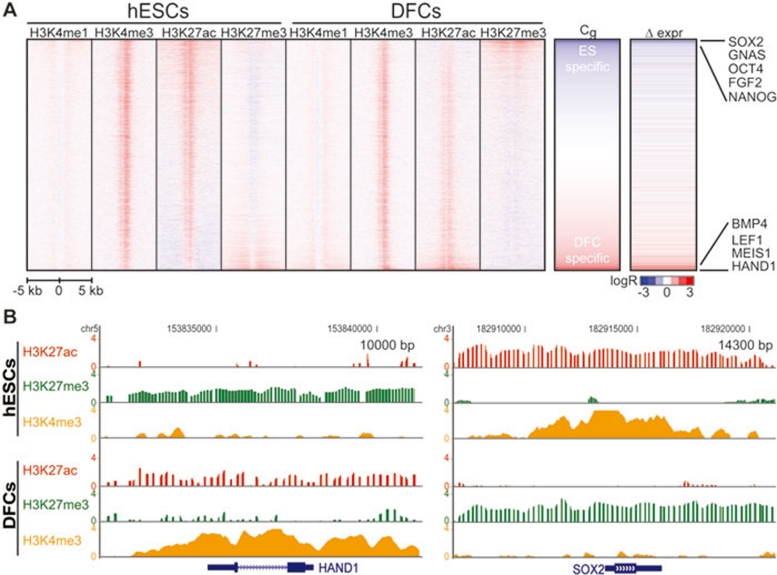
Figure 1.
Dynamic switch of H3K27 modifications at promoters. (A; left) Heat map of histone modifications H3K4me1, H3K4me3, H3K27ac and H3K27me3 within 5 kb of 22 047 TSSs, before and after differentiation. Middle: for each gene and cell type, we calculate the difference (H3K27ac – H3K27me3), and rank genes by comparing the difference of this value between the cell types (DFC – hESC). A negative value represents hESC enrichment of H3K27ac and DFC enrichment of H3K27me3 (blue Cg). A positive value represents DFC enrichment of H3K27ac and hESC enrichment of H3K27me3 (red Cg). Right: difference in gene expression (DFC/hESC); blue is hESC-specific expression while red is DFC-specific expression. Representative genes are noted on the far right. (B) UCSC Genome Browser snapshots showing the log2 ratio enrichment for H3K27ac (red), H3K27me3 (green) and H3K4me3 (orange) compared to input. Gene names are listed at the 5′ end of the gene structure. Left: a 10-kb window around the HAND1 gene illustrating the presence of H3K27me3 in hESCs that switches to H3K27ac following differentiation. (right) A 14.3-kb window around the SOX2 gene illustrating the presence of H3K27ac in hESCs that switches to H3K27me3 following differentiation.
To quantify how these modifications switch upon differentiation, we ranked TSSs by the change in levels of active H3K27ac and repressive H3K27me3: Cg = (H3K27acDFC – H3K27achESC) – (H3K27me3DFC – H3K27me3hESC) (Figure 1A and Supplementary information, Data S1). Genes with low Cg exhibit a combination of H3K27ac loss and H3K27me3 gain after differentiation. Examination of gene expression reveals that in general these genes are actively transcribed in hESCs and repressed in DFCs (Supplementary information, Table S1). This class of genes is of particular interest, as it contains the key stem cell transcription factors OCT4 (POU5F1), SOX2 and NANOG. For example, SOX2 shows hyperacetylation at H3K27 in hESCs that is lost following differentiation and becomes marked by H3K27me3 (Figure 1B). Additional genes showing the same active to repressive switch include notable transcription factors and signaling molecules likely important in the regulation of ESC pluripotency and self-renewal (Table 1 and Supplementary information, Table S2). For example, of just the few gene promoters included, a number of WNT signaling factors are revealed, including TCF7L1, FZD7, FZD8 and SFRP2. Also, targeted deletion of one gene on the list, FOXD3, resulted in a decreased ability of ES cells to self-renew and an increased tendency to differentiate to trophectoderm, endoderm, and mesendoderm 51. In addition, knockdown of another gene on the list, ZIC3, induced differentiation of ES cells to the endodermal lineage 52. These observations support the hypothesis that genes undergoing dynamic H3K27ac / H3K27me3 switch may play roles in pluripotency and self-renewal. Finally, we observe that based on the Cg metric of change in chromatin structure, OCT4, SOX2 and NANOG ranked 30, 1 and 155, respectively, among the top 1% of 22 047 genes. However, based on changes in gene expression, these genes would have ranked 2 591, 13 and 637, respectively, only among the top 12% of all genes. Thus, change in chromatin structure is a powerful method for categorizing functionally related genes.
Table 1. Representative transcription factors and signaling molecules associated with H3K27me3 following differentiation.
| Gene Name: TRANSCRIPTION FACTORS | Symbol | Gene Name: SIGNALING MOLECULES | Symbol |
|---|---|---|---|
| ATONAL HOMOLOG 1 | ATOH1 | ADENYLATE CYCLASE ACTIVATING POLYPEPTIDE 1 (PITUITARY) RECEPTOR TYPE I | ADCYAP1R1 |
| CHROMOBOX HOMOLOG 6 | CBX6 | BRAIN-SPECIFIC ANGIOGENESIS INHIBITOR 3 | BAI3 |
| CAMP RESPONSIVE ELEMENT | CREB5 | CHROMOSOME 21 OPEN READING FRAME 29 | C21ORF29 |
| BINDING PROTEIN 5 | |||
| CUT-LIKE 2 | CUTL2 | CONTACTIN 2 (AXONAL) | CNTN2 |
| FORKHEAD BOX B1 | FOXB1 | CONTACTIN ASSOCIATED PROTEIN-LIKE 3 | CNTNAP3 |
| FORKHEAD BOX D3 | FOXD3 | COMPLEMENT COMPONENT (3D/EPSTEIN BARR VIRUS) RECEPTOR 2 | CR2 |
| HOMEOBOX HB9 | HLXB9 | CUB AND SUSHI MULTIPLE DOMAINS 1 | CSMD1 |
| IROQUOIS HOMEOBOX PROTEIN 1 | IRX1 | EPH RECEPTOR A8 | EPHA8 |
| IROQUOIS HOMEOBOX PROTEIN 2 | IRX2 | V-ERB-B2 ERYTHROBLASTIC LEUKEMIA VIRAL ONCOGENE HOMOLOG 2 | ERBB2 |
| LIM HOMEOBOX 9 | LHX9 | V-ERB-B3 ERYTHROBLASTIC LEUKEMIA VIRAL ONCOGENE HOMOLOG 3 | ERBB3 |
| MLX INTERACTING PROTEIN-LIKE | MLXIPL | FAMILY WITH SEQUENCE SIMILARITY 19 (CHEMOKINE (C-C MOTIF)-LIKE), MEMBER A4 | FAM19A4 |
| V-MYC MYELOCYTOMATOSIS VIRAL | MYCN | FIBROBLAST GROWTH FACTOR 19 | FGF19 |
| RELATED ONCOGENE | |||
| NANOG HOMEOBOX | NANOG | FIBROBLAST GROWTH FACTOR 4 (HEPARIN SECRETORY TRANSFORMING PROTEIN 1) | FGF4 |
| NEUROGENIN 3 | NEUROG3 | FIBROBLAST GROWTH FACTOR 8 (ANDROGEN-INDUCED) | FGF8 |
| NK3 TRANSCRIPTION FACTOR | NKX3-1 | FRIZZLED HOMOLOG 7 | FZD7 |
| RELATED, LOCUS 1 | |||
| OLIGODENDROCYTE | OLIG3 | FRIZZLED HOMOLOG 8 TRANSCRIPTION FACTOR 3 | FZD8 |
| ORTHODENTICLE HOMOLOG 1 | OTX1 | GLUTAMATE RECEPTOR, IONOTROPIC, DELTA 2 | GRID2 |
| ORTHODENTICLE HOMOLOG 2 | OTX2 | GLUTAMATE RECEPTOR, METABOTROPIC 7 | GRM7 |
| PROTOCADHERIN 1 (CADHERIN-LIKE 1) | PCDH1 | HEDGEHOG INTERACTING PROTEIN | HHIP |
| POU DOMAIN, CLASS 3, | POU3F1 | KALLMANN SYNDROME 1 SEQUENCE TRANSCRIPTION FACTOR 1 | KAL1 |
| POU DOMAIN, CLASS 3, | POU3F3 | LIPOPROTEIN LIPASETRANSCRIPTION FACTOR 3 | LPL |
| POU DOMAIN, CLASS 3, | POU3F4 | LEUCINE RICH REPEAT TRANSMEMBRANE NEURONAL 3 | LRRTM3 |
| TRANSCRIPTION FACTOR 4 | |||
| POU DOMAIN, CLASS 5, | POU5F1 | PROTOCADHERIN 1 (CADHERIN-LIKE 1) | PCDH1 |
| TRANSCRIPTION FACTOR 1 | |||
| PC4 AND SFRS1 INTERACTING PROTEIN 1 | PSIP1 | PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | PCSK9 |
| SAL-LIKE 2 | SALL2 | PRO-PLATELET BASIC PROTEIN (CHEMOKINE (C-X-C MOTIF) LIGAND 7) | PPBP |
| SINE OCULIS HOMEOBOX HOMOLOG 3 | SIX3 | PROTEIN TYROSINE PHOSPHATASE, RECEPTOR-TYPE, Z POLYPEPTIDE 1 | PTPRZ1 |
| SRY (SEX DETERMINING REGION Y)-BOX 1 | SOX1 | POLIOVIRUS RECEPTOR-RELATED 1 (HERPESVIRUS ENTRY MEDIATOR C; NECTIN) | PVRL1 |
| SRY (SEX DETERMINING REGION Y)-BOX 2 | SOX2 | SECRETOGLOBIN, FAMILY 3A, MEMBER 2 | SCGB3A2 |
| SRY (SEX DETERMINING REGION Y)-BOX 21 | SOX21 | SECRETED FRIZZLED-RELATED PROTEIN 2 | SFRP2 |
| SRY (SEX DETERMINING REGION Y)-BOX 3 | SOX3 | STANNIOCALCIN 2 | STC2 |
| TRANSCRIPTION FACTOR 7-LIKE 1 | TCF7L1 | TERATOCARCINOMA-DERIVED GROWTH FACTOR 1 | TDGF1 |
| (T-CELL SPECIFIC, HMG-BOX) | |||
| THYMUS HIGH MOBILITY GROUP BOX | TOX | THROMBOSPONDIN 2 | THBS2 |
| PROTEIN TOX | |||
| ZIC FAMILY MEMBER 3 HETEROTAXY 1 | ZIC3 | TUMOR NECROSIS FACTOR RECEPTOR SUPERFAMILY, MEMBER 8 | TNFRSF8 |
| (ODD-PAIRED HOMOLOG) | |||
| ZINC FINGER PROTEIN 649 | ZNF649 | VASOACTIVE INTESTINAL PEPTIDE RECEPTOR 2 | VIPR2 |
In contrast, genes with high Cg show gain of H3K27ac and loss of H3K27me3 upon differentiation (Supplementary information, Table S2). These genes show the opposite expression pattern to that of low Cg genes, illustrating the close correlation between epigenetic modifications and gene expression. For example, the transcription factor gene HAND1 shows no H3K27ac in the hESC epigenome but is enveloped by H3K27me3-marked chromatin. Following differentiation, HAND1 undergoes a complete switch: losing H3K27me3, gaining H3K27ac and becoming actively expressed (Figure 1B). These results agree with recent findings examining H3K27me3 loss at developmentally important gene promoters 15, 19, 21, 44. Overall, ∼5.7% of all promoters exhibit at least a 2-fold change in H3K27 chromatin state during hESC differentiation, defining a set of genes differentially marked and expressed between these cells. The change in chromatin state during a change in cell fate distinguishes this set of genes amongst the 12% that are differentially expressed. Given that only a fraction of genes are epigenetically repressed following differentiation, it may suggest these genes are at the top of the hierarchy of regulatory factors in the prior hESC state. Therefore, assessing changes in H3K27 acetylation and trimethylation may prove more advantageous than simply monitoring how H3K4/27me3 bivalent genes change knowing that monovalent H3K4me3 genes are not always expressed 15, 19, 43, 45, 46.
Genome-wide identification of enhancers in hESCs and early differentiation
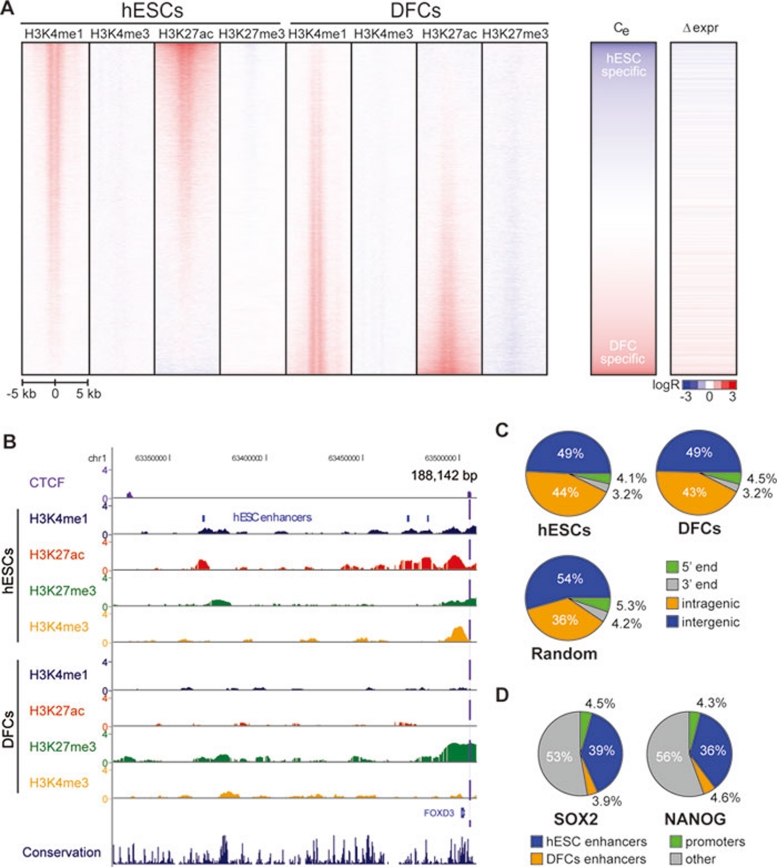
Recent studies have suggested that enhancers play important roles in cell-type-specific and tissue-specific gene expression. To identify enhancers that regulate stem cell gene expression during differentiation, we employ a computational method that identifies potentially active enhancers based on chromatin modification patterns of H3K4me1 and H3K4me3 23, 24. This method predicts 28 809 enhancers in hESCs and 33 369 in DFCs (Figure 2A, Supplementary information, Data S1, Figure S2A and Table S3). This definition of enhancers is used throughout. A number of predicted hESC enhancers were near important regulators of pluripotency. For example, we predicted three hESC-specific enhancers upstream of FOXD3, a gene that is important for pluripotency and known to activate Nanog and Oct4 expression in mouse ESCs 51 (Figure 2B). The distribution of the chromatin-predicted enhancers is primarily distal to the TSSs, with ∼50% lying in intergenic regions for each cell type and just over 40% falling in intragenic regions, above what is expected at random (Figure 2C and Supplementary information, Data S1). Additionally, these enhancers tend to be clustered, indicating that multiple enhancers may act together to drive gene expression (Supplementary information, Figure S2B; see below).
Figure 2.
Enhancer features and cell-type specificity. (A; left) A heat map of histone modifications ± 5 kb of predicted enhancers, ranked based on differences in H3K27ac (DFC – hESC). Middle: the cell-type specificity of chromatin modifications at enhancers, Ce = (H3K27acDFC – H3K27achESC). Right: changes in gene expression of neighboring genes. (B) UCSC Genome Browser snapshots of a 188-kb window at the FOXD3 locus showing the log2 ratio enrichment for H3K27ac (red), H3K27me3 (green), H3K4me3 (orange), H3K4me1 (blue) and CTCF (purple-dashed line indicates binding outside gene) compared to input. Predicted enhancers (blue bars above H3K4me1 peaks) lose their chromatin signature after differentiation. (C) Distribution of enhancers in each cell type relative to 5′ and 3′ ends of genes, as well as intragenic and intergenic regions. (D) Overlap of ChIP-Seq binding sites for transcription factors SOX2 and NANOG, compared to promoters, predicted hESC enhancers and predicted DFC enhancers.
To identify the common themes of enhancer sequences and further elucidate the transcriptional regulatory mechanisms guiding ES cells and differentiation, we investigated if known transcription factor binding sites (TFBS) from the JASPAR and TRANSFAC databases were enriched at predicted enhancers in a cell-type-specific manner. We identified both hESC-specific motifs and DFC-specific motifs (Table 2 and Supplementary information, Tables S4). The high-confidence hESC-specific motifs include those that are recognized by KLF4 and c-MYC, two transcription factors that are capable of reprogramming human fibroblasts to become iPS cells when transduced with OCT4 and SOX2 31, 53, 54, 55. Also included in this list is a motif for FOXD3, which is known to be involved in maintaining mouse ESCs and in the hESC pluripotency gene regulatory network 56, 57. A joint OCT4:SOX2 motif in the TRANSFAC database was identified, consistent with the role of these two factors in regulating ES cell gene expression 58, 59. Additionally, a number of motifs are consistently found from both databases (Supplementary information, Table S4). In contrast to the hESC-specific motifs, the high-confidence DFC-specific enhancer motifs represent several transcription factors known to be involved in early development or differentiation, including Brachyury (mesoderm gene expression), FOXC1 (heart field specification), the Myf family (myogenesis) and ZEB1 (epithelial-mesenchymal transitions; Table 2) 60, 61, 62, 63. Of the transcription factor motifs that we classify at DFC-specific enhancers, none of the corresponding factors are known to play a role in human ESC maintenance or in reprogramming to an induced pluripotent state.
Table 2. Transcription factor binding site motifs enriched in hESC or DFC enhancers.
| Motif TF | P-value (ESC) | # of enhancers (ESC) | % of hESC enhancers with TF sites | Known Role in Stem Cell Biology | Reference* |
|---|---|---|---|---|---|
| MZF1_5-13 | <0.001 | 127 | 0.1235 | Inhibits haematopoietic differentiation in ES cells | (Perrotti et al., 1995) |
| PAX5 | <0.001 | 360 | 0.3502 | ||
| FOXD3 | <0.001 | 223 | 0.2169 | Known pluripotency gene | See Text |
| OCT4:SOX2 | <0.001 | 382 | 0.2918 | Known pluripotency genes | See Text |
| KLF4 | <0.001 | 104 | 0.1012 | Role in induced pluripotency | See Text |
| GABPA | <0.001 | 147 | 0.1430 | Regulates Oct3/4 expression in mouse ES cells | (Kinoshita et al., 2007) |
| FOXI1 | <0.001 | 35 | 0.0340 | ||
| HNF1A | <0.001 | 145 | 0.1411 | ||
| MYC:MAX | <0.001 | 290 | 0.2821 | Myc plays a role in inducing pluripotency | See Text |
| PPARG | <0.001 | 334 | 0.3249 | ||
| NFKB1 | <0.001 | 105 | 0.1021 | ||
| Gfi1 | 0.013 | 1091 | 0.4085 | Role in haematopoietic stem cell maintenance | (Hock et al., 2004) |
| Egr-1 | 0.021 | 790 | 0.3044 | Controls haematopoietic stem cell proliferation | (Min et al., 2008) |
| HFH4 (FOXJ1) | 0.024 | 533 | 0.2159 | ||
| OCT1 (POU2F1) | 0.010 | 294 | 0.1381 | Binds Nanog and Rex-1 promoters in vitro | (Rosfjord and Rizzino, 1994; Wu da and Yao, 2005) |
| Motif TF | P-value (DFC) | # of enhancers (DFC) | % of DFC enhancers with TF sites | Known Role in Development | Reference* |
|---|---|---|---|---|---|
| RELA | <0.001 | 414 | 0.1285 | Regulates apoptosis/proliferation in developing organs | (Barkett and Gilmore, 1999) |
| FOXC1 | <0.001 | 95 | 0.0295 | Role in cardiac and renal morphogenesis | (Lehmann et al., 2003) |
| MYF | <0.001 | 44 | 0.0137 | Role in muscle development | (Rudnicki and Jaenisch, 1995) |
| ZEB1 | <0.001 | 19 | 0.0059 | Role in smooth muscle cell differentiation | (Nishimura et al., 2006) |
| Brachyury | <0.001 | 35 | 0.4965 | Role in mesoderm and notochord development | (Smith, 1999) |
| HIF1 | <0.001 | 64 | 0.4954 | Role in placental vascularization | (Withington et al., 2006) |
| TEF | 0.0321 | 201 | 0.4912 | Expressed in the pituitary gland during embryogenesis | (Drolet et al., 1991) |
| Pitx2 | 0.0165 | 310 | 0.4879 | Role in left-right asymmetry during development | (Shiratori et al., 2001) |
| C/EBP | 0.0047 | 373 | 0.4834 | These factors have roles in differentiation & proliferation | (Nerlov, 2007) |
| ICSBP (IRF8) | 0.0393 | 342 | 0.4892 | Role in myeloid development | (Holtschke et al., 1996) |
| GR | 0.0357 | 385 | 0.4889 | Knockout mice sustain liver, medulla, & lung defects | (Cole et al., 1995) |
| SRF | <0.001 | 424 | 0.4843 | Role in smooth muscle cell differentiation | (Wang et al., 2004) |
| STAT5A | 0.0212 | 465 | 0.4881 | Role in mammary development & erythropoiesis | (Hennighausen and Robinson, 2008) |
| MEF-2 | <0.001 | 521 | 0.4862 | Role in cardiogenesis | (Mohun and Sparrow, 1997) |
*References are listed in the Supplementary information, Data S1.
If the predicted enhancers function in vivo, we expect significant binding of transcription factors. In order to test this hypothesis, we employed high-throughput sequencing coupled with chromatin immunoprecipitation (ChIP-Seq) to determine the binding sites for SOX2 and NANOG. We identified 4 818 SOX2 and 20 973 NANOG binding sites (FDR = 1%) using the MACS peak finding software 6 against a background of input hESC DNA. Comparing to putative hESC enhancers, 39.1% and 35.5% of the SOX2 and NANOG binding sites were recovered, respectively, compared with 3.9% and 4.6% at putative DFC enhancers (Figure 2D). Additionally, a number of binding sites not recovered by hESC enhancer predictions show a weak enrichment of H3K4me1 in hESCs but not DFCs, which may reflect enhancers missed by the prediction algorithm (Supplementary information, Figure S3). The presence of these key stem cell regulators at enhancers suggests a central role of enhancers in defining the ES cell gene expression program. These results indicate that other transcription factors with motifs enriched in hESC enhancers, such as KLF4, MYC and FOXD3, likely bind to the predicted hESC enhancers.
Dynamics of chromatin state at enhancers reveal cell-type-specific usage
Since promoters that undergo dynamic changes in chromatin structure generally belong to important stem cell and developmental genes, we wondered if chromatin dynamics at enhancers would identify potential sequences regulating the same processes. To assess the dynamics of chromatin modifications at human enhancers, we clustered H3K4me1, H3K4me3, H3K27me3 and H3K27ac at the H3K4me1-marked enhancers. Most predicted enhancers exhibit dramatic gains or losses of H3K4me1 and H3K27ac during differentiation (Supplementary information, Figure S2A). Of particular note is the general absence of H3K27me3 at these sequences, suggesting that this repressive modification is mainly found at promoters. In contrast, a significant number of enhancers are associated with H3K27ac. We ranked the predicted enhancers by the change in levels of acetylation between hESCs and DFCs: Ce = (H3K27acDFC – H3K27achESC) (Figure 2A). Just as individual enhancers studies have shown the presence of hyperacetylation 48, 49, 64, 65, 66, 67, 68, we find that hyperacetylated enhancers tend to be cell-type specific. In addition, hyperacetylated enhancers are nearer to upregulated genes than enhancers lacking acetylation (Figure 2A, expression heat map), suggesting a role of H3K27ac in modulating enhancer activity.
CTCF-organized regulatory domains predict enhancer targets
Genes regulated by enhancers marked in a cell-type-specific manner likely contribute to defining the unique abilities of stem cells. However, to find these target genes, we first need to link H3K4me1-marked enhancers to the genes they regulate. To do this, we focused on the vertebrate insulator binding protein CTCF 69, 70, which is known for its enhancer-blocking activity when bound between enhancers and promoters (for review, see 71, 72, 73). Focusing on 1% of the human genome, we had previously found that 90% of the CTCF-binding sites in hESCs are shared in DFCs, and others have made similar observations on the cell-type invariance of CTCF binding 23, 73, 74 (Supplementary information, Figure S4). Thus, as a map of CTCF binding in hESCs can be used as a proxy for CTCF binding in DFCs, we completed our cis-regulatory map by performing ChIP-chip to map 33 302 CTCF-binding sites genome wide (FDR = 1%) in the hESCs (Supplementary information, Table S5). Supporting the accuracy of this map, 70% of the hESC CTCF sites previously mapped in 1% of the human genome are recovered here.
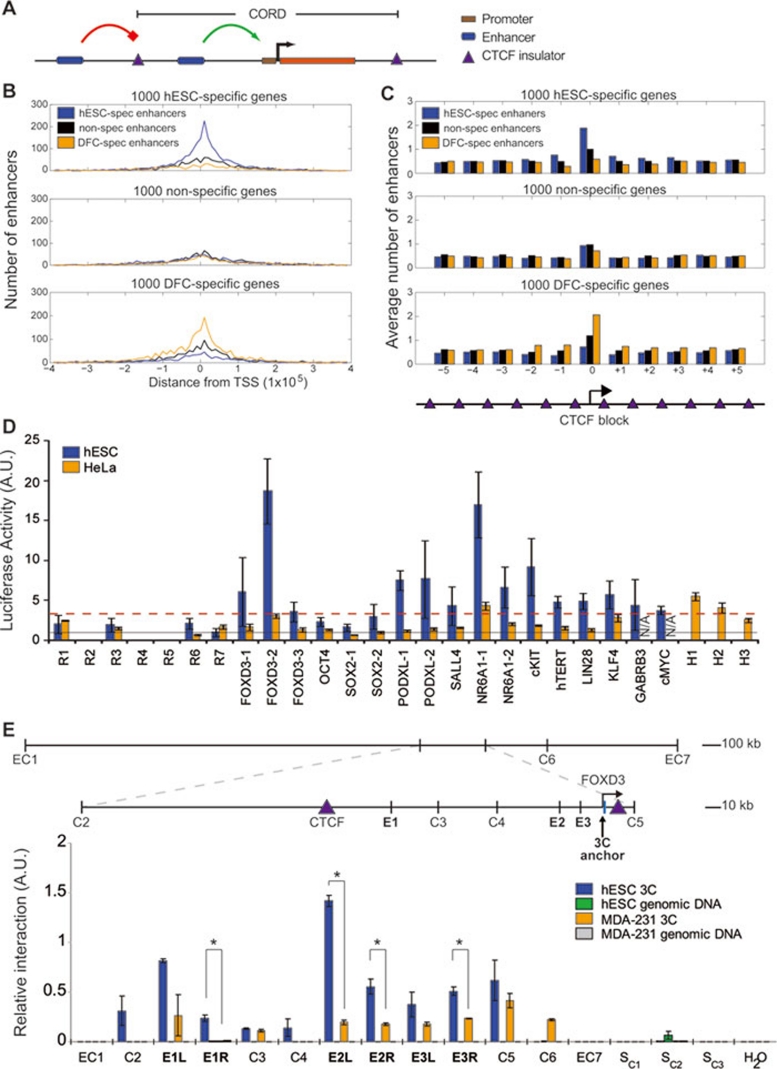
We then partitioned the genome into CTCF-organized regulatory domains (CORDs). Each CORD is a cis-regulatory block containing at least one gene, and its outer boundaries are defined by CTCF-binding sites (Figure 3A). The median size of a CORD is 83 kb (Supplementary information, Figure S4C), the majority (57%) of CORDs contain only one promoter (Supplementary information, Figure S4D), and about half of all CORDs (51.3% in hESC and 50.9% in DFC) contain no predicted enhancers (Supplementary information, Figure S4E). From our observations above, we then assigned predicted enhancers to the promoters in the same CORD. If the model of CTCF function is true, then we expect hESC-specific enhancers to be highly enriched in CORDs containing hESC-specific genes compared to DFC-specific genes and vice versa. Using the Ce ranking from Figure 2A, we divided the predicted enhancers into three equal-sized groups that are hESC specific, nonspecific and DFC specific. We observed that hESC-specific enhancers are highly enriched within the CORDs containing the 1000 most hESC-specific genes. Similarly, DFC-specific enhancers are enriched within CORDs containing the 1000 most DFC upregulated genes (Figure 3B). In contrast, neighboring CORDs do not show enrichment of cell-type-specific enhancers (Figure 3C). These results suggest enhancers may play an important role in regulating gene expression from promoters in the same CORD.
Figure 3.
Enrichment of cell-specific enhancers within CTCF-organized regulatory domain (CORDs) and enhancer validation. (A) Diagram of CORDs. Regions bounded by CTCF containing promoters and enhancers. (B) Distribution of hESC specific, DFC specific and nonspecific enhancers within CTCF-defined domains containing promoters of hESC-specific, DFC-specific, and nonspecific genes. (C) As in B, but expanded to neighboring CTCF-defined domains. (D) Reporter assays of enhancer function at predicted hESC enhancers and randomly chosen genomic regions, cloned downstream of a luciferase gene. The dashed red line indicates a P-value cutoff of 1%. hESC-specific enhancers were selected from within CORDs for known ES-related genes and tested in H1 and HeLa cells. H1-H3, enhancers specifically marked in HeLa cells; N/A, failed transfection in HeLa. (E) 3C was performed to assess the interactions between the FOXD3 promoter and three predicted enhancers in its CTCF-defined domain (E1, E2 and E3). The interaction strength is compared to predicted enhancers outside the CTCF-defined domain (CE1 and CE7), to loci lacking the enhancer chromatin signature (C2-C6), control regions on a different chromosome (SC1-SC3) and water.
Through the examination of enhancer enrichment relative to all genes within their respective CORD (See Supplementary information, Data S1), we observe that CORDs containing differentially expressed genes are enriched with cell-type-specific enhancers, while non-differentially expressed genes remain static for enhancer enrichment (Figure 4A). The dynamics of chromatin reorganization upon differentiation also reveals that enhancers are generally weak and act synergistically, as the number of enhancers increases within CORDs, differential expression also increases linearly on a log scale (Pearson correlation = 0.82; Supplementary information, Figure S5).
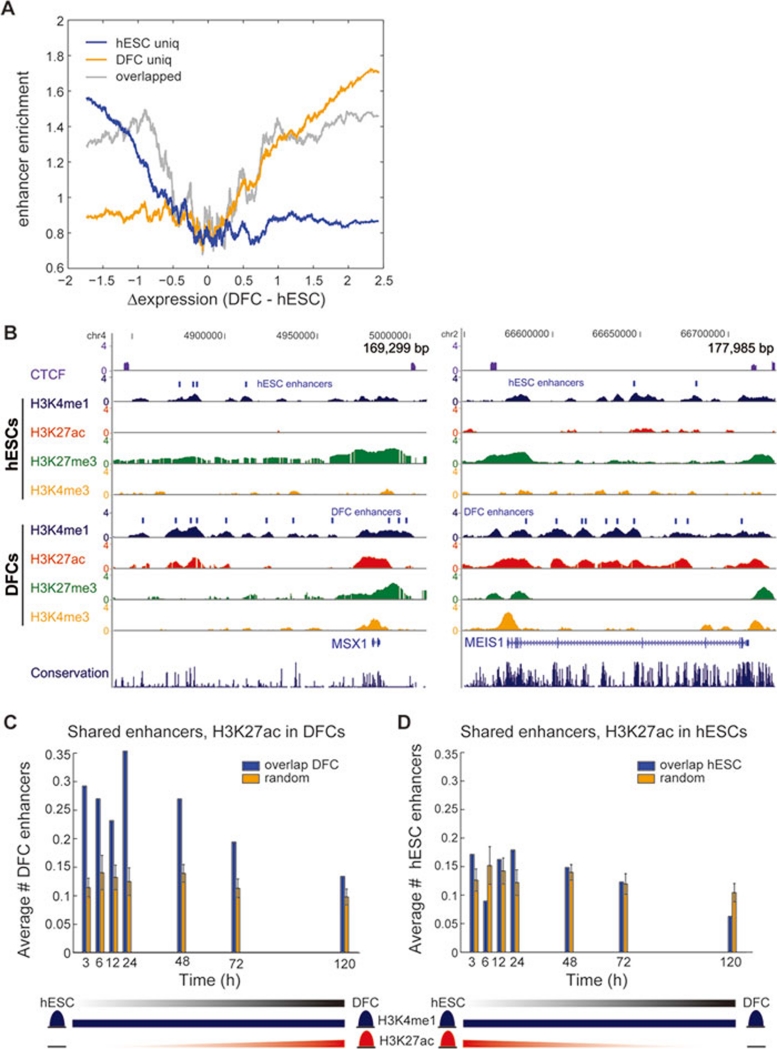
Figure 4.
Subset of shared enhancers are poised for early response. (A) Enhancer enrichment relative to gene expression for three subsets of enhancers: those uniquely marked in hESCs (blue), those uniquely marked in DFCs (orange) and the remaining 8 863 that are marked in both (grey). A subset of shared enhancers is enriched at differentially expressed genes in both cell types (see Supplementary information, Data S1). (B) UCSC Genome Browser snapshots of MSX1 and MEIS1 gene loci. These genes are specifically expressed in DFCs, but have H3K4me1-marked enhancers in hESCs (blue). These enhancers lack H3K27ac (red) in hESCs, which is highly enriched following differentiation. (C, D) We measured gene expression at 3, 6, 12, 24, 48, 72 and 120 h after BMP4/bFGF treatment of hESCs. For differentially expressed genes at each time point, we counted the average number of acetylated enhancers with cell-type specificity, defined as the 2 000 shared enhancers with the most H3K27ac in (C) DFCs and (D) hESCs. Random is described in the Supplementary information, Data S1.
With this framework in place, we next set out to validate the function of enhancers potentially regulating key pluripotent genes. We cloned 17 enhancers downstream of the Luciferase gene in a reporter construct and measured the luciferase activity in hESCs after transient transfection. Of the 17 putative enhancer constructs tested in this assay, 14 (82%) showed higher level of enhancer activity (P = 0.01) compared to random genomic regions that showed no significant reporter activity (Figure 3D; see Materials and Methods). To examine the cell-type specificity of these predicted enhancers, we next tested them in different cell lines. Of the 15 successfully transfected into HeLa cells, only 1 (6.7%) tested positive (Figure 3D). To further assess enhancer cell-type specificity, we also tested 10 DFC-specifically predicted enhancers in hESCs, and only 2 (20%) showed activity (Supplementary information, Figure S4F). Together, these results support the accuracy and cell-type specificity of these predicted enhancers.
While reporter activity in a luciferase assay implies that an enhancer can function, it does not necessarily mean that an enhancer acts on an endogenous gene. To provide further evidence of endogenous gene activation, we examined the three-dimensional structure of chromatin near the FOXD3 gene. The FOXD3 CORD contains three hESC predicted enhancers: E1, E2 and E3 in decreasing distance from the TSS. We performed chromosome conformation capture (3C) 76 to measure the interaction strength between the FOXD3 promoter and these three predicted enhancers. We find that all three predicted enhancers showed significant interaction compared to (1) control regions not predicted to be enhancers (C2-C4 and C6), (2) hESC predicted enhancers outside the FOXD3 CORD (EC1 and EC7) and (3) the same regions in the MDA-MB-231 breast cancer cell line (Figure 3E). Interestingly, the enhancer displaying the strongest interaction strength with the FOXD3 promoter is E2, which also shows the strongest luciferase reporter activity, suggesting that it may be a key regulatory element regulating FOXD3 activity in hESCs. Together, these results suggest that enhancers within a CORD contribute to regulating the expression of genes within the CORD.
In light of the 3C validation of promoter-enhancer interactions, we extended our analysis to predict promoter targets within the CORDs of the top 1% of hESC-specific enhancers and DFC-specific enhancers based on Ce to identify additional genes contributing to the hESC and DFC expression program (Supplementary information, Table S6). These lists provide additional candidates for genes important in defining each cellular state. As confirmation, we discovered several putative enhancers in CORDs containing genes important for hESC regulation. A view of the SOX2 locus reveals a number of predicted enhancers downstream of the gene. To date, only a single enhancer has been identified in mouse ESCs, ∼4 kb downstream of the TSS 77, which is also epigenetically marked as a predicted human enhancer in this region, and is one of several predicted enhancer elements downstream of the gene (Supplementary information, Figure S6A). We also predict several hESC-specifically marked enhancers in the CORDs containing OCT4 and NANOG (Supplementary information, Figure S6B and S6C), as well as a number of other genes required for ES cell pluripotency.
Genes regulated by cell-type-specific enhancers may contribute to defining each cellular state. Further examination of potential enhancer gene targets reveals JMJD2C, JARID2, LEFTY1, as well as other transcription factors, and MAP kinase signaling molecules in hESCs, while DFC enhancer targets reveal genes, such as several HOX and GATA factors (Supplementary information, Table S6). By linking enhancers to target promoters, our results allow for the expansion of regulatory networks and provide a more precise depiction of regulatory pathways in ES cells.
Chromatin dynamics at poised enhancers correlate with cell fate commitment
One of the most intriguing aspects of ES cells is their ability to differentiate into a variety of other cell types in the body in response to different environmental cues. Our analysis shows that there are three classes of H3K4me1-marked enhancers: those marked specifically in hESCs, those marked specifically in DFCs and those marked in both. While the first and second groups are enriched near genes specifically expressed in hESCs and DFCs, respectively, enhancers marked in both cell types are enriched near both hESC- and DFC-specific genes (Figure 4A and Supplementary information, Figure S7A). To further examine hESC differentiation, we examined this class of 8 863 shared enhancers that are marked before and after differentiation, reasoning that extracellular signaling may act through some of these sequences to activate a group of key regulators for cell fate determination.
Particularly interesting among these enhancers are those that are enriched in CORDs containing DFC-specific genes (Figure 4A). Many of these shared enhancers are only marked by H3K4me1 in ES cells, but upon differentiation they gain H3K27ac (Figure 4A and Supplementary information, Figure S7A). Since H3K27ac is a mark of activity that we have previously shown to overlap with H3K4me1 23, we hypothesized that these enhancers may be inactive in ES cells but poised and awaiting a regulatory signal to activate them, therefore giving rise to acetylation and differentiation. If true, then we expect these enhancers to be enriched near genes induced early during differentiation. When we examined the enrichment of shared enhancers near genes differentially upregulated at various time points during BMP4 treatment (3, 6, 12, 24, 48 and 120 h), we indeed observed that this set of poised enhancers is significantly enriched in CORDs containing early response genes (Figure 4C). This is in contrast to the most DFC-specific acetylated enhancers from Figure 3A (Supplementary information, Figure S7B) or the shared enhancers that lose acetylation and show no enrichment near the same genes (Figure 4D).
Interestingly, the enhancers in this category can be found near genes coding for the developmental transcription factors MSX1 and MEIS1, which are upregulated at 3 and 48 h, respectively. Each of these genes is highly expressed in DFCs, and their CORDs contain numerous shared enhancers, but H3K27ac only marks the enhancers in DFCs (Figure 4B). In addition, BMP4 itself as well as downstream factors SMAD3, SMAD6, SMAD7 and ID2 are also found in this category at 3 h. This set of genes contains a number of additional transcription factors, including HAND1, GATA3, CDX2, FOXO4, LEF, JUN and SOX9 (Supplementary information, Table S7). These seven factors along with SMAD3 all have TFBS motifs enriched in DFC-specific enhancers (Supplementary information, Table S4), suggesting these factors go on to establish the cell fate through transcriptional regulation at enhancers. Thus, our results suggest that poised enhancers may contribute to ES cell differentiation by pre-marking enhancers for genes likely responsible for early steps in differentiation. Our results are supported by two recent studies that also noted H3K27ac can distinguish active from poised enhancers in mouse and human ESCs 28, 29. However, distinct differences do exist. Besides species differences in the study by Creyghton et al., the study by Rada-Iglesias et al. in hESCs assesses activation by H3K27ac acquisition during differentiation to neuroectoderm. Additionally, our method excludes promoter-proximal H3K4me1 that partially overlaps with H3K4me3, defined as class II enhancers by Rada-Iglesias et al., as it can be challenging to distinguish where the enhancer ends and promoter begins. Finally, we have predicted the target genes of poised enhancers based on CORDs. In conclusion, in a manner similar to bivalent promoters, poised enhancers may provide one additional means by which pluripotency and developmental competence are maintained in ES cells.
Conclusions
In summary, we have analyzed chromatin modification landscapes in an in vitro model of hESC differentiation to identify potential genes and regulatory sequences contributing to pluripotency and lineage specification. We provide a global view of chromatin dynamics upon differentiation of hESCs along a largely mesendodermal lineage, laying the foundation for understanding how chromatin state is involved in regulating pluripotency and commitment to specific lineages. By assessing how chromatin state changes at promoters and enhancers during differentiation, we made several observations. First, a large number of promoters undergo a chromatin switch between methylation and acetylation at H3K27, which is strongly correlated with changes of gene expression. Importantly, such a chromatin state switch is most pronounced at promoters of genes involved in regulating pluripotency or cell fate determination, implicating the corresponding chromatin remodeling process in the regulation of pluripotency. Second, we show that the majority of enhancers mapped exhibit dramatic gain or loss of chromatin modifications H3K4me1 and H3K27ac during differentiation, and the dynamic chromatin modifications correlate with cell-type-specific gene expression within CORDs. The cell-type-specific enhancer regulation of genes within CORDs expands the potential of an ESC regulatory network. Third, we also identify a set of enhancers marked by H3K4me1 in hESCs and DFCs that become acetylated upon differentiation. This subset of enhancers bears the characteristic chromatin signature of poised enhancers 29. The poised enhancer state may allow for activation of early response genes important for the initial steps in differentiation (Figure 5). Our results therefore provide additional evidence supporting the role of epigenetic processes in regulating pluripotency and cell fate determination.
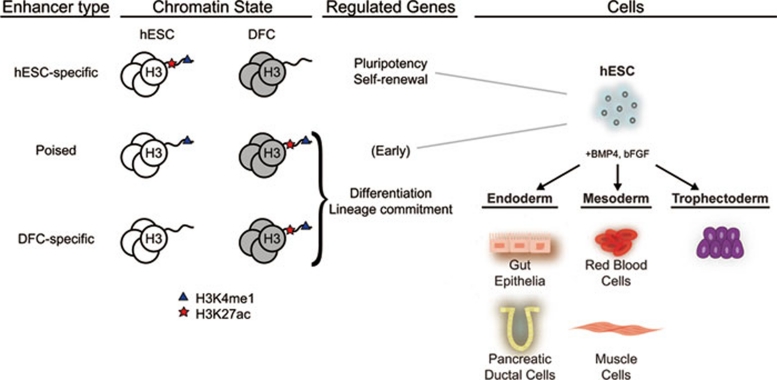
Figure 5.
Model of cell-type-specific enhancers and poised enhancers in cell fate. This model illustrates the role of poised enhancers in hESC pluripotency and cell fate commitment. ES cells grown in the presence of BMP4 and bFGF give rise to three of four possible lineages (ectoderm excluded). Poised enhancers contribute to initiation of lineage determination by activating early response genes that go on to establish the cell fate.
Materials and Methods
Cell culture
For ChIP-chip experiments, passage 32 H1 cells were grown in mTeSR1 medium 30 on Matrigel (BD Biosciences, San Jose, California) for 5 passages. A total of 15 × 10 cm2 dishes were grown using standard mTeSR1 culture conditions and 20 × 10 cm2 dishes were cultured in mTeSR1 supplemented with 200 ng/ml BMP4 (RND systems, Minneapolis, MN) for 4-6 days post passage. When cells were ∼70% confluent, they were crosslinked. To crosslink, 2.5 ml of crosslinking buffer (5 M NaCl, 0.5 M EDTA, 0.5 M EGTA, 1 M HEPES, pH 8, 37% fresh formaldehyde) was added to 10 ml culture medium and incubated at 37 °C for 30 min; 1.25 ml of 2.5 M glycine was added to stop the crosslinking reaction. Cells were removed from culture dish with a cell scraper, and collected by centrifugation for 10 min at 2 500 rpm. at 4 °C. Cells were washed three times with cold PBS. After the final spin, cells were pelleted and flash frozen using liquid nitrogen. BMP4-treated cells were subjected to the same procedure after 6 days of exposure.
For non-time course expression experiments, cells were grown in the same conditions as for ChIP-chip above for 4 days, and then RNA was harvested for analysis on NimbleGen microarrays (NimbleGen Systems). For the time course expression experiments, cells were grown in conditions similar to above, except that the BMP4 concentration was 50 ng/ml, and the cells were cultured for 5 days.
ChIP-chip and ChIP-Seq
ChIP-chip procedure and antibodies against H3K4me1, H3K4me3 and CTCF were previously described 24, 74, 78. Additional antibodies are commercially available (α-H3K27ac, ab4729, Abcam; α-H3K27me3, 07-44919, Upstate). ChIP-DNA samples were hybridized to NimbleGen HD microarrays (NimbleGen Systems). DNA was labeled according to NimbleGen Systems' protocol. Samples were hybridized at 42 °C for 16 h on a MAUI 12-bay hybridization station (BioMicro Systems). The GeneChip Microarray Core on the UCSD campus hybridized Affymetrix genome-wide tiling arrays according to the manufacturer's protocol. We used the Mpeak program to determine binding sites of CTCF peaks as previously described 74, 79 with the following modifications. Peaks consisted of at least three consecutive probes having a signal threshold above 1.5 standard deviations at a false discovery rate of 1%.
For ChIP-Seq, see Supplementary information, Data S1. All data are available through GEO, accession number: GSE30434.
Enhancer predictions
The procedure used to predict enhancers follows closely to that in 24. Specifically, we first bin the tiling ChIP-ChIP data into 100 bp bins, averaging multiple probes that fall into the same bin. Empty bins are interpolated if the distance between flanking non-empty bins is less than 1 kb, and set to 0 otherwise. We scan this binned data, keeping only those windows (1) in the top 10% of the intensity distribution and (2) having H3K4me1 and H3K4me3 profiles in the top 1% of all windows using the same training set of sites as in 24 (Figure 1A and 1B). We use a discriminative filter on H3K4me1 and H3K4Me3 to keep only those sites that correlate with the averaged enhancer training set more than the promoter training set. Finally, we apply a descriptive filter on H3K4me1 and H3K4me3, keeping only those remaining predictions having a correlation of at least 0.5 with an averaged training set.
Gene expression analysis for hESCs and differentiated cells
For the non-time course gene expression analysis, we isolated the total RNA from H1 ES cells or BMP4-treated cells using Trizol (Life Technologies Inc., Carlsbad, CA) according to the manufacturer's recommendations. For the time course gene expression analysis, RNA was isolated using the Qiagen RNeasy (Qiagen) with the column DNase I digestion step included. PolyA RNA was then isolated using the Oligotex mRNA Mini Kit (Qiagen). The mRNA were then reversed transcribed, labeled, mixed with differently labeled sonicated genomic DNA and hybridized to a single array that tiled transcripts from ∼36 000 human loci from the hg17 assembly (Roche NimbleGen). Detailed descriptions of array design, labeling, hybridization and data analysis are provided in the supplementary section (Supplementary information, Data S1). We set the expression level of genes in undifferentiated cells as 1 and calculated the relative fold change of individual genes in the differentiated cells.
Acknowledgments
This work is supported, in part, by funding from the California Institute of Regenerative Medicine (to BR; RS1-00292-1 and RN2-00905-1), the NIH (U01 ES017166-01) and the Ludwig Institute for Cancer Research.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Table S1, related to Figure 1 Figure 1 Sorted Gene List and Expression Values Ranked by Cg.
Table S2, related to Figure 1 Top 1% genes exhibiting H3K27 switch following differentiation.
Table S3, related to Figure 2 Enhancers Predicted by Chromatin Signatures in hESC and DFC.
Table S4, related to Figure 2 Complete Motif List for Enhancers.
Table S5, related to Figure 3 CTCF Bound Sites.
Table S6, related to Figure 3 Genes within CTCF-defined domains with top enhancers (top 1% Ce ranking)
Table S7, related to Figure 4 Genes Up-regulated at 3hrs with Poised Enhancer.
Figure S1, related to Figure 1 k-means cluster of modifications at 22 047 gene transcription start sites (center of 10-kb window).
Figure S2, related to Figure 2
Figure S3, related to Figure 2 Clustering of histone modifications at distal SOX2 and NANOG binding sites not predicted as enhancers.
Figure S4, related to Figure 3 Additional analysis of CTCF binding site analysis and CTCF-defined domains.
Figure S5, related to Figure 3 Changes in enhancer numbers correspond to changes in gene expression.
Figure S6, related to Figure 3 UCSC Genome browser shots of histone modifications and enhancer predictions in CTCF-defined blocks (CORDs) containing (A) SOX2, (B) OCT4(POU5F1), (C) NANOG, and the (D) HOXB locus.
Figure S7, related to Figure 4 Shared Enhancers May be Poised.
Supplemental Figures, Tables, Legends and Procedures for Hawkins, Hon et al.
References
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Chan YS, Yang L, Ng HH. Transcriptional regulatory networks in embryonic stem cells. Prog Drug Res. 2011;67:239–252. doi: 10.1007/978-3-7643-8989-5_12. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AS, Bernstein BE. Developmental biology. Pluripotent chromatin state. Science. 2009;323:220–221. doi: 10.1126/science.1166261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, et al. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, et al. DNA methylation and SETDB1/H3K9ME3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Ren B, Natarajan R. PU.1 and C/EBP(alpha) synergistically program distinct response to NF-kappaB activation through establishing monocyte specific enhancers. Proc Natl Acad Sci USA. 2011;108:5290–5295. doi: 10.1073/pnas.1017214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, et al. From the cover: histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Afouda BA, Ciau-Uitz A, Patient R. GATA4, 5 and 6 mediate TGFbeta maintenance of endodermal gene expression in Xenopus embryos. Development. 2005;132:763–774. doi: 10.1242/dev.01647. [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Weber H, Symes CE, Walmsley ME, Rodaway AR, Patient RK. A role for GATA5 in Xenopus endoderm specification. Development. 2000;127:4345–4360. doi: 10.1242/dev.127.20.4345. [DOI] [PubMed] [Google Scholar]
- Nelson TJ, Faustino RS, Chiriac A, Crespo-Diaz R, Behfar A, Terzic A. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008;26:1464–1473. doi: 10.1634/stemcells.2007-0808. [DOI] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Hyslop L, Stojkovic M, Armstrong L, et al. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells. 2005;23:1035–1043. doi: 10.1634/stemcells.2005-0080. [DOI] [PubMed] [Google Scholar]
- Deb K, Sivaguru M, Yong HY, Roberts RM. Cdx2 gene expression and trophectoderm lineage specification in mouse embryos. Science. 2006;311:992–996. doi: 10.1126/science.1120925. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh T, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, O'Geen H, Komashko VM, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Labosky PA. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem Cells. 2008;26:2475–2484. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LS, Loh YH, Zhang W, et al. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol Biol Cell. 2007;18:1348–1358. doi: 10.1091/mbc.E06-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006;20:1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Botquin V, Hess H, Fuhrmann G, et al. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddington RS, Rashbass P, Wilson V. Brachyury--a gene affecting mouse gastrulation and early organogenesis. Dev Suppl. 1992. pp. 157–165. [PubMed]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Braun T, Bober E, Winter B, Rosenthal N, Arnold HH. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. EMBO J. 1990;9:821–831. doi: 10.1002/j.1460-2075.1990.tb08179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–724. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ricciardi RP. E1A is the component of the MHC class I enhancer complex that mediates HDAC chromatin repression in adenovirus-12 tumorigenic cells. Virology. 2006;352:338–344. doi: 10.1016/j.virol.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Palmer MB, Majumder P, Green MR, Wade PA, Boss JM. A 3′ enhancer controls snail expression in melanoma cells. Cancer Res. 2007;67:6113–6120. doi: 10.1158/0008-5472.CAN-06-4256. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Dunn KL, Davie JR. The many roles of the transcriptional regulator CTCF. Biochem Cell Biol. 2003;81:161–167. doi: 10.1139/o03-052. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, et al. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Barrera LO, Ren B, Wu YN. ChIP-chip: Data, model, and analysis. Biometrics. 2007;63:787–796. doi: 10.1111/j.1541-0420.2007.00768.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 1 Figure 1 Sorted Gene List and Expression Values Ranked by Cg.
Table S2, related to Figure 1 Top 1% genes exhibiting H3K27 switch following differentiation.
Table S3, related to Figure 2 Enhancers Predicted by Chromatin Signatures in hESC and DFC.
Table S4, related to Figure 2 Complete Motif List for Enhancers.
Table S5, related to Figure 3 CTCF Bound Sites.
Table S6, related to Figure 3 Genes within CTCF-defined domains with top enhancers (top 1% Ce ranking)
Table S7, related to Figure 4 Genes Up-regulated at 3hrs with Poised Enhancer.
Figure S1, related to Figure 1 k-means cluster of modifications at 22 047 gene transcription start sites (center of 10-kb window).
Figure S2, related to Figure 2
Figure S3, related to Figure 2 Clustering of histone modifications at distal SOX2 and NANOG binding sites not predicted as enhancers.
Figure S4, related to Figure 3 Additional analysis of CTCF binding site analysis and CTCF-defined domains.
Figure S5, related to Figure 3 Changes in enhancer numbers correspond to changes in gene expression.
Figure S6, related to Figure 3 UCSC Genome browser shots of histone modifications and enhancer predictions in CTCF-defined blocks (CORDs) containing (A) SOX2, (B) OCT4(POU5F1), (C) NANOG, and the (D) HOXB locus.
Figure S7, related to Figure 4 Shared Enhancers May be Poised.
Supplemental Figures, Tables, Legends and Procedures for Hawkins, Hon et al.