Abstract
Methylation of cytosine is a DNA modification associated with gene repression. Recently, a novel cytosine modification, 5-hydroxymethylcytosine (5-hmC) has been discovered. Here we examine 5-hmC distribution during mammalian development and in cellular systems, and show that the developmental dynamics of 5-hmC are different from those of 5-methylcytosine (5-mC); in particular 5-hmC is enriched in embryonic contexts compared to adult tissues. A detectable 5-hmC signal appears in pre-implantation development starting at the zygote stage, where the paternal genome is subjected to a genome-wide hydroxylation of 5-mC, which precisely coincides with the loss of the 5-mC signal in the paternal pronucleus. Levels of 5-hmC are high in cells of the inner cell mass in blastocysts, and the modification colocalises with nestin-expressing cell populations in mouse post-implantation embryos. Compared to other adult mammalian organs, 5-hmC is strongly enriched in bone marrow and brain, wherein high 5-hmC content is a feature of both neuronal progenitors and post-mitotic neurons. We show that high levels of 5-hmC are not only present in mouse and human embryonic stem cells (ESCs) and lost during differentiation, as has been reported previously, but also reappear during the generation of induced pluripotent stem cells; thus 5-hmC enrichment correlates with a pluripotent cell state. Our findings suggest that apart from the cells of neuronal lineages, high levels of genomic 5-hmC are an epigenetic feature of embryonic cell populations and cellular pluri- and multi-lineage potency. To our knowledge, 5-hmC represents the first epigenetic modification of DNA discovered whose enrichment is so cell-type specific.
Keywords: epigenetics, 5-hydroxymethylcytosine, 5-methylcytosine, CpG-methylation, stem cells, mammalian development, pluripotency
Introduction
The methylation of cytosines in a CpG context is a common modification in mammalian genomes, and is generally associated with gene repression in vivo and in vitro 1. Alteration of 5-methylcytosine (5-mC) patterns during development contributes to the regulation of gene expression and cell specification 1, 2, 3. In addition to 5-mC, a novel cytosine modification, 5-hydroxymethylcytosine (5-hmC), has recently been found in mouse brain and murine embryonic stem cells (mESCs) 4, 5. The conversion of 5-mC to 5-hmC is catalysed by Tet (Ten-eleven translocation) oncogene family member proteins 4, 6. Notably, as 5-hmC is interpreted as 5-mC in bisulphite sequencing 7, 8, 9, 10, the routine method of mC identification, these two cytosine modifications are indistinguishable from each other in the vast majority of currently available experimental results. Therefore, there is a need to re-evaluate many DNA methylation data, taking into account the existence of this novel cytosine modification with its (probable) distinct functional role. Since it has been already reported that methyl-CpG binding proteins do not interact with 5-hmC-containing DNA substrates 7, 11, these two modifications are likely to play distinct roles in biological systems. Although a recent report suggested the importance of Tet1 in mESC self-renewal and inner cell mass (ICM) specification in early embryos 6, the biological functions and developmental distribution of genomic 5-hmC levels have not been studied. Here we assessed 5-hmC distribution throughout mammalian development, and in adult tissues in vivo and in in vitro cell systems using immunochemical methods.
Results
Genomic 5-hmC is enriched in embryonic and induced pluripotent stem cells compared to differentiated cells
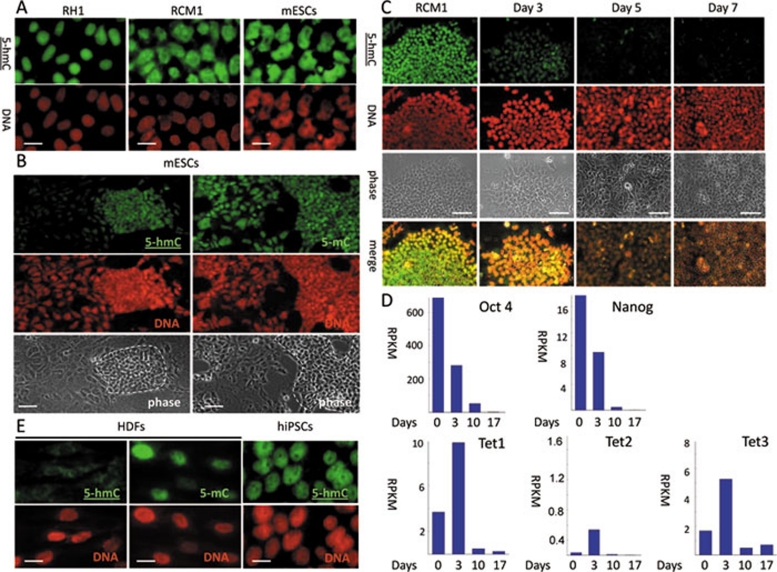
We used two commercially available anti-5-hmC antibodies produced by Diagenode and Active Motif for our analysis. Since the Diagenode antibody has not been characterised in immunochemistry previously, we first confirmed its specificity in dot blot assays using PCR-produced DNA fragments with all of the cytosines replaced by either 5-hmC or 5-mC, and total genomic human ESC and human dermal fibroblast (HDF) DNA (Supplementary information, Figure S1A). The anti-5-hmC antibody specifically recognised the 5-hmC-enriched PCR fragment and hESC genomic DNA, but not 5-mC-containing or unmodified PCR fragments or HDF DNA. The dot blot assay exhibited relatively low sensitivity, and only the equivalent of 500 ng of total genomic DNA produced a detectable signal with the anti-5-hmC antibody. Since the estimated genomic proportion of 5-hmC in mESCs is relatively low (<1% total cytosine content) 4, we used a peroxidase-conjugated secondary antibody coupled with a tyramide signal enhancement system for 5-hmC detection in subsequent immunochemical staining experiments. Under these conditions, the anti-5-hmC antibody produced distinctive nuclear staining patterns on mESCs and hESCs (Figure 1A and Supplementary information, Figure S1B), but not on human and mouse cancer and immortalised cell lines (Supplementary information, Figure S1C). These results were consistent with previously reported data obtained by thin layer chromatography (TLC) 4, 5 and with our dot blot results (Supplementary information, Figure S1A). We obtained essentially identical results using the Active Motif anti-5-hmC antibody, which has been successfully used in dot blots, immunochemistry and other applications previously 6, 9, 12, 13. In our experiments 5-hmC was strongly enriched in hESCs, compared to a very weak 5-hmC signal in HDFs (Supplementary information, Figure S2). Both antibodies also behaved similarly in all further experiments. We concluded that the Diagenode anti-5-hmC antibody specifically recognises 5-hmC, but not 5-mC, in immunocytochemical assays. Notably, the distribution of 5-hmC staining in ESCs was strikingly different from that of 5-mC. The intensity of the 5-hmC signal was strong in the cells forming the colonies of ESCs, and was considerably weaker in surrounding differentiating fibroblast-like “stromal” cells, whereas 5-mC was detectable in both cell types at similar levels (Figure 1B and Supplementary information, Figure S1D). This is consistent with the observation that 5-hmC content decreases upon differentiation of murine ESCs, as reported previously 4, 14. We decided to examine the dynamics of this process using a protocol to differentiate hESCs to hepatic endoderm 15. The 5-hmC signal dropped significantly following a 3-day treatment with Wnt3A and Activin to induce definitive endoderm, and was virtually undetectable after an additional 2 days of differentiation (Figure 1C). Concurrent analysis of transcriptome changes by global quantitative deep sequencing revealed a parallel reduction in the number of Oct4 and Nanog transcripts (Figure 1D). Unexpectedly, we observed an increase in the numbers of transcripts of all three Tet genes at 3 days post-induction at the time point where the 5-hmC signal dropped significantly (Figure 1D). Possible explanations for this discrepancy include the presence of mechanisms regulating Tet proteins at either the post-transcriptional/post-translational levels, or the inhibition of their oxygenase enzymatic activity. Over 5 days of differentiation, the numbers of Tet1, Tet2 and Tet3 transcripts also decreased (Figure 1D). To determine the correspondence of 5-hmC with acquisition of a pluripotent state we carried out 5-hmC immunostaining on human induced pluripotent stem cells (iPSCs) 16 and their parental somatic cell lines. hiPSC lines generated from both normal and Huntington's disease tissues (Supplementary information, Figure S3), but not from their parental HDFs, exhibited a strong 5-hmC signal (Figure 1E and Supplementary information, Figure S1D). In summary, our screening of in vitro cell systems for the presence of immunochemically detectable levels of 5-hmC revealed a strong correlation of high levels of this modification with a pluripotent stem cell state.
Figure 1.
The presence of immunochemically detectable 5-hydroxymethylcytosine (5-hmC) levels correlates with pluripotency in mammalian cells. 5-hmC is detectable in mouse and human (RH1 and RCM1) ESCs. (A) Lines stained with 5-hmC antibody. (B) 5-hmC and 5-mC staining of mouse ESC colonies (defined morphologically on the basis of phase images and indicated by dotted lines) and surrounding fibroblast-like differentiating cells. 5-hmC Immunostaining (C) and quantitative transcriptome deep sequencing (Solexa) data for Oct4, Nanog, Tet1, 2 and 3 (D) of RCM1 cells differentiating towards hepatic endoderm. Identical exposure times were used for different time points. Days post-induction are shown. RPKM – reads per kilobase per million reads. (E) High levels of 5-hmC are detectable in human iPSCs but not in human dermal fibroblasts (HDFs). DNA signal is indicated. Scale bars are 20 μm in A and E, 50 μm in B and 100 μm in C. The experiments were performed using the Diagenode antibody.
Global hydroxymethylation of the paternal genome coincides with the loss of 5-methylcytosine signal in zygote
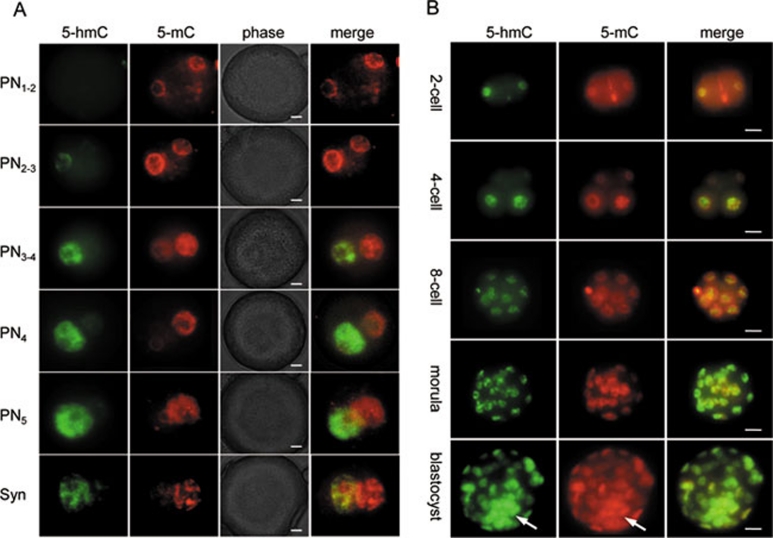
Several hours after fertilisation, chromatin of both maternal and paternal pronuclei of the mammalian zygote is subjected to extensive genome-wide reprogramming as the embryo makes a transition from a germ cell to a somatic developmental programme 2, 17. One notable reprogramming event is a dramatic loss of the 5-mC signal detectable by immunochemistry only in the paternal pronucleus after fertilisation 18. It has been suggested that conversion of 5-mC to 5-hmC may represent an intermediate step in the process of cytosine demethylation 4. In agreement with a recent study 19, we could not detect any 5-hmC staining in the embryos before 2.5 h post-fertilisation, when a weak 5-hmC signal started to appear in the paternal pronucleus (Figure 2A, PN1-2, PN2-3). The intensity of the 5-hmC signal increased dramatically during the next 2 h (Figure 2A, PN3-4, PN4), peaking at 5-8 h post-fertilisation (Figure 2A, PN5, syngamy) coincident with a significant decrease and subsequent loss of 5-mC staining in the male pronucleus. The majority of embryos exhibited a strong 5-hmC signal, exclusively in the paternal pronucleus, whereas the maternal pronuclear 5-hmC staining was weak or undetectable (Figures 2A, Supplementary information, S4A, S4B). This corresponded to a strong 5-mC signal in the maternal pronucleus and its loss in the paternal pronucleus. Some embryos (11.4% at PN4-5) showed comparable levels of strong 5-hmC staining in both male and female pronuclei concurrently with equally low or absent 5-mC signals in both pronuclei (Supplementary information, Figure S4A, S4B). Nonparametric statistical analysis showed a significant negative correlation (r = −0.9834, p < 0.0001) between 5-hmC and 5-mC immunostaining scores (Supplementary information, Figure S4A-S4C), suggesting that a decrease in 5-mC staining intensity is related to an increase in 5-hmC content in the mouse zygote at PN4-5. This most likely represents genome-wide 5-mC conversion into 5-hmC. Since 5-hmC is not detectable using an anti-5-mC antibody 4 and accumulates in the paternal genome at significant levels during the embryo's first cell cycle, together with a previous report, 19 these results suggest that the loss of 5-mC signal in the zygote at least partially represents a consequence of genome-wide 5-mC conversion into 5-hmC. 5-hmC-Independent demethylation of several genes has indeed been detected using bisulphate sequencing in the zygote 20, but bisulphite sequencing has never been performed on a genome-wide scale in preimplantation embryos. It is most likely that hydroxylation-independent demethylation of certain genome regions and global 5-mC hydroxylation occurs simultaneously in mouse embryos after fertilisation.
Figure 2.
5-Hydroxymethylcytosine (5-hmC) distribution in mouse pre-implantation development. (A) Distribution of 5-hmC and 5-mC during indicated pronuclear stages in mouse zygote. Merged and phase views, where pronuclei are distinguishable at PN3-PN4 stages, are shown. The exposure times were identical for each channel. 5-hmC antibodies produced by Active Motif were used. Scale bar, 10 μm. (B) 5-hmC Distribution at the indicated later stages of mouse pre-implantation development. 5-hmC Antibodies produced by Active Motif were used. 5-mC signal and merge view are indicated. ICM is arrowed. Scale bar, 10 μm.
High levels of 5-hmC colocalise with the ICM in blastocysts and with nestin-expressing cell populations in post-implantation mouse embryos
To determine the dynamics of 5-hmC content in later mammalian development, we carried out whole-mount immunostaining of pre-implantation and sections of post-implantation mouse embryos. A detectable 5-hmC signal was present throughout all the stages of pre-implantation development, slightly decreasing by the 8-cell stage (Figure 2B). Strong 5-hmC staining started to appear at the morula stage and was observed in mouse blastocysts with the maximum signal intensity in the ICM (Figure 2B and Supplementary information, Figure S5A). In contrast, the 5-mC signal showed less perceptible variation between the ICM and trophoblast cells of blastocysts than did the 5-hmC signal (Figure 2B and Supplementary information, Figure S5B).
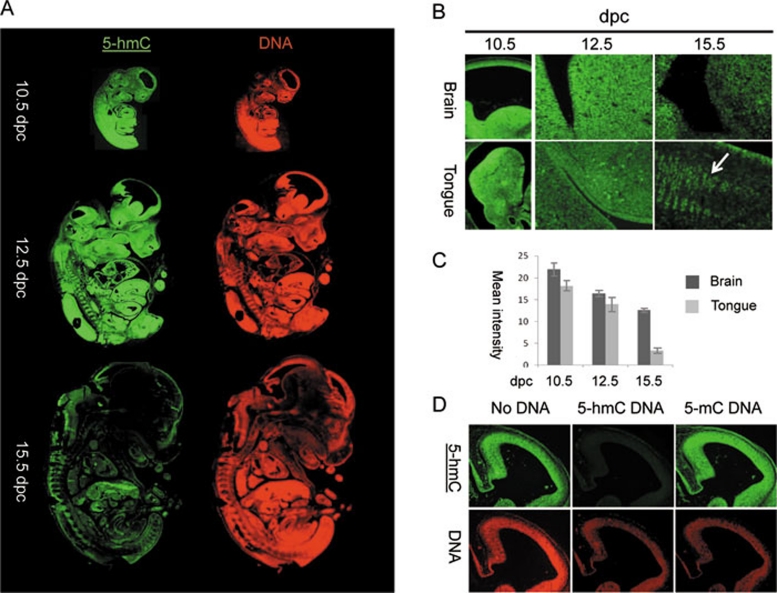
The 5-hmC enrichment in the ICM of mouse blastocysts is consistent with our immunochemical detection of 5-hmC in mESCs and previous TLC data 4, 5. It is also consistent with the expression of Tet proteins in both mESCs and the blastocyst ICM 4, 6. Interestingly, the study of murine post-implantation embryos revealed a pattern of regional enrichment of 5-hmC nuclear staining correlated with the presumptive location of multi-potent progenitor or stem cells, becoming increasingly restricted with development (Figure 3A). At 10.5 days post-coitum (dpc) most embryonic tissues were highly enriched for 5-hmC (Figure 3A). At 12.5 dpc, mouse embryos exhibited intense 5-hmC staining in a range of embryonic tissues. Significantly higher levels of 5-hmC were observed in the brain, spinal cord, tailbud and liver but not in heart, skin or mesenchymal tissue (Figure 3A and 3B, Supplementary information, Figure S5C-S5G, Figure 4A-4D). By 15.5 dpc, 5-hmC staining was generally less intense and limited to the brain and peripheral neural tissue (Figure 3A and 3B). According to 5-hmC signal quantification, the intensity of 5-hmC staining gradually decreased in brain tissue over time and fell dramatically in embryonic tongue between 12.5 dpc and 15.5 dpc (Figure 3B and 3C). Whereas high levels of 5-hmC were detected in most of the cells of embryonic tongue at 10.5 dpc and 12.5 dpc, 5-hmC distribution in this organ at 15.5 dpc was limited to certain cell populations (Figure 3B). As expected, over the same period of development, the global level of DNA methylation was not restricted to any particular embryonic cell populations and did not vary significantly between embryonic stages (Supplementary information, Figures S5C-S5G, S6).
Figure 3.
The dynamics of 5-hydroxymethylcytosine (5-hmC) in mouse post-implantational embryonic development. (A) Sagital sections of 10.5 days post coitum (dpc), 12.5 dpc and 15.5 dpc mouse embryos stained with Diagenode antibody to 5-hmC. Immunostaining for single-stranded DNA is shown in red. (B) Fragments of brain and tongue (indicated) embryonic sagital sections at 10.5 dpc, 12.5 dpc and 15.5 dpc stages (indicated). Corresponding slides were immunostained for 5-hmC in parallel with identical conditions and imaged with the same exposure times. Cell populations of embryonic tongue enriched in 5-hmC at 15.5 dpc are arrowed. (C) A schematic showing of the results of quantification of 5-hmC signal in embryonic brain and tongue at 10.5 dpc, 12.5 dpc and 15.5 dpc. Corresponding slides were immunostained for 5-hmC in parallel and imaged with the same exposure times. Mean values for mean signal intensities of 10 random measurements of parts of images shown in B are presented. The same central part of developing brain was quantified for all the stages analysed. Error is expressed as s.e.m. (D) The results of 5-hmC immunostaining of 3 adjacent sections of 12.5 dpc embryo brain region using primary antibody mix pre-incubated with PCR-produced DNA fragments where all the cytosines were replaced with either 5-hmC (5-hmC DNA) or 5-mC (5-mC DNA). Control staining without pre-incubation with any DNA is shown (no DNA). The 5-hmC immunostaining is specifically competed by 5-hmC- but not by 5-mC-containing DNA. Genomic DNA was visualised using single-stranded DNA antibody (indicated) used at a high titre. The experiments were performed using the Diagenode antibody.
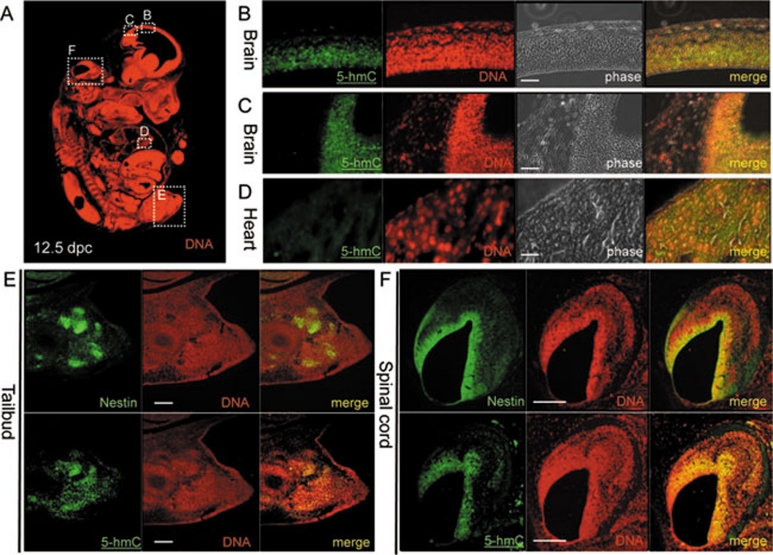
Figure 4.
5-Hydroxymethylcytosine (5-hmC) localises with nestin-expressing stem cell populations in 12.5 dpc mouse embryos. (A) Sagital section of 12.5 dpc mouse embryo with DNA visualised and B, C, D, E and F views indicated with dotted squares. Immunohistochemical detection of 5-hmC in sagital sections of mouse embryonic brain (B, C) and heart (D). Localisation of 5-hmC and nestin in the sagital adjacent sections of tailbud (E) and a part of the spinal cord (F). DAPI signal in nestin immunostaining was colourised as red. Anti-single-stranded DNA antibody was used for DNA visualisation in 5-hmC immunostaining experiments. Scale bars are 150 μm in B, C and D and 300 μm in E and F. The experiments were performed using the Diagenode antibody.
To identify these 5-hmC-rich cell populations, we performed immunostaining for nestin, an established marker of multi-potent progenitors and stem cells 21, 22, in 12.5 dpc mouse embryos. Adjacent serial sections were used for nestin and 5-hmC or 5-mC staining, as immunodetection of 5-hmC and 5-mC requires treatment of tissues with 4 N HCl, which is incompatible with the preservation of most protein epitopes. We observed that nestin staining closely correlates with the intense 5-hmC signal in brain, peripheral neural tissue and the tailbud, suggesting that a high genomic content of 5-hmC is a feature of nestin-expressing multi-potent fetal cells (Figure 4E and 4F). Consistent with our observed association of high levels of 5-hmC with multi-lineage potency, we also observed a highly localised 5-hmC signal in the olfactory epithelium of 17.5 dpc mouse embryos (Supplementary information, Figure S7). The olfactory epithelium is known to contain multi-potent stem cells constantly differentiating into supporting and olfactory cells in the embryo and adult 23.
The presence of high 5-hydroxymethylcytosine levels is restricted to neuronal tissue and bone marrow in adult human and mouse
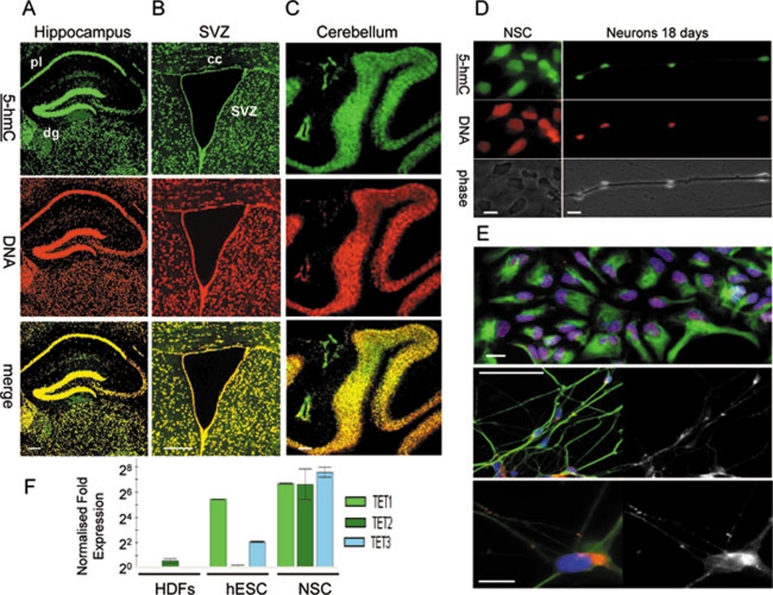
Finally, we evaluated adult murine and human tissues for genomic 5-hmC distribution. In agreement with previously reported data obtained by TLC 5, we could not detect any significant 5-hmC signal in mouse liver, skin, ovary, spleen or heart (Supplementary information, Figure S8), nor in any human tissue tested, except for brain and bone marrow (Figure 5 and Supplementary information, Figure S9). In mouse brain, strong 5-hmC nuclear staining was found not only in the hippocampal region and subventricular zone, which have previously been reported as regions of active neurogenesis containing populations of neural stem cells (NSCs) and neural progenitors 24, 25, 26, 27, 28, but also in the neuronal tissue of the cortex and other parts of the brain and cerebellum (Figure 6A-6C, Supplementary information, Figure S8B). In particular, in the hippocampus, higher levels of 5-hmC were observed in the dentate gyrus than the adjacent pyramidal layer (Figure 6A), but this difference may be due to the relatively higher cell density of the former. Brain white matter did not display an intense 5-hmC signal (Figure 6A, 6B and Supplementary information, Figure S8B). Consistent with these results and in agreement with already published data 29, NSC derived in vitro from hESCs also exhibit strong 5-hmC staining, which does not disappear or decline after 18 days of neuronal differentiation (Figure 6D and 6E), but differentiation of progenitors towards oligodendrocyte lineages is characterised by the progressive loss of 5-hmC staining (data not shown). Notably, the transcripts of all 3 Tet genes are highly expressed in human NSCs, which make them distinct from human ES cells where only Tet1 and Tet3 transcripts are present at significant levels (Figure 6F).
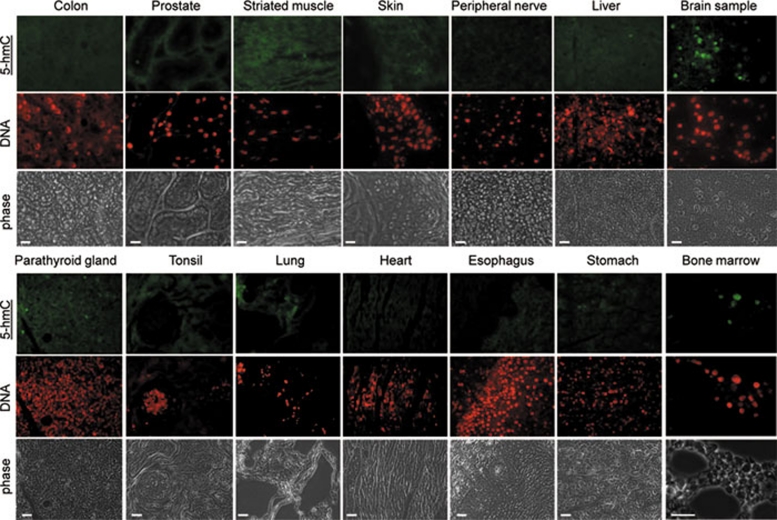
Figure 5.
The presence of immunochemically detectable 5-hydroxymethylcytosine (5-hmC) levels is restricted to bone marrow and neuronal tissues in adult mammals. A range of indicated normal adult human tissues from a Folio histological tissue array were immunostained with anti-5-hmC antibody. Phase views are shown. An identical maximal exposure of the images made using the 488 nm (5-hmC, tyramide) filter is shown for all the samples, except brain and bone marrow, to illustrate the pattern of background staining, which does not correspond to the DNA signal (indicated) in the majority of tissues. All the triplicate tissue samples on the array exhibited identical staining patterns. Six different bone marrow specimens were tested and gave essentially the same results. Representative views are shown. Scale bars are 20 μm, except 30 μm in bone marrow sample. Essentially the same results were obtained with samples of ovary, testis, kidney, small intestine, salivary gland, uterus, uterine cervix, mesothelium, eye muscle, adrenal gland, pancreas and breast tissue (see Supplementary information, Figure S9). The experiments were performed using the Diagenode antibody.
Figure 6.
5-Hydroxymethylcytosine (5-hmC) is dramatically enriched in both neuronal progenitors and differentiated post-mitotic cells of neuronal lineages. (A) 5-hmC Is visualised by immunochemistry in hippocampal A and ventricular (B) regions of mouse brain and cerebellar folia (C). Pyramidal layer (pl), dentate gyrus (dg), subventricular zone (svz) and corpus colossum (cc) are indicated. Scale bars are 250 μm. The experiments were performed using the Diagenode antibody. (D) Human neural stem cells (NSCs) exhibit high levels of 5-hmC staining, which does not disappear after 18 days of differentiation into neurons. hESC-derived NSCs (indicated) and neurons derived from them after 18 days (neurons 18 days) of differentiation were immunostained for 5-hmC. 5-hmC, DNA and phase views are indicated. Scale bars are 20 μm. The experiments were performed using the Diagenode antibody. (E) The upper panel shows a representative field of human NSC culture used for 5-hmC immunostaining in D stained for Sox1 (red nuclear signal), DAPI (blue) and nestin (green cytoplasmic signal). The majority of cells co-express both markers. Scale bar is 50 μm. The middle panel shows a representative field of the neuronal culture obtained after 18 days of differentiation and used for 5-hmC immunostaining in D, stained for β-III tubulin (green), Synapsin I (red in the colour image and presented as single channel in the grayscale image on the right) and DAPI (blue). The majority of cells co-express β-III tubulin and Synapsin (scale bar is 50 μm). The lower panel shows the same stained population at a higher magnification (scale bar is 10 μm). (F) The results of real-time RT-PCR analysis of Tet1/2/3 transcripts (indicated) in HDFs, human H1 ES cells and obtained from them human NSCs (indicated). The transcripts of all three Tet genes are highly expressed in human NSCs. The data were normalised with relative to GAPDH. Error is expressed as s.e.m.
Discussion
Our study shows a correlation between high levels of genomic 5-hmC and multi-lineage-potential during mammalian development and in a number of adult tissues. High 5-hmC level is a feature of induced or embryonic pluripotent stem cells. Nestin-expressing fetal stem cells in post-implantation development, and stem cell-rich tissues including adult bone marrow and nasal epithelium of the late development mouse embryo also exhibit high 5-hmC levels, whereas the actively proliferating unipotent cells of adult mammalian skin or liver do not. Sometimes it appears that a strong 5-hmC signal coincides with stronger DNA staining (SVZ, olfactory epithelium), but it is worth noting that a higher DNA content is an expected feature of some rapidly proliferating stem cell populations, wherein a significant proportion of cells is present in the S-phase of the cell cycle. A more detailed analysis will be required to determine if high 5-hmC is being utilised by other adult tissue-specific stem cells. According to our study, neuronal lineages seem to be exceptional in their maintenance of high 5-hmC levels in differentiated, post-mitotic cells. In agreement with this finding it has been previously reported that differentiation of mESCs into neuronal progenitors and subsequently into mature neurons has been shown to be accompanied by relatively few changes in DNA methylation, most of which occur during the first stage of differentiation 30, 31. Thus, neurons may represent a unique mammalian post-mitotic differentiated cell type, with a chromatin organisation similar in some aspects (5-hmC enrichment) to that of stem cells. Our analysis of 5-hmC distribution in adult mouse tissues is supported by other reports on 5-hmC tissue quantification showing that 5-hmC is strongly enriched in the central nervous system, compared to other adult organs 5, 12, 14. Our data are also generally in agreement with TLC-based data 5, an isotope-based chromatographic method for quantification of 5-hmC 12, and with immunolocalisation of 5-hmC in mouse brain presented in the same study 12. In contrast to Globisch and colleagues, our results do not reveal immunochemically detectable 5-hmC in adult mouse liver and kidney, but the resolution of the corresponding figures in that report does not allow us to draw conclusions on the specificity of the observed staining in these tissues. It is also important to note that there is a slight discrepancy between the study of enzymatic quantification of genomic 5-hmC 14, where its levels in mouse lung and, especially, in kidney were similar to those in brain, and both our immunochemical data and the other published data, based on biochemical methods 5, 12, where the 5-hmC content of lung and kidney was reported to be much lower than that of neural tissues. It is possible that these differences stem from the nature and sensitivity of the methods used to quantify genomic 5-hmC in the corresponding reports.
Previous studies have established that 5-mC- and 5-hmC-containing DNAs have different protein binding properties; for example, the interaction of MBD proteins with 5-mC is abolished by hydroxymethylation 7, 11. This implies that these two epigenetic modifications perform distinct biological functions. It is possible that an elevated genomic content of 5-hmC could contribute both to the general “openness” of the chromatin in stem cell and neuronal nuclei, and to the regulation of expression of specific target genes during stem cell self renewal and differentiation. Further work is necessary to reveal the identity and roles of any 5-hmC binding proteins, and identify genes affected by hydroxymethylation of cytosine residues in tissue-specific contexts.
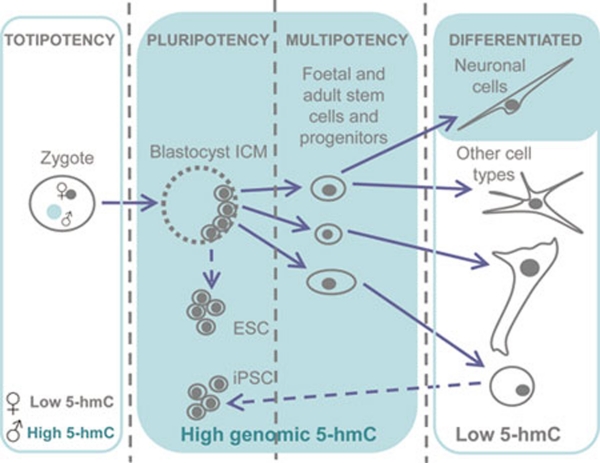
In conclusion, we show that unlike 5-mC, genomic 5-hmC is significantly enriched in embryonic contexts, cells with multi-lineage potency and neurons in mammals (Figure 7). To our knowledge, 5-hmC is the first epigenetic modification shown to possess such a cell-type-specific enrichment in its distribution.
Figure 7.
A graphical summary illustrating 5-hydroxymethylcytosine (5-hmC) distribution in mammalian development. High levels of 5-hmC are marked with green shading.
Materials and Methods
ES cell culture and iPSC generation
RH1 and RCM1 hESCs have been described previously 32, 33. Mouse HM1 ESCs were maintained on gelatin-coated dishes in Glasgow's Minimum Essential medium (GIBCO), supplemented with 15% fetal bovine serum, 55 μM β-mercaptoethanol (GIBCO), 2 mM l-glutamine, 0.1 mM MEM non-essential amino acids, 5 000 units ml−1 penicillin/streptomycin and 1 000 units ml−1 of murine LIF (Chemicon) under feeder-free conditions. hESCs and hiPSCs were cultured on matrigel-coated plates and slides in feeder-free mTeSR1 medium (STEMCELL Technologies). Cells were passaged using collagenase IV (200 units ml−1 in DMEM, GIBCO) and mechanical dissociation. hESC differentiation to hepatic endoderm was performed according to the published protocol 15. hiPSCs were produced as described previously 16 and validated by flow cytometry, embryoid body formation, differentiation into endoderm, mesoderm and ectoderm and RT-PCR as described 32.
Immunocytochemistry, immunohistochemistry, pre-implantation embryo culture, imaging and dot blot assay
For immunocytochemistry, cells were fixed in 4% formaldehyde for 15 min. Paraffin-embedded formaldehyde-fixed sections of wild-type CD1 mouse embryos and adult tissues were used for immunohistochemistry. A normal human tissue histological array (Folio) was used for 5-hmC detection in human tissues. Tissue sections were de-waxed according to standard procedures. Cells and tissue sections were permeabilised for 15 min with PBS containing 0.5% Triton X-100. For 5-hmC and 5-mC staining, permeabilised cells and tissue sections were incubated in 4 N HCl for 1 h at 37 °C and then neutralised in 100 mM Tris-HCl (pH 8.5) for 10 min, followed by a standard immunostaining protocol.
Anti-5-hmC (Diagenode, 1:1 000 dilution; Active Motif, 1:5 000; 1:50 000 dilutions), anti-5-mC (Eurogentec) and anti-nestin clone 401 (DSHB) primary antibodies were used. DNA was visualised using an anti-single-stranded DNA antibody (Zymo research). Peroxidase-conjugated anti-rat secondary antibody (Dako) and the tyramide signal enhancement system (Perkin Elmer) were employed for 5-hmC detection.
Mouse embryos were produced by mating superovulated F1 females with CD1 males, and cultured according to standard procedures. Anti-5-hmC (Active Motif, 1:5 000 dilution) and anti-5-mC (Eurogentec, 1:200 dilution) antibodies were used for immunochemistry. Pre-implantation embryos were immunostained as described 34 with the use of peroxidase-conjugated secondary antibody and the additional tyramide (Perkin-Elmer) signal enhancement step for 5-hmC staining. Control staining without primary antibody produced no detectable signal. Images were acquired using a Zeiss Axiovert 200 immunofluorescence microscope and Axiovision software. Dot blot assays were performed as reported previously 10.
Image quantification
Image quantification was performed using Fiji software. Corresponding slides were processed in identical conditions and were imaged at the same exposure settings. Mean intensities were measured for 10 random squares on each region of interest for each sample. The 5-hmC signal was standardised to the total DNA signal. Mean values of the mean intensities are presented. Experimental error is expressed as s.e.m.
Statistical analysis
We used a numerical grading system 0 (absent), 1 (low) and 2 (high) to account for differences in 5-hmC and 5-mC staining intensities in maternal and paternal pronuclei of the mouse zygotes (the examples of the categories are arrowed on Supplementary information, Figure S4A). The statistical analysis was performed using GraphPad Prism software. The individual 5-hmC and 5-mC immunostaining scores (total number of xy pairs = 70) of both maternal (n = 35) and paternal pronuclei (n = 35) were correlated using Spearman's nonparametric test. The P value was considered significant for P < 0.05.
Solexa deep sequencing
Total RNA was isolated from hESCs at the times indicated after induction of differentiation and sequenced directly using a Solexa 1G system. The Solexa results were normalised as follows: the raw count reads of each gene were first divided by the total number of aligned reads multiplied by 1 million to take into account the depth of sequencing. The results were then divided by the length of each gene multiplied by 1 000 to get an RPKM value (read per kilo bases per million reads).
Neural stem cells
Neural stem cells were generated from p57 H9 hESCs and cultured as described 35. NSCs' identity was confirmed by immunostaining for Pax6, Sox2, Sox1 and Nestin. H9 hESCs were cultured on Matrigel with TeSR2 (STEMCELL Technologies) medium. Differentiation was performed in Neurobasal medium (N2 supplement 1:100).
Real time RT-PCR
Total RNA was extracted with TriReagent (Sigma). Samples were reverse transcribed using random primers (Promega) and Superscript II RT (Invitrogen). Products were detected using SYBR Green PCR Mastermix (Applied Biosystems) and a PTC-200 cycler with a Chromo-4 detection system (MJ Research). Data were normalised with relative to GAPDH. Error is expressed as s.e.m. Primer sequences for human Tet proteins are available upon request.
Immunostaining competition experiment
Immunostaining competition experiments were performed as described 12 using PCR-produced 100-bp DNA fragments, where all the cytosines were replaced with either 5-hmC or 5-mC.
Acknowledgments
We thank Adrian Bird (University of Edinburgh) for encouragement, Paz Freile, Sebastian Greenhough, Sharmin Haideri, Heidi Mjoseng, Jane Taylor (MRC Centre for Regenerative Medicine, University of Edinburgh) and Britt Tye (Roslin Cells) for technical support; the Histology team of MRC Human Reproductive Sciences Unit for excellent service and John Iredale (MRC Centre for Regenerative Medicine, University of Edinburgh) for access to Solexa data. DH is an RCUK fellow.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
5-hmC detection in human ESCs (RH1) and human dermal fibroblasts (HDFs) using a 5-hmC antibody produced by Active Motif.
hiPSC validation.
Immunochemistry on mouse blastocysts performed using anti-5-hmC antibody produced by Diagenode.
Lateral section of the nasal cavity region of a mouse 17.5 dpc embryo showing the olfactory epithelial tissue stained with anti-5-hmC antibody.
Related to figure 5.
Supplementary Experimental Procedures
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu H, Shiragami M. Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry. 1979;18:632–637. doi: 10.1021/bi00571a013. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulphite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48:317–319. doi: 10.2144/000113403. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Fletcher J, Payne C, et al. Highly efficient differentiation of hESC to functional hepatic endoderm requires ActivinA and Wnt3a signalling. Proc Natl Acad Sci USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in mammalian development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Wislet-Gendebien S, Wautier PF, Leprince B. Astrocytic and neuronal fate of mesenchymal stem cells expressing nestin. Brain Res Bull. 2005;68:95–102. doi: 10.1016/j.brainresbull.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Wiese C, Rolletschek A, Kania G, et al. Nestin expression-a property of multi-lineage progenitor cells. Cell Mol Life Sci. 2004;61:2510–2522. doi: 10.1007/s00018-004-4144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell W, Féron F, Wetzig A, et al. Multipotent stem cells from adult olfactory mucosa. Dev Dyn. 2005;233:496–515. doi: 10.1002/dvdy.20360. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci USA. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Alvarez-Buylla A. Neural stem cells in mammalian development. Curr Opin Cell Biol. 2006;18:704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Seri JM, García-Verdugo BS, McEwen A, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory. Nat Rev Neurosci. 2010;11:339–350. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Fletcher JM, Ferrier PM, Gardner JO, et al. Variations in humanized and defined culture conditions supporting derivation of new human embryonic stem cell lines. Cloning Stem Cells. 2006;8:319–334. doi: 10.1089/clo.2006.8.319. [DOI] [PubMed] [Google Scholar]
- De Sousa PA, Gardner J, Sneddon S, et al. Clinically failed eggs as a source of normal human embryo stem cells. Stem Cell Res. 2009;2:188–197. doi: 10.1016/j.scr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Beaujean N, Taylor J, Gardner J, et al. Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biol Reprod. 2004;71:185–193. doi: 10.1095/biolreprod.103.026559. [DOI] [PubMed] [Google Scholar]
- Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
5-hmC detection in human ESCs (RH1) and human dermal fibroblasts (HDFs) using a 5-hmC antibody produced by Active Motif.
hiPSC validation.
Immunochemistry on mouse blastocysts performed using anti-5-hmC antibody produced by Diagenode.
Lateral section of the nasal cavity region of a mouse 17.5 dpc embryo showing the olfactory epithelial tissue stained with anti-5-hmC antibody.
Related to figure 5.
Supplementary Experimental Procedures