Abstract
Brachydactyly type A1 (BDA1), the first recorded Mendelian autosomal dominant disorder in humans, is characterized by a shortening or absence of the middle phalanges. Heterozygous missense mutations in the Indian Hedgehog (IHH) gene have been identified as a cause of BDA1; however, the biochemical consequences of these mutations are unclear. In this paper, we analyzed three BDA1 mutations (E95K, D100E, and E131K) in the N-terminal fragment of Indian Hedgehog (IhhN). Structural analysis showed that the E95K mutation changes a negatively charged area to a positively charged area in a calcium-binding groove, and that the D100E mutation changes the local tertiary structure. Furthermore, we showed that the E95K and D100E mutations led to a temperature-sensitive and calcium-dependent instability of IhhN, which might contribute to an enhanced intracellular degradation of the mutant proteins via the lysosome. Notably, all three mutations affected Hh binding to the receptor Patched1 (PTC1), reducing its capacity to induce cellular differentiation. We propose that these are common features of the mutations that cause BDA1, affecting the Hh tertiary structure, intracellular fate, binding to the receptor/partners, and binding to extracellular components. The combination of these features alters signaling capacity and range, but the impact is likely to be variable and mutation-dependent. The potential variation in the signaling range is characterized by an enhanced interaction with heparan sulfate for IHH with the E95K mutation, but not the E131K mutation. Taken together, our results suggest that these IHH mutations affect Hh signaling at multiple levels, causing abnormal bone development and abnormal digit formation.
Keywords: Indian hedgehog, BDA1, diffusion, heparin, crystal structure, degradation
Introduction
The Hedgehog (Hh) family of secreted proteins regulates a variety of developmental processes, maintains adult tissue homeostasis, and functions as a morphogen gradient 1, 2. The events leading to the transduction of the Hh signal involve protein synthesis and processing, intracellular trafficking and release, extracellular movement, receptor binding, and subsequent activation of the target genes. In Hh-producing cells, Hh protein is synthesized in the endoplasmic reticulum as a precursor of ∼46 kDa. Following cleavage of the signal peptide, the precursor undergoes an autocatalytic process to yield the functional N-terminal domain (about 20 kDa), to which cholesterol and palmitoyl moieties are also added (HhN) 3, 4, 5. Secretion is thought to involve lipid rafts and a transmembrane protein Dispatched (Disp) 6.
Hedgehog signaling occurs via binding to the receptor Patched (PTC) on the receiving cells, and the signal is transduced through Smoothened (SMO) and the GLI complex, resulting in the activation of target genes, such as Ptc1, Gli1, and Hip1 7, 8. The range of Hh signaling is thought to be facilitated by its interaction with heparan sulfate proteoglycans (HSPGs) in the extracellular matrix 9, 10, 11 and the formation of a soluble multimeric complex where the lipid moieties are buried to facilitate movement 12, 13, 14. A defining feature of Hh signaling is that small changes in the concentration of Hh ligand can elicit different cellular responses. As a result, the processing, release, and trafficking of Hh ligands must be tightly regulated to ensure the proper signaling gradient, changes in which could lead to dysregulation of cellular processes.
In mammals, there are three members of the Hh family, Sonic (Shh), Indian (Ihh), and Desert (Dhh) Hh, each of which has tissue-specific functions. Mutations in SHH are known to cause holoprosencephaly (HPE) in humans 15, 16. More recently, IHH was shown to be a disease locus for brachydactyly type A1 (BDA1) 17, 18. It is the first recorded example of a Mendelian autosomal dominant inheritance disorder in humans 19 and is characterized by a pronounced shortening of the middle phalanges or their fusion with the terminal phalanges and by a shortening of the proximal phalanges (the thumbs and big toes).
IHH is produced by prehypertrophic chondrocytes during endochondral ossification, and it has a role in chondrocyte maintenance, proliferation, and hypertrophic differentiation 20. Ihh−/− mice die at birth because of respiratory failure caused by restrictive underdevelopment of the rib cage, but mice heterozygous for the null allele were normal, with no observable skeletal or digital abnormalities. This suggests that the Ihh mutations causing BDA1 are not likely to result into a simple loss of function.
Since the discovery of the first three heterozygous missense mutations, c.G283A (p.E95K), c.C300A (p.D100E), and c.G391A (p.E131K) (c. represents cDNA and p. represents protein) in the region encoding the amino-terminal signal domain of the Ihh gene (Figure 1A) 17, additional heterozygous and homozygous missense Ihh mutations that cause BDA1 21, 22, 23, 24 and acrocapitofemoral dysplasia (ACFD) 25 have been reported. Notably, multiple mutations in codons 95 and 100 indicated that these codons might be mutation hot spots. Moreover, when mapped onto an X-ray crystal structure of the SHH protein, a number of these mutations are within a calcium-binding groove that interacts with the PTC receptor and other interacting partner proteins, including HIP and CDO 26. These mutations affected the strength of binding to PTC, reducing the signaling capacity 26. In a mouse model for BDA1, with a human-equivalent p.E95K mutation, signaling potency was reduced, and the range of signaling was enhanced. This mutation affected chondrocyte differentiation in the growth plate, delaying endochondral bone formation, and impaired recruitment of mesenchymal cells into the distal cartilage of the developing digit, causing the brachydactyly phenotype 27. However, detailed biochemical characterization of the three key mutations that line the surface of this calcium-binding interaction groove has not been performed, and whether these mutations lead to a similar physical and biochemical outcome is not clear.
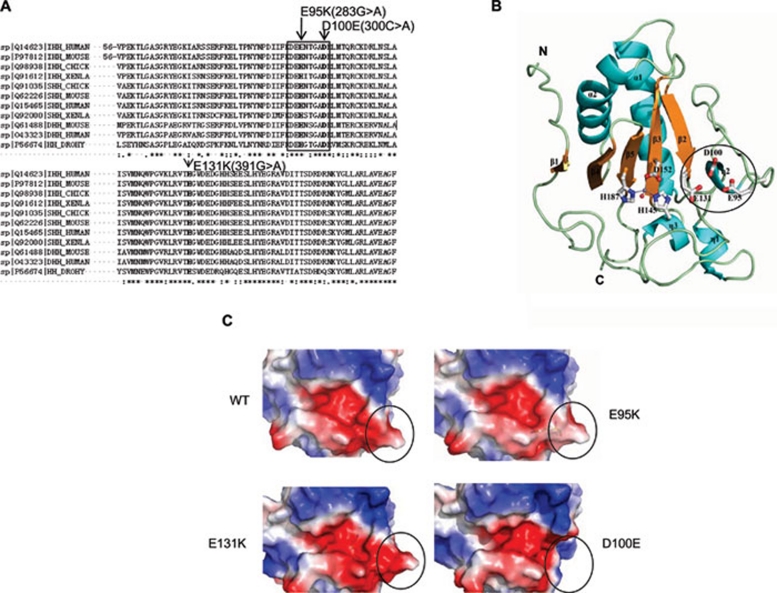
Figure 1.
Crystal structure and electrostatic potentials of human IhhN protein. (A) Comparison of the amino acid sequences of Drosophila, Xenla, chick, mouse, and human Hedgehog proteins. The mutations are shown in bold, and the eight-amino acid motif is indicated within the black pane. (B) Schematic representation of the overall structure of human IhhN. Secondary structures are labeled, with the α-helix and η-helix colored in cyan and the β-strand in yellow. The zinc ion is shown as the magenta dot, and the side chains of H145, D152, and H187 are shown in white with red oxygen atoms and blue nitrogen atoms. The zinc-bound water molecule is shown as a red dot. The N- and C-termini are individually labeled. The missense mutations at positions E95, D100, and E131, linked to BDA1, are labeled and shown as sticks. The eight-amino acid motif is indicated as a black circle. (C) Electrostatic potentials of human IhhN. Compared with WT, the E95K mutant changes the negatively charged area to positive. The E131K mutant exists in two alternative conformations, one of which is consistent with the WT, and the other forms a salt bridge between the carboxyl oxygen atom of E95 and the Nɛ atom of K131. The D100E mutant crystal structure showed high flexibility and local structure changes in the region comprising residues 93-98, indicated by a black circle. The electrostatic potentials of protein surfaces were scaled to the range of −5 (red) and 5 kT (blue), calculated using PyMOL 31 and APBS 32.
In this study, we report the crystal structure of the functional N-terminal domain of human IHH and show that the E95K mutation changes the charge, from negative to positive, of a surface involved in the interaction with partners, without changing the vicinal tertiary structure; however, the D100E mutation changes the local tertiary structure. Nevertheless, these subtle structural changes in the calcium-binding domain impaired protein stability, and the mutant proteins were subject to intracellular degradation. The changes also impair the interaction with the receptor PTC1, reducing signaling potency in cell-based assays. Notably, the formation of a multimeric complex appeared unaffected, but an enhanced interaction with HS by p.E95K might explain the longer range of signaling observed in a mouse model for BDA1.
Results
Structural analysis of WT and mutant IhhN
To better understand the effect of the Ihh mutations, we determined the crystal structures of human wild type (WT) and three BDA1-related (E95K, E131K, and D100E) mutant IhhN protein domains (Supplementary information, Table S1). The overall structures of the WT and mutant IhhN are almost identical, consisting of an α+β sandwich core with two α-helices and a mixed six-stranded β-sheet surrounded by a two-stranded anti-parallel β-sheet and three short helices (Figure 1B). At the bottom of a solvent-accessible cleft, a zinc ion is coordinated by the side chains of H145, D152, and H187 and a water molecule, which is similar to the situation seen in murine ShhN (92% sequence identity) 28. The main chain superimposition between human IhhN and murine ShhN gave a root mean square deviation (RMSD) of 0.202 Å, indicating a significant similarity. Furthermore, two recent papers have solved the structures of IhhN complexes and demonstrated that IhhN is capable of binding calcium, which is most significant for the interactions with its partners 29, 30. However, IhhN shows a relatively low binding affinity for calcium, and bound calcium was not observed in either the WT IhhN or the mutant forms in our study, without the addition of calcium during the purification and crystallization process.
In the structure of murine ShhN, a crystal-packing interaction was found between two symmetry-related subunits, in which the C-terminus of one subunit is buried at the zinc-binding site of the other subunit, as previously described 28. In our study, we found that the main interactions between the C-terminus structures and the binding clefts consist of hydrogen bonds, rather than electrostatic interactions. Surface electrostatic potentials show that the binding cleft is fully negatively charged and is located at the solvent-accessible side of the mixed β-sheet. The overall structure and charge distribution are almost identical between the WT and the mutant proteins, although variations are found at the negatively charged binding cleft in mutant proteins (Figure 1C). Compared with WT, the E95K substitution changes a negatively charged area to positive, without altering the vicinal tertiary structure. Notably, the E95 residue in the E131K structure exists in two alternative conformations; one is consistent with the WT protein and the other forms a salt bridge between the carboxyl oxygen atom of E95 and the Nɛ atom of K131. Although the side chain of K131 is buried without changing the local charges in both conformations, the surface in the vicinity of E95 is obviously changed in the latter conformation (Figure 1C). In the structure of the D100E mutant, the charge distribution in the vicinity of the zinc-binding site is barely affected. Notably, the region comprising residues 93-98 shows high flexibility, as suggested by the weak electron density in this part of the structure and shown by the black circle in Figure 1C.
Enhanced degradation of IhhN proteins with BDA1 mutations via the lysosomal pathway
To assess how the mutant proteins are processed and maintained within mammalian cells, the expression and processing of the IHH proteins were studied in transiently transfected EcR-CHO cells. Western blot analysis of the cell extracts showed that the level of mature IhhN fragments from the E95K- and D100E-transfected cells was significantly reduced (Figure 2A), although the levels of their corresponding IhhC fragments were comparable to that of cells transfected with the WT construct (Figure 2B). This result suggests that the E95K and D100E IhhN fragments are unstable and subjected to proteolytic degradation, rather than to impaired autoprocessing of the IHH precursors. This observation was supported by immunofluorescence staining of transfected cells in which the mutant IhhN proteins were found to be localized at the cell surface, although the signals for E95K and D100E were weaker than cells expressing WT proteins (Supplementary information, Figure S1). The tethered IhhN proteins might have undergone an autocatalytic modification with cholesterol, which is essential for Hh membrane association 33. Furthermore, we showed that IhhN proteins are degraded through the lysosomal pathway, as incubation with two lysosome inhibitors, chloroquine and bafilomycin, significantly reduced the degradation of IhhN proteins, but treatment with proteasome inhibitor MG132 did not (Figure 2C).
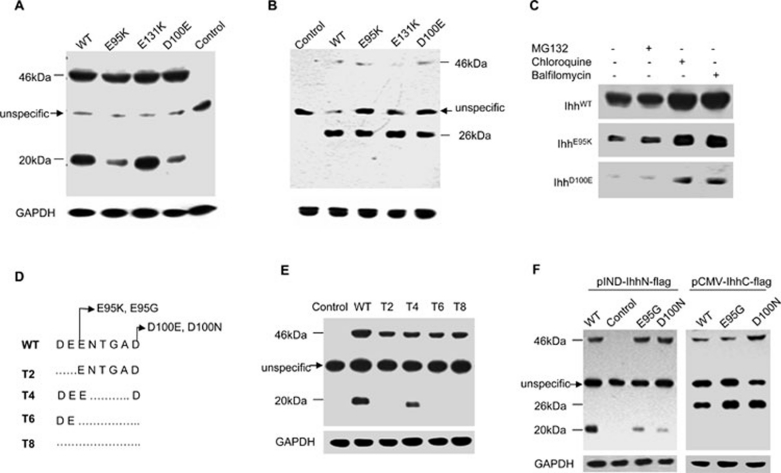
Figure 2.
Analysis of the processing and intracellular maintenance of BDA1 mutant IhhN proteins. (A, B) Cell homogenates of EcR-CHO cells transfected with the WT or mutant Ihh in which double FLAG-epitope tags were introduced into the N- (A) or C-terminus (B) of the IhhN fragment were analyzed by western blot. Molecular masses (kDa) are indicated on the left, where 46 kDa refers to the full-length IHH protein and 26 kDa and 20 kDa indicate IhhC and IhhN proteins, respectively. The arrow indicates a nonspecific band. Control refers to the cell homogenates transfected with an empty vector. Protein loading was monitored by western blotting of the mouse anti-GAPDH monoclonal antibody. The levels of E95K and D100E IhhN fragments (20 kDa) are reduced significantly (A), while the levels of IhhC fragments are comparable (B). (C) The degradation of WT and two mutant IhhN proteins is suppressed by lysosome inhibitors (chloroquine and balfilomycin), but not by a proteasome inhibitor (MG132). (D) Sketch map showing the truncation constructs. The deleted amino acids are shown as a broken line. (E) Stability of the truncated proteins in EcR-CHO cells. The IhhN fragments of T2, T6, and T8 are barely detectable, but the T4 fragment is comparable to that of WT. (F) Stability of E95G and D100N mutant proteins in EcR-CHO cells. E95G and D100N IhhN fragments are degraded, and the corresponding IhhC fragments are stable, with levels comparable to WT.
Multiple mutations in codons 95 and 100 indicate that these codons might be mutation hot spots. Moreover, their proximity to one another suggests that they might reside in a limiting region of the IHH protein. Protein sequence alignment across species indicates that the amino acids flanking E95 or D100 are highly conserved (Figure 1A). From the crystal structure, there appears to be an eight-amino acid motif that participates in the formation of a cleft at the surface of the IhhN protein (Figure 1B). To investigate the role of this motif and the key amino acids that might be involved, we generated truncated constructs (T2, T4, T6, and T8), based on the polarization of amino acids, as shown in Figure 2D, for expression in transfected EcR-ECHO cells. Immunoblot analysis of cell lysates transfected with these constructs indicated that aspartate (D) and glutamate (E) are essential for IhhN-peptide stability because the IhhN fragments of T2 and T6, but not T4, were readily degraded (Figure 2E). Overall, our findings suggest that the negatively charged calcium-binding groove, and the D and E amino acids at the surface of the structure are crucial for stability. Consistent with this notion, we showed that the IhhN fragments of other BDA1 mutations at these positions (p.E95G and p.D100N) were also unstable (Figure 2F).
The degradation of mutant IhhN proteins is temperature-sensitive
Holoprosencephaly-related mutations in SHH impair the capacity of Hh signaling because of the induction of a temperature-dependent conformational change of the SHH protein 34. Although the change is subtle, based on the observed changes in the crystal structure of the mutant IhhN proteins, a study of the stability of mutant IHH proteins, at different temperatures and durations of temperature exposure, revealed that WT and mutant IHHs are all stable at 30 °C but become unstable at 37 °C. As shown in Figure 3A, the E95K and D100E mutants degraded more quickly than the WT protein. In particular, D100E was almost completely undetectable after a 3-h treatment with puromycin (Figure 3A and Table 1). In fact, the D100E IhhN fragment was barely detectable in the conditioned medium (data not shown). We hypothesized that this result might be due to a rapid degradation of the D100E protein that impaired its membrane localization and subsequent secretion. This hypothesis was supported by the immunoblot assay of the D100E proteins with and without cholesterol modification. The D100E IhhN fragment with cholesterol modification was barely detectable in the conditioned medium, whereas fragments without cholesterol modification can be readily detected (Figure 3B).
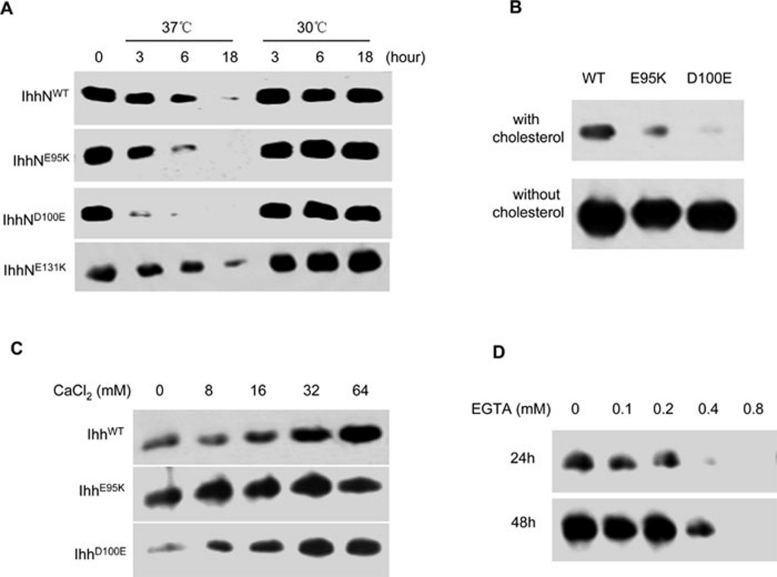
Figure 3.
Temperature-sensitive and calcium-dependent degradation of IhhN proteins. (A) Stability of WT or mutant IhhN proteins in EcR-CHO cells cultured for various durations at 30 °C and 37 °C. D100E mutant IhhN protein is degraded more rapidly than the others at 37 °C. All proteins are stable at 30 °C. (B) D100E IhhN is detectable in the conditioned medium from transiently transfected EcR-CHO cells at 37 °C, when the cholesterol modification is disrupted. The primary antibody indicated above is an anti-FLAG monoclonal antibody. (C) Wild type, E95K, and D100E mutant protein expressions were analyzed in the presence of increasing calcium concentrations (8-64 mM). Their relative stabilities were significantly enhanced. (D) Wild type IHH protein expression was assayed in the presence of increasing EGTA concentrations (0.1-0.8 mM). The level of IhhN protein was markedly reduced in the presence of 0.4 mM EGTA and was barely detectable in the presence of 0.8 mM EGTA.
Table 1. Properties of mutant IhhN proteins.
| Protein | Autoprocessing | Stability | Cholesterol modification | Palmitoylation | Multimer formation | Relative AKP induction activity at 500 nM (750 nM) in C3H10T1/2 assay | Dissociation constant (Kd) for Ptc-CTD, nM* | Heparin-binding affinity, salt elution concentration, M |
|---|---|---|---|---|---|---|---|---|
| WT | + | +++ | + | + | + | 1.0 (1.0) | 20.6 | 0.32-0.49 |
| E95K | + | + | + | + | + | 0.47 (0.5) | 40.6WΩ※ | 0.32-0.66 |
| E131K | + | +++ | + | + | + | 0.50 (0.52) | 30.5 | 0.32-0.49 |
| D100E | + | − | + | ND | ND | 0.44 (0.38) | >100 | ND |
Calcium is required for the stability of the Indian Hedgehog protein
A recent paper showed that BDA1 mutations in IHH are clustered in a calcium-binding groove that is involved in the interaction with the receptor and binding partners 26. Given that we have shown an enhanced degradation of the Hh proteins associated with the BDA1 mutations, we hypothesize that the rapid degradation of mutant IhhN is related to impaired calcium binding. Indeed, the intracellular level of E95K and D100E IhhN was markedly increased in the presence of calcium (Figure 3C). We further investigated the stability of WT IhhN using increasing concentrations of EGTA, a calcium chelator. The amount of WT IhhN was markedly reduced in the presence of 0.4 mM EGTA and was barely detectable at 0.8 mM EGTA (Figure 3D). Furthermore, we analyzed the protein stability at different concentrations of ionomycin, a calcium ionophore, and found that the intracellular level of E95K and D100E IhhN increased in the presence of ionomycin, demonstrating a requirement for calcium in the stability of the IhhN protein (Supplementary information, Figure S2A). These results suggest that calcium binding might be affected in the E95K and D100E mutant proteins, which could contribute to the enhanced intracellular degradation.
BDA1 mutations impair IHH-PTC1 binding
Hedgehog protein signals receive cells by binding to the PTC1 receptor. We have previously shown that the E95K mutation affected the affinity of this binding 27. Thus, we performed the same cell-based competition assay for the D100E and E131K mutants, and compared the results with those obtained from the WT and E95K IHH proteins. Quantitative analysis of 32P-IhhN binding to PTC1 in a Ptc-CTD140 expression cell line revealed that the interaction of D100E and E131K IHH proteins with PTC1 was significantly reduced compared with WT. We observed a higher dissociation constant (Kd) for E131K (30.5 nM) than for WT (20.6 nM), with the Kd for D100E binding exceeding 100 nM (Figure 4A and Table 1). The Kd of E95K was previously determined to be 40.6 nM 27. The amount of mutant IhhN protein that was added to the medium was identical to that used for WT, as determined using BCA protein quantification. The recombinant mutant proteins added to the medium were stable at the assay temperature of 4 °C for 2 h and 4 h, showing no degradation (Supplementary information, Figure S2B).
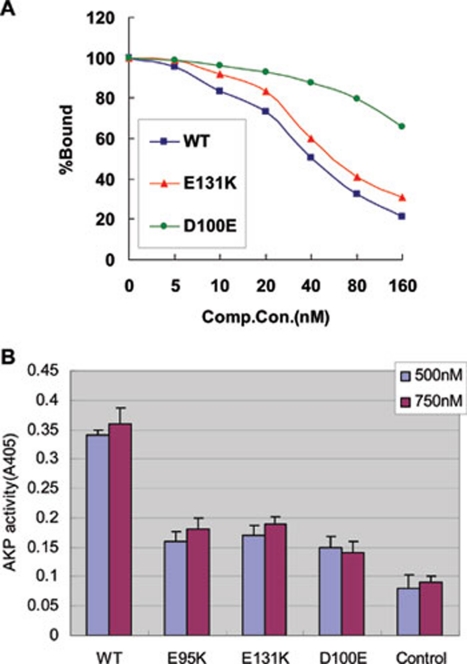
Figure 4.
Patched1-binding ability and activity of three BDA1 mutant IhhN proteins. (A) Binding of altered IhhN proteins to the PTC1 receptor. Binding of 32P-IhhN to EcR-CHO cells stably expressing Ptc-CTD140 is measured in the presence of unlabeled recombinant proteins; binding is normalized to the amount (100%) bound in the absence of any competitors. (B) Alkaline phosphatase expression is analyzed in C3H10T1/2 cells after a 5-day incubation with purified WT IhhN protein or three mutants at 500 or 750 nM.
BDA1 mutations impair Hedgehog activity
Hedgehog pathway activation is mainly triggered by the IhhN-PTC1 interaction. The reduced binding affinity between mutant IhhN and PTC1 prompted us to examine whether these BDA1-related missense mutations affect Hh activity. We expressed and purified the WT and mutant recombinant IhhN (E95K, E131K, and D100E) peptides and compared their in vitro biological activities by measuring their ability to induce alkaline phosphatase (AKP) activity in the mouse embryonic fibroblast line C3H10T1/2. This line is a mesenchymal stem cell line that has been extensively used as an in vitro model of differentiation, and it is responsive to Hh, as assessed by induction of AKP, a marker of mature differentiated osteoblasts. Incubation of C3H10T1/2 cells with 500- or 750-nM IhhN protein for 5 days induced robust osteoblast differentiation, indicated by the level of AKP activity (Figure 4B and Table 1). However, this activity was significantly impaired for the mutant IHH proteins. The impaired activity was not due to enhanced degradation, as WT and mutant IhhN proteins were stable in the medium throughout the 5 days at 37 °C (Supplementary information, Figure S2C). This finding suggests that the IHH proteins might not be internalized and are thereby protected from the intracellular degradation observed in transfected cells, as shown in Figures 2A and 3A. Notably, the level of purified E95K and D100E IhhN was significantly reduced in the presence of EGTA after incubation at 37 °C for 5 days, but no change was observed when CaCl2 was added in WT and E131K IhhN culture (Supplementary information, Figure S2D). These results indicated that the baseline calcium level is important for the stability of E95K and D100E IhhN proteins in the extracellular culture medium and might imply that these mutant proteins could be directly degraded because they are more sensitive to some proteases at lower calcium concentrations.
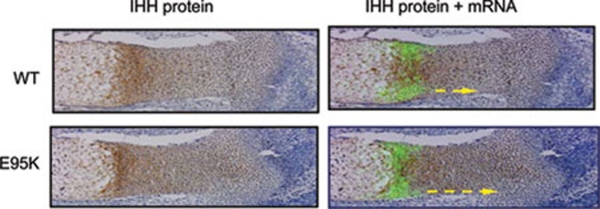
Extended distribution of E95K IHH in vivo
Binding to PTC1 is thought to be one of the processes regulating Hh movement and gradient formation 8, 35. Previously, we have shown a potential change in the IHH gradient in the developing growth plate in homozygous IhhE95K/E95K BDA1 mice using in situ hybridization analysis of downstream Hh targets, including Ptc1 and Gli1 27. Here, we showed by immunostaining that the Hh gradient was indeed longer than the normal in the growth plate of the BDA1 mouse (Figure 5). At E15.5, we detected a decreasing IHH gradient from the source of Ihh expression (prehypertrophic chondrocytes) toward the proliferative and reserve zones. This expression pattern was demonstrated by merging consecutive sections stained for the IHH protein (Figure 5, brown signal) with Ihh mRNA patterns detected by in situ hybridization (Figure 5, green signal). The distribution of E95K IhhN protein is clearly farther away from the source compared with WT IhhN.
Figure 5.
Morphogen gradient of WT and E95K mutant IHH proteins in vivo. Immunohistochemistry (brown signal) and in situ hybridization (green signal) signals from consecutive sections are merged, showing an extended distribution of E95K IHH protein in the tibia growth plate at E15.5 (yellow dotted line).
Multimeric formation and lipid modification are not affected by IHH-E95K and IHH-E131K
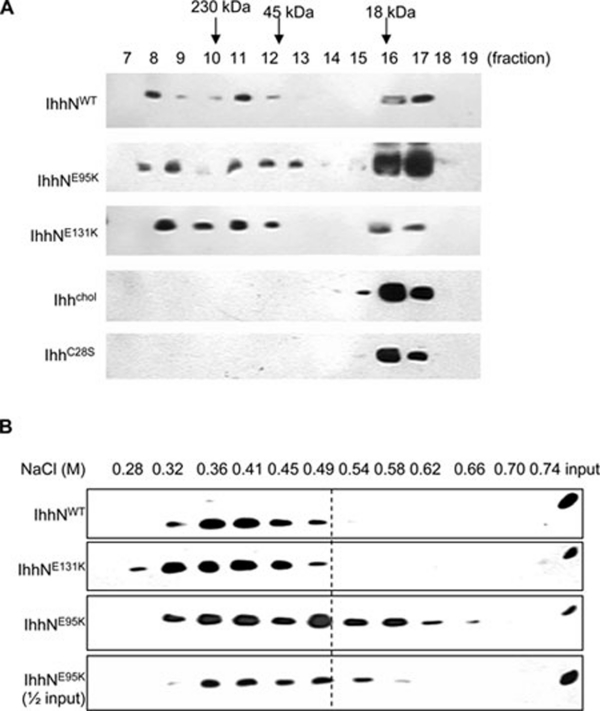
The formation of Hh multimers controls its diffusion 12, 13, 14. We investigated the multimeric formation of IhhN proteins by using gel-filtration chromatography 12. E95K and E131K IhhN from the conditioned medium of transfected EcR-CHO cells showed similar elution profiles compared with that of WT IhhN, with the majority of IHH proteins eluting at a position consistent with a multimer (Figure 6A, fractions 8-12, and Table 1). A minor peak consistent with a cholesterol-modified monomeric form (IhhNp) was also observed in fractions 16 and 17. These observations suggest that the formation of Hh multimers is not affected by the E95K and E131K mutations. Moreover, cholesterol modification and palmitoylation of these mutant proteins are unlikely to be affected, as formation of multimeric Hh requires modification by these lipids 13, 14. Multimer formation of D100E IhhN was not examined because it is rapidly degraded intracellularly and is not sufficiently present in the conditioned medium for further analysis.
Figure 6.
Formation of multimeric complexes and heparin-binding affinity assay of WT and mutant IhhN proteins. (A) N-terminal fragment of Indian Hedgehog (IhhN) present in the conditioned medium of EcR-CHO cells was fractionated by gel-filtration chromatography to determine the presence of IHH multimers. The majority of IHH migrates as a multimer at fractions 8-12. A minor peak consistent with the monomeric form of IhhNp was observed in fractions 16 and 17. IHHChol and IHHC28S represent IHH without cholesterol and palmitate modifications, respectively. (B) The interaction between IHH and heparin by affinity chromatography. Wild type and E131K IHH showed a similar profile, eluting from the column between 0.32 and 0.49 M NaCl. However, the elution profile for E95K IhhN was extended considerably to between 0.32 and 0.66 M NaCl.
Enhanced interaction between E95K-IhhN and heparin
Another extracellular regulator of Hh diffusion in the extracellular matrix is the interaction with HSPGs 9, 10, 11. We tested this interaction using affinity chromatography to assess the binding of WT and mutant (E95K and E131K) IHH to a heparin-agarose column. Elution from the columns with a salt gradient allowed us to characterize the relative binding affinity of WT and mutant IHH to heparin. Wild type and E131K IHH showed a similar profile, eluting from the column between 0.32 and 0.49 M NaCl. However, the elution profile for the E95K IhhN was considerably extended to between 0.32 and 0.66 M NaCl (Figure 6B and Table 1), indicating an enhanced affinity for heparin. This effect appeared to be independent of E95K IhhN concentration, as it was eluted in a similar NaCl concentration range of 0.32–0.58 M NaCl, even when half the amount of protein was loaded onto the column (Figure 6B and Table 1). This result suggests that the phenotypic outcome of the E95K mutation could in part be due to an enhanced interaction with HSPGs.
Discussion
In a mouse model for BDA1, with a human-equivalent E95K mutation in IHH, there is a change in the potency and range of Hh signaling. This mutation affected chondrocyte differentiation in the growth plate, delaying endochondral bone formation, and impaired the recruitment of mesenchymal cells into the distal cartilage in the developing digit, causing brachydactyly 27. The E95K mutation maps to a calcium-binding groove in the X-ray crystal structure of murine SHH, a region that interacts with the PTC receptor and other partner proteins, including HIP and CDO 26, 30. There are three BDA1 mutations that line the surface of this calcium-binding groove. Here, we provide the first X-ray crystal structures for the human WT and the three mutant IhhN proteins, and analyzed their structural and biochemical consequences.
The structure of the human IhhN protein shares significant similarities with murine ShhN, with a RMSD value of only 0.202 Å. We confirmed that the three critical BDA1 mutations (p.E95K, p.D100E, and p.E131K) were within a negatively charged calcium-binding groove, as shown previously when mapped to the murine SHH structure 28, 36. Furthermore, a recent paper about the complex structure between IhhN and CDOFn3 has demonstrated that this calcium-binding site is important for the interactions with Ihh partners 30. The main-chain superimposition between IhhN and its complex with CDOFn3 gave an RMSD of 0.268 Å, which was mainly contributed by the calcium-binding region. In our structures, the three mutations within this groove did not significantly disrupt the overall structure, but subtle changes were confined within the calcium-binding groove. Clearly, p.E95K altered the surface charge within this groove, changing it from negative to positive, without altering the vicinal structure. This alteration could impair the interaction with partner molecules through this groove and would be consistent with impaired PTC1, CDOFn3, and HIP1 interactions, as previously demonstrated 27, 30, as it would alter the electrostatic interactions that are required. The E95 residue is likely to have an important role in the interaction with partners, as a p.E95G mutation and a specific deletion of this amino acid also leads to a BDA1 phenotype 21, 22. This is further supported by our finding that in the E131K structure there are two alternative conformations of E95, one consistent with the WT and another that forms a salt bridge between the carboxyl oxygen atom of E95 and the Nɛ atom of K131. This changes the surface in the vicinity of E95 in this conformation. Thus, the effect of the E131K mutation might, in part, result from this change at E95. Biochemically, the K95 and K131 mutant proteins share a similar degree of impaired interaction with PTC1, with Kd values of 40.6 and 30.5 nM, respectively, compared with the Kd value of 20.6 nM for WT proteins.
The D100E mutation did not significantly alter the charge distribution. However, in the D100E structure, the region between residues 93 and 98 showed higher flexibility than normal, again suggesting the possible contribution of disrupted interactions via E95. In fact, residues 93-98 might contribute to a strict complementarity requirement for proper protein interactions, as the interaction of D100E IHH with PTC1 (Kd > 100 nM) is significantly lower than for E95K and E131K mutant proteins. In all cases, the impaired interaction with PTC1 leads to a reduced signaling capacity for activating downstream targets, such as Gli1 and Ptc1 27, 37, and impaired induction of osteogenic differentiation of C3H10T1/2 cells, as shown in the present study. This impaired interaction appeared to be a common feature of the three mutations lining the surface of the calcium-binding groove.
We have shown previously that the E95K mutation also altered the range of IHH signaling in vivo 27, with reduced signaling closer to the source, but enhanced signaling farther away. This effect was demonstrated by the activation of downstream targets, such as Ptc1, Gli1, and Hip1 27. Here, we provide a view of the IHH gradient in the growth plate using immunostaining, which showed that the E95K IHH proteins diffused farther away from the source, the prehypertrophic chondrocytes. Unfortunately, mouse models for the E131K and D100E mutations are not available for comparison, but are most likely similar, given the similarity in the effect on PTC1 binding.
The factors contributing to the regulation of extracellular diffusion of Hh proteins are complex and not fully understood. Reduced binding to PTC1 is a contributing factor because the sequestering of IHH by cells close to the source was reduced 8, 35, which would allow the mutant IHH to diffuse farther. This effect was also a compounding effect as the subsequent induction of Ptc1 expression was reduced because of the impaired signaling 27. Studies of the Drosophila gene Tout-velu (ttv) clearly demonstrated a contribution of HSPG in facilitating the Hh gradient 9, 10, 11. Although genetic inactivation of the EXT1 gene (the mammalian homolog of ttv), coding for glycosyltransferase for the synthesis of HS, in mice also supported a role for HSPG in shaping the IHH gradient, it appears to have a restrictive role 38. Thus, mice carrying a hypomorphic mutation in Ext1 demonstrate an elevated range of IHH signaling caused by reduced HS synthesis during embryonic chondrocyte differentiation, which suggested that HS restricts IHH propagation in mice 38. The reason for this discrepancy is unclear, but it might be related to the different mode of Hh movement between the different tissues studied. On the basis of the mouse studies of IHH signaling in the growth plate 38, the enhanced binding of E95K IHH protein to heparin suggested a stronger interaction with HSPGs, which should reduce the diffusion of the E95K IHH proteins. The overall effect on diffusion could still be positive because of a reduced interaction with PTC1. However, it is also possible that both factors might have contributed to the longer signaling range seen in the BDA1 mouse 27.
HSPGs contain a core protein with attached HS glycosaminoglycans (GAG), and have a wide distribution in the extracellular matrix and the cell surface. They also have a close relationship with many important signaling pathways, including Hh signaling 9, 10, 11. Hedgehog can interact with the negatively charged sulfated GAG side chains of proteoglycans through a highly conserved positively charged Cardin-Weintraub sequence of the type XBBBXXBX, a motif characterized by a cluster of basic amino acids (B) that allows for electrostatic interactions between the positive charges on the protein and the negatively charged sulfates of HSPGs 39. At present, it is not clear how the E95K mutation alters binding to HS. It might act through the Cardin-Weintraub sequence near the N-terminus, or the change in charge density on the calcium-binding groove might allow such an interaction within this region. The latter is implicated by E95K structure analysis and a previous ShhN structural study in which it was suggested that amino acids 25-45 within the N-terminal part of ShhN coordinate the intermolecular interaction of adjacent Shh molecules 36. The E95K mutation significantly changes the negative charge to a positive charge, possibly reducing the interactions between the N-terminus of E95K IhhN and the binding cleft and, consequently, enhancing the binding of heparin to the positively charged N-terminus.
Whether this enhanced interaction of the E95K IHH with HS contributes to the phenotypic outcome is not known. Glypican-3, one of six mammalian glypicans, is a cell-surface HSPG and acts as a competitive inhibitor of Hh binding to PTC1 at the cell membrane 40. Thus, an enhanced interaction with glypican-3 would further reduce the signaling capacity via PTC1. Studies have suggested that HSPGs can facilitate Hh movement by means of lipoprotein binding in the extracellular matrix and can antagonize PTC-mediated Hh sequestration, thereby promoting long-range movement 41, 42. Furthermore, a recent study has shown that Hh monomers can form nanoscale oligomers that are selectively enriched in visible clusters containing HSPGs 43. The association of Hh oligomers with HSPG clusters promoted the dispatch of Hh for long-range transport (possibly via lipoprotein particles) and the activation of Hh target-gene expression in distant receiving cells 43. However, enhanced binding to HS is not a common finding and was not demonstrated for the E131K mutant protein. The reason for the difference in HS-binding between E95K and E131K is not clear, nor is the discrepancy with the finding of the Ext1 hypomorphic mouse 38. Studies on a BDA1 mouse with the E131K mutation would be informative, but are outside the scope of this current project.
Lipid modifications are unlikely to be affected by these mutations as they are distant from the palmitoylation and cholesterol attachment sites at the N- and C-termini, respectively 5, 13. These lipid modifications are needed for the formation of freely diffusible and soluble multimeric IHH protein complexes for long-range signaling 12, 13, 14. Indeed, the E95K and E131K mutations did not affect the formation of these multimeric complexes. We were unable to assess complex formation for the D100E mutation because insufficient proteins were secreted from transfected cells because of intracellular degradation.
All three mutations resulted in enhanced intracellular degradation to varying extents via the lysosomal pathway. This result suggests that these mutations may also affect IHH biosynthesis or secretion as part of the disease mechanism. Mutations that introduce the misfolding of proteins could lead to instability, and such misfolded proteins are normally targeted for degradation via the proteasome or lysosomal pathways or both 44, 45. In the case of these three mutations, degradation is primarily through the lysosomal pathway. Our structural data showed minimal disruption of the overall folding of the mutant proteins. The intracellular stability of the proteins, however, might be related to subtle changes within the calcium-binding groove. Calcium binding changes the local charge and leads to the ordering of the calcium-coordinating loop 26. This notion is supported by the observation that chelation of calcium using EGTA reduces the level of WT IHH in transfected cells, whereas the addition of calcium increases the level of mutant IHH proteins. A higher concentration of calcium could push the equilibrium toward calcium binding of the mutant proteins 46.
The fact that this lysosomal degradation is temperature-sensitive in mammalian cells provides support for the protein instability hypothesis, as E95K and D100E IhhN proteins were more stable when transfected cells were grown at 30 °C than at 37 °C. It is possible that lysosomal enzymes are less active at 30 °C, contributing to the increased level. However, we favor the hypothesis that at a lower temperature, the interactions surrounding the calcium-binding groove can be stabilized. Such temperature-regulated stabilization has been shown for SHH mutants (W117G and W117R) that cause HPE, where the mutant proteins are inactive at 37 °C but display some activity at 32 °C, presumably due to conformational changes or stabilization 34. Another possibility for defects caused by these mutations is impaired intracellular trafficking, as secretion of Hh proteins requires interaction with Disp 6, 47. It is also likely that this interaction is mediated by the calcium-binding site, a disruption of which would target the mutant proteins into the lysosome. Degradation of the mutant proteins is confined to intracellular processes during IHH biosynthesis, as purified WT or mutant IhhN proteins were highly stable when added to the culture medium at 37 °C over long periods. This finding suggests that the amount of IHH proteins internalized after binding to PTC1 is minimal, as endocytosis could lead to subsequent lysosomal degradation.
Our data reveal that the three mutations lining the calcium-binding groove affect the function of IHH at multiple levels. The changes within this groove as a result of the mutations include a change in the surface charge and the flexibility of the helix loop in this region, affecting Ihh binding to its receptor and, most likely, other interacting partners, including HIP, CDO, and GAS1. An altered interaction with Disp is also possible, as well as the interaction with HSPG. All would contribute to modulating the overall levels of IHH in the extracellular space, its capacity to signal via interaction with its receptor, and the range of binding with partners and the extracellular matrix, causing abnormal bone development and abnormal distal digit formation.
Materials and Methods
Site-directed mutagenesis and IHH expression constructs
Human Ihh cDNA-encoding full-length protein was cloned by PCR. Three mutations (E95K, D100E, and E131K) were introduced through PCR employing the primers 5′-CTTCAAGGACGAGAAGAACACAGGCG-3′, 5′-CACAGGCGCCGAACGCCTCATGACCC-3′, and 5′-GCTGCGGGTGACCAAGGGCTGGGACG-3′, respectively. The bold and underlined nucleotide represents the single-nucleotide substitution in each of the primers. The mutations were confirmed by automated DNA-sequence analysis employing the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, CA) and the ABI Prism 3100 genetic analyzer. The WT and mutated Ihh cDNAs were subcloned into the pIND(Sp1)-based inducible expression vector (Invitrogen) (pIND-Ihh-WT, pIND-Ihh-E95K, pIND-Ihh-D100E, and pIND-Ihh-E131K). Double FLAG-epitope-tags were introduced into the Ihh N-terminal fragment, as described previously 48. The pIND(Sp1)-based constructs of IhhChol and IhhC28S were all produced as described above from primers that are available upon request. Meanwhile, the WT and mutated Ihh cDNAs were subcloned into the pCMV-based expression vector (pCMV-Ihh-WT, pCMV-Ihh-E95K, pCMV-Ihh-D100E, and pCMV-Ihh-E131K), and the double FLAG-epitope tags were at the C-terminus of the IHH. The pIND(Sp1)-based constructs and the pCMV-based constructs of Ihh-E95G, Ihh-D100N, IhhChol, IhhC28S, T2, T4, T6, and T8 were all produced as described above from primers that are available upon request. The fragment encoding the N-terminal domain of human IHH (amino acid residues 28-202) was amplified, and the PCR products were inserted into the pGEX-2T vector (Amersham) and the pET28b vector (Novagen). The extra sequence encoding MGHHHHHHM was added at the N-terminus of IhhN in pET28b-based constructs and subjected to crystallization (see below). Fragments encoding amino-terminal FLAG-tagged Ptc-CTD140 (PTC1 with a truncation, resulting in a 140-residue carboxyl-terminal deletion, amino acids nos. 1-1 309 and the double copy of the flag-epitope tag introduced as above) were inserted into the pIND(Sp1) vector (pIND-Ptc-CTD140).
Cloning, expression, and purification of recombinant IhhN for crystallization
The pET28b-based recombinant protein was expressed by the BL21 (DE3) strain of E. coli induced with 0.2 mM IPTG at 37 °C for 5 h when the OD600 reached 0.6. Pellets were collected by centrifugation at 8 000× g for 10 min at 4 °C and re-suspended in 200 mM NaCl and 20 mM Tris-HCl, pH 7.5, followed by sonication. After centrifugation at 25 000× g for 30 min at 4 °C, the supernatant was collected and applied to a zinc-chelating sepharose column (Amersham Biosciences). The target protein was eluted with 300 mM imidazole in 200 mM NaCl and 20 mM Tris-HCl, pH 7.5, and loaded on a Superdex 75 16/60 column that was equilibrated with 200 mM NaCl and 20 mM Tris-HCl, pH 7.5. Fractions containing the recombinant IhhN were pooled, desalted, concentrated to 20 mg/ml in a final buffer of 100 mM NaCl and 20 mM Tris-HCl, pH 7.5, and subjected to crystallization. Protein concentration was determined by its absorbance at 280 nm with a theoretical molar extinction coefficient of 27 055 M−1 cm−1 (http://www.expasy.org/tools/protparam.html).
IHH activity assay in C3H10T1/2 cells
The induction of AKP by IHH activity in C3H10T1/2 cells (obtained from ATCC) was measured by growing cells to full confluency in 24-well plates in a growth medium containing DMEM, 10% fetal bovine serum (FBS), and 2% penicillin/streptomycin. WT or mutant IhhN was added to the growth medium, and after 4-5 days of further incubation at 37 °C the cells were washed in cold PBS and lysed in passive lysis buffer (Promega) containing a protease inhibitor (protease inhibitor cocktail set III, Calbiochem) at room temperature for 15 min. The lysate was mixed with chromogenic substrate p-nitrophenyl phosphate (Sigma), dissolved in lysis buffer, and incubated at room temperature for 15 min, after which the absorbance at 405 nm was measured. The stability of IhhN proteins in the medium was tested by western blotting with the Shh antibody. All induction assays were performed in triplicate.
PTC1-binding assay of recombinant IHH
The pGEX-2T constructs containing WT and mutant Ihh genes were expressed in E. coli and were purified according to the Bulk and RediPack GST Purification Modules, followed by cleavage of the GST tag using thrombin. The proteins were subjected to PTC1 binding or Hh-signaling induction (see below). 32P-labeled IhhN protein was prepared as described previously 7, using the catalytic subunit of protein kinase A (Sigma), [γ-32P] ATP, and recombinant IhhN protein containing a protein kinase A site (RRASV) at the C-terminus of IhhN. To generate stable cell lines, EcR-CHO cells (Invitrogen) were transfected with the pIND-Ptc-CTD140 construct, and several independent clones were isolated for growth in 10% FBS and 800 μg/ml G418. G418-resistant colonies were cloned using limiting dilution and screened for expression by immunoblot analysis of cell lysates with an anti-FLAG monoclonal antibody. Stock cultures of the stable-expression PTC1 EcR-CHO cells were maintained in complete medium supplemented with 400 μg/ml G418. Assays of 32P-IhhN binding to cells expressing Ptc-CTD140 with competition by IhhN variants were conducted as described by Fuse et al. 7. PTC1 expression was induced in cloned stable derivatives of the cell line EcR-CHO by the addition of ponasterone A. After induction, 3 × 105 cells were mixed at increasing concentrations (0.1-100 nM) of 32P-IhhN (for Scatchard analyses) or with a fixed concentration (20 nM) of 32P-IhhN and various concentrations of competitors (for competitive binding assays). After incubation at 4 °C, the cells were collected and the bound 32P-IhhN was determined.
Transfection and expression of IHH variants in mammalian cells
For the expression of IHH protein, recombinant plasmids were transiently transfected into EcR-CHO cells using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, ponasterone A (5 μg/ml, Invitrogen) 36 was added to the cell culture medium, and after another 24 h, the cells were washed with cold PBS. The cells were homogenized in a passive lysis buffer (Promega) that contained a protease inhibitor (Calbiochem). After lysis, cell homogenates were cleared by centrifugation, separated using 12% SDS-PAGE, and immunoblotted using an anti-FLAG monoclonal antibody (Stratagene) for detection. Similarly, the pCMV-based Ihh constructs containing the WT and mutants were transiently transfected into EcR-CHO cells and 24 h later, the cells were lysed and immunoblotted by using the same antibody.
For the stability analysis of the three mutations at different temperatures and durations, the pIND-based Ihh constructs containing the WT and mutations were transiently transfected into EcR-CHO cells. Twenty-four hours later, ponasterone A (5 μg/ml) was added to the cell culture medium and the cells were cultured at 30 °C for 24 h before the cell extracts of one group (WT, E95K, D100E, and E131K) were harvested. This time point was denoted as 0 h. Meanwhile, the cells in culture were treated with puromycin (10 μg/ml), which inhibits protein synthesis. Incubation continued for 3, 6, and 18 h at 30 and 37 °C before the harvesting of cell extracts and immunoblotting.
For the stable expression of IHH protein, the pIND-based Ihh constructs were transfected into EcR-CHO cells. Stable clones were generated and amplified in a medium containing F12, 10% FBS, 1% penicillin/streptomycin, Zeocin (250 μg/ml), and G418 (800 μg/ml), and the generation of the processed N-terminal domain (IhhNp) was measured by immunoblotting of the cell-free extract using an anti-FLAG monoclonal antibody.
For the protein-degradation analysis, a lysosome inhibitor (chloroquine, 200 μM and balfilomycin, 40 nM, Sigma) or a proteasome inhibitor (MG132, 0.1 μM, Sigma) was added to the EcR-CHO cell culture medium while ponasterone A was added, and the generation of WT, E95K, and D100E IhhN proteins was measured by using immunoblotting.
For the stability analysis of IhhN proteins at different concentrations of EGTA and calcium, the pIND-based Ihh constructs containing the WT and two mutations (E95K and D100E) were transiently transfected into EcR-CHO cells. Twenty-four hours later, ponasterone A (5 μg/ml) was added to the cell-culture medium, and the cells were cultured at 30 °C for 24 h. The WT IHH cultured cells were treated with EGTA (0.1–0.8 mM) for 24 and 48 h at 37 °C. Meanwhile, WT, E95K, and D100E IHH cultured cells were treated with calcium (8-64 mM) for 6 h at 37 °C. The cell extracts were harvested and immunoblotted with an anti-FLAG antibody.
Heparin-binding assay of IHH variants derived from mammalian cells
Conditioned supernatants (2 ml) from transfected EcR-CHO cells stably expressing WT and mutant Ihhs were applied to a 1-ml heparin-agarose column (HEP-II-5, Sigma-Aldrich) in equilibration buffer (10 mM Tris, pH 7.5, with 150 mM NaCl). After loading, the column was washed with at least five column volumes of equilibration buffer until the A280 had dropped below 0.01. Twenty milliliters of a salt gradient from 0 to 2 M NaCl in 10 mM Tris, pH 7.5 was applied. Fractions (0.5 ml) were obtained from the time of protein application until the end of the gradient. Elution of WT and mutant IhhN was detected by western blot.
Size analysis of IHH proteins in conditioned medium
Conditioned medium from transfected EcR-CHO cells stably expressing WT and mutant Ihhs was collected and centrifuged at 9 000× g for 60 min. The supernatants were concentrated using a Centriprep YM-10 (Amicon) and loaded onto a Superdex 200 (Amersham Biotech) gel-filtration column that had been equilibrated with PBS/0.001% NP-40. Then, 1-ml fractions were collected and precipitated using TCA, resolved by 12% SDS-PAGE, and then immunoblotted with an anti-FLAG monoclonal antibody.
Immunohistochemistry and in situ assay
WT and mutant embryos were fixed and processed using standard procedures 49. Immunohistochemistry was carried out using the Envision+ System (DAKO) and HRPDAB colorimetric detection. Slides containing comparable paraffin sections from E15.5 mouse tibia were treated with 0.8% hyaluronidase (Sigma) at 37 °C for 30 min before incubation with a polyclonal SHH antibody (Santa Cruz, sc-9024). In situ hybridization on dewaxed sections was performed as previously described 27, using a [35S]-UTP-labeled Riboprobe for Ihh 50.
Crystallization, data processing, and structure refinement of IhhN
The crystals were grown at 16 °C in a hanging drop of 1.0-μl protein sample at 20 mg/ml in 100 mM NaCl and 20 mM Tris-HCl, pH 7.5 with an equal volume of reservoir solution. Crystals of WT IhhN protein were crystallized in 0.1 M Tris-HCl, pH 8.5, and 2.0 M (NH4)2SO4. Crystals of the E95K and E131K mutants were crystallized in 0.1 M sodium cacodylate, pH 6.5, 30% PEG 8000, and 0.2 M (NH4)2SO4. Crystals of the D100E mutant were crystallized in 30% PEG4000, 0.2 M (NH4)2SO4. All crystals were obtained after 3-4 weeks. The crystals were transferred to the cryoprotectant of the reservoir solution supplemented with 25% glycerol and flash cooled using liquid nitrogen. The X-ray diffraction data were collected at 100 K in a liquid nitrogen stream by using a Rigaku MM007 X-ray generator (λ = 1.5418 Å) with a MarResearch 345 image-plate detector at the School of Life Sciences, University of Science and Technology of China (USTC, Hefei, PR China). The data were processed using iMosflm, version 0.6.1 51. The structures were resolved using the molecular replacement method in the program Phaser 52, and the atomic coordinates of murine ShhN as the search model (PDB code 1VHH). Structures were fitted and rebuilt using the program Coot 53, and refined with REFMAC5 54 and CNS 55. The overall assessments of model quality were performed using PROCHECK 56. In all structures, there is one IhhN molecule with coordinated zinc in one crystallographic asymmetric unit. The crystal packing of WT IhhN is the same as that of E95K and E131K mutants, but different than that of D100E mutant. The atomic coordinates of human IhhN have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) under the entries of 3K7G, 3K7H, 3K7I, and 3K7J Collections and refinement statistics of the IHH structure are summarized in Table 1. All figures were prepared using PyMOL (http://www.pymol.org).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30800613), the 973 Program (2010CB529600, 2007CB947300), the 863 Program (2009AA022701), the Shanghai Municipal Commission of Science and Technology Program (09DJ1400601), the Natural Science Foundation of Shanghai, China (Grant No. 08ZR1411000), the National Key Project for the Investigation of New Drugs (2008ZX09312-003), the Shanghai Leading Academic Discipline Project (B205), and the General Research Fund of Hong Kong (HKU760608M). The co-author, Xizhi Guo, was supported by the “Pujiang Talent” Project (08PJ1407200).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Data collection and refinement statistics of IHH structure
The subcellular localization of WT and mutant IhhN in the EcR-CHO cells.
The stability of WT and mutant IhhN proteins.
References
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Autoproteolysis in hedgehog protein biogenesis. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- Porter JA, von Kessler DP, Ekker SC, et al. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374:363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- Porter JA, Ekker SC, Park WJ, et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Fuse N, Maiti T, Wang B, et al. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc Natl Acad Sci USA. 1999;96:10992–10999. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona JP, Lee JH, Robertson CP, Enga K, Kapur RP, Roelink H. Receptor-mediated endocytosis of soluble and membrane-tethered Sonic hedgehog by Patched-1. Proc Natl Acad Sci USA. 2000;97:12044–12049. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394:85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- Takei Y, Ozawa Y, Sato M, Watanabe A, Tabata T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development. 2004;131:73–82. doi: 10.1242/dev.00913. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X, Lin X. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- Zeng X, Goetz JA, Suber LM, Scott WJ, Jr, Schreiner CM, Robbins DJ. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 2001;411:716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, White B, Tyurina OV, et al. Synergistic and antagonistic roles of the Sonic hedgehog N- and C-terminal lipids. Development. 2004;131:4357–4370. doi: 10.1242/dev.01301. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nature Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, et al. Mutations in the C-terminal domain of Sonic Hedgehog cause holoprosencephaly. Hum Mol Genet. 1997;6:1847–1853. doi: 10.1093/hmg/6.11.1847. [DOI] [PubMed] [Google Scholar]
- Gao B, Guo J, She C, et al. Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A-1. Nat Genet. 2001;28:386–388. doi: 10.1038/ng577. [DOI] [PubMed] [Google Scholar]
- Gao B, He L. Answering a century old riddle: brachydactyly type A1. Cell Res. 2004;14:179–187. doi: 10.1038/sj.cr.7290218. [DOI] [PubMed] [Google Scholar]
- Farabee WC.Hereditary and sexual influence in meristic variation: a study of digital malformations in manThesis, Harvard University, 1903
- Lanske B, Karaplis AC, Lee K, et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick TJ, Au KS, Mastrobattista JM, McCready ME, Bulman DE, Northrup H. Identification of a mutation in the Indian hedgehog (IHH) gene causing brachydactyly type A1 and evidence for a third locus. J Med Genet. 2003;40:42–44. doi: 10.1136/jmg.40.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready ME, Sweeney E, Fryer AE, et al. A novel mutation in the IHH gene causes brachydactyly type A1: a 95-year-old mystery resolved. Hum Genet. 2002;111:368–375. doi: 10.1007/s00439-002-0815-2. [DOI] [PubMed] [Google Scholar]
- Giordano N, Gennari L, Bruttini M, et al. Mild brachydactyly type A1 maps to chromosome 2q35-q36 and is caused by a novel IHH mutation in a three generation family. J Med Genet. 2003;40:132–135. doi: 10.1136/jmg.40.2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder EM, Hoogeboom AJM, Coert JH, de Graaff E. Deletion of 1 amino acid in Indian hedgehog leads to brachydactylyA1. Am J Med Genet Part A. 2008;146:2152–2154. doi: 10.1002/ajmg.a.32441. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Coucke PJ, Giedion A, et al. Homozygous mutations in IHH cause acrocapitofemoral dysplasia, an autosomal recessive disorder with cone-shaped epiphyses in hands and hips. Am J Hum Genet. 2003;72:1040–1046. doi: 10.1086/374318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008;455:979–983. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Hu JX, Stricker S, et al. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature. 2009;458:1196–1200. doi: 10.1038/nature07862. [DOI] [PubMed] [Google Scholar]
- Hall TM, Porter JA, Beachy PA, Leahy DJ. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signaling domain of sonic hedgehog. Nature. 1995;378:212–216. doi: 10.1038/378212a0. [DOI] [PubMed] [Google Scholar]
- Bishop B, Aricescu AR, Harlos K, O'Callaghan CA, Jones EY, Siebold C. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat Struct Mol Biol. 2009;16:698–703. doi: 10.1038/nsmb.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ. All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J Biol Chem. 2010;285:24584–24590. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL.The PyMOL Molecular Graphics SystemDeLano Scientific, San Carlos, CA, USA. 2002
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Wolf A, Wagner M, Kuhlmann J, Waldmann H. The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc Natl Acad Sci USA. 2004;101:8531–8536. doi: 10.1073/pnas.0308449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity T, Fuse N, Beachy PA. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc Natl Acad Sci USA. 2005;102:17026–17031. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Goetz JA, Singh S, Suber LM, Kull FJ, Robbins DJ. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J Biol Chem. 2006;281:4087–4093. doi: 10.1074/jbc.M511427200. [DOI] [PubMed] [Google Scholar]
- Guo S, Zhou J, Gao B, et al. Missense mutations in IHH impair Indian Hedgehog signaling in C3H10T1/2 cells: implications for brachydactyly type A1, and new targets for Hedgehog signaling. Cell Mol Biol Lett. 2010;15:153–176. doi: 10.2478/s11658-009-0040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev Cell. 2004;6:801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Rubin JB, Choi Y, Segal RA. Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development. 2002;129:2223–2232. doi: 10.1242/dev.129.9.2223. [DOI] [PubMed] [Google Scholar]
- Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev Cell. 2008;14:700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Panáková D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signaling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- Eugster C, Panáková D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13:57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Vyas N, Goswami D, Manonmani A, et al. Nanoscale organization of Hedgehog is essential for long-range signaling. Cell. 2008;133:1214–1227. doi: 10.1016/j.cell.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Knecht E, Aguado C, Cárcel J, et al. Intracellular protein degradation in mammalian cells: recent developments. Cell Mol Life Sci. 2009;66:2427–2443. doi: 10.1007/s00018-009-0030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Costelli P, Reffo P, Penna F, Autelli R, Bonelli G, Baccino FM. Ca(2+)-dependent proteolysis in muscle wasting. Int J Biochem Cell Biol. 2005;37:2134–2146. doi: 10.1016/j.biocel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Ma Y, Erkner A, Gong R, et al. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- Guy RK. Inhibition of sonic hedgehog autoprocessing in cultured mammalian cells by sterol deprivation. Proc Natl Acad Sci USA. 2000;97:7307–7312. doi: 10.1073/pnas.97.13.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah KS, Levy A, Trainor PA, et al. Human COL2A1-directed SV40 T antigen expression in transgenic and chimeric mice results in abnormal skeletal development. J Cell Biol. 1995;128:223–237. doi: 10.1083/jcb.128.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell–cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data collection and refinement statistics of IHH structure
The subcellular localization of WT and mutant IhhN in the EcR-CHO cells.
The stability of WT and mutant IhhN proteins.