Abstract
The virulence-attenuated Leptospira interrogans serovar Lai strain IPAV was derived by prolonged laboratory passage from a highly virulent ancestral strain isolated in China. We studied the genetic variations of IPAV that render it avirulent via comparative analysis against the pathogenic L. interrogans serovar Lai strain 56601. The complete genome sequence of the IPAV strain was determined and used to compare with, and then rectify and reannotate the genome sequence of strain 56601. Aside from their highly similar genomic structure and gene order, a total of 33 insertions, 53 deletions and 301 single-nucleotide variations (SNVs) were detected throughout the genome of IPAV directly affecting 101 genes, either in their 5′ upstream region or within their coding region. Among them, the majority of the 44 functional genes are involved in signal transduction, stress response, transmembrane transport and nitrogen metabolism. Comparative proteomic analysis based on quantitative liquid chromatography (LC)-MS/MS data revealed that among 1 627 selected pairs of orthologs, 174 genes in the IPAV strain were upregulated, with enrichment mainly in classes of energy production and lipid metabolism. In contrast, 228 genes in strain 56601 were upregulated, with the majority enriched in the categories of protein translation and DNA replication/repair. The combination of genomic and proteomic approaches illustrated that altered expression or mutations in critical genes, such as those encoding a Ser/Thr kinase, carbon-starvation protein CstA, glutamine synthetase, GTP-binding protein BipA, ribonucleotide-diphosphate reductase and phosphate transporter, and alterations in the translational profile of lipoproteins or outer membrane proteins are likely to account for the virulence attenuation in strain IPAV.
Keywords: genome, proteome, Leptospira, virulence, mutation
Introduction
Leptospira belong to spirochetes, a group of bacteria primitive in the evolution of prokaryotes 1. Leptospires are thin, coiled microbes with a hook at one or both ends. They are highly motile, obligate aerobe organisms that share features of both Gram-positive and Gram-negative bacteria 2. Leptospirosis, caused by a diversity of pathogenic species within the genus Leptospira, has emerged as a globally important zoonotic disease 3, 4. A wide variety of mammalian hosts, such as rodents, horses, cattle and pigs, may serve as reservoirs. Bacteria persistently colonize the proximal renal tubules of carrier animals; and humans and other animals become infected through exposure to infected urine, contact with urine-contaminated soil and water 3, 5 or through the host-to-host transmission cycle 6. Hosts infected by pathogenic Leptospira display diverse manifestations that range from subclinical infection, a mild influenza-like febrile illness, to severe systemic disease with renal and hepatic failure, extensive vasculitis, pulmonary hemorrhage and death.
With the availability of the whole genome sequence of Leptospira interrogans serovar Lai strain 56601, a highly virulent strain isolated in Sichuan province, China, in 1958 7, functional genomic research has been carried out in recent years 8, 9, 10. However, key factors that contribute to pathogenesis still remain to be systematically elucidated. The L. interrogans serovar Lai strain IPAV was originally isolated from a patient in China, but details of the isolation record were unavailable. After it was transferred to the N H Swellengrebel Laboratory of Tropical Hygiene, the Royal Tropical Institute, Amsterdam, by Hans Korver, this strain lost its virulence through the process of high-frequency passage as tested in the Institute Pasteur after 1987 (Mathieu Picardeau, personal communication). It was confirmed to belong serologically to the Lai serovar of serogroup Icterohemorrhagiae after transfer to the Department of Medical Microbiology and Parasitology, Institutes of Medical Sciences, Shanghai Jiao Tong University School of Medicine, in 2001. Therefore, the currently available IPAV strain is a candidate suitable for comparative analysis against the Lai-type strain 56601, with the aim of understanding the mechanism of pathogenesis and virulence.
In this study, we first confirmed the avirulent characteristics of strain IPAV and then determined its entire genomic sequence using high-throughput pyrophosphate sequencing 11. In addition, proteomic MS/MS data were integrated into the genome annotation of IPAV and reannotation of strain 56601. Comparative genomic analysis indicated that the two closely related strains shared highly similar genome structure and gene-order features. Therefore, the biased distribution of a limited number of genetic variations, including insertions, deletions and single-nucleotide variations (SNVs), in certain groups of genes may account for the virulence attenuation in IPAV. Quantitative proteomic analysis also revealed a detectable functional bias in proteomic profiles between the two strains cultured under laboratory conditions, which is likely caused by the genetic variations. These findings were further analyzed with a focus on individual genes, which was important for a better understanding of the molecular pathogenic mechanism of leptospirosis.
Results
The avirulent character of the L. interrogans strain IPAV as tested on experimental animal models
The virulence of the L. interrogans strain 56601 on experimental animal models with a lethal dose infection was described previously 12, 13. The avirulence property of the strain IPAV was tested on leptospirosis animal models as a negative control for strain 56601 infection experiments in our recent work 14, where all experimental animals survived intraperitoneal infection with strain IPAV (5 × 108), whereas an approximate 90% mortality rate was observed with the complete leptospirosis course for the group of animals inoculated with strain 56601 (5 × 108).
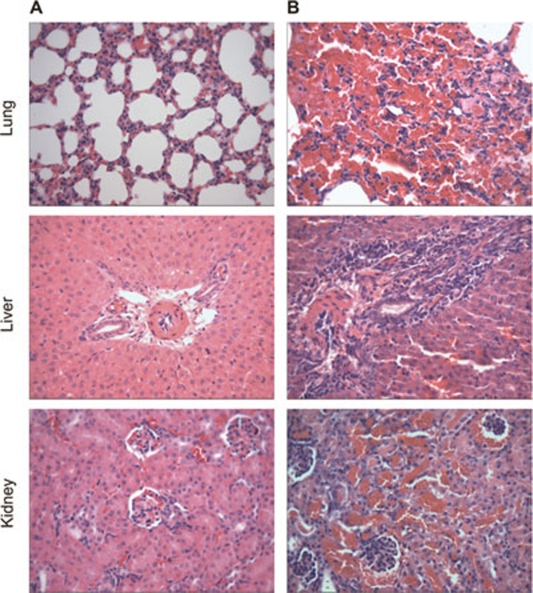
Meanwhile, 6 days post inoculation, sectioned lung, liver and kidney from guinea pigs infected by strains IPAV and 56601 were subjected to histological analysis via hematoxylin and eosin (H&E) staining (Figure 1). Tissue slices from guinea pigs injected with IPAV did not present conspicuous pathologic changes (Figure 1A). In contrast, tissue slices from animals inoculated with strain 56601 produced histopathologic characteristics typical of severe leptospirosis. These included intra-alveolar hemorrhage, edema of alveolar septa and inflammatory infiltrates in the lung; hepatocyte swelling, necrosis, defective hepatic cord formation and increase of monocytes and neutrophils in portal tracts; and glomerular capsules and the lumen of renal tubules pervaded with erythrocytes in kidney (Figure 1B).
Figure 1.
Histological sections in the lung, liver and kidney of guinea pigs infected with L. interrogans strain IPAV (A) and strain 56601 (B) (H&E, magnification × 400).
General features of the IPAV genome are highly similar to that of strain 56601
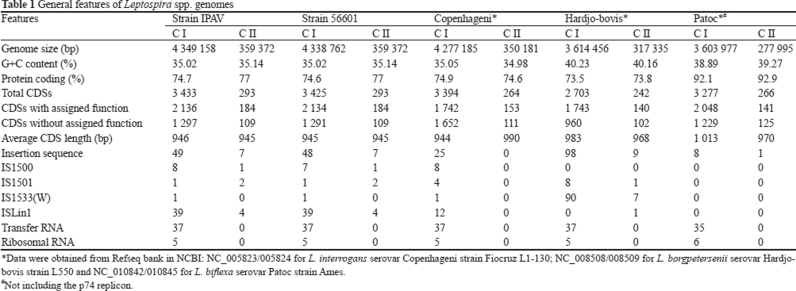
The complete genome of L. interrogans strain IPAV was sequenced along with the rectification of strain 56601. The genome of strain IPAV consists of two circular chromosomes (Figure 2). The large chromosome (CI) consists of 4 349 158 bp with an average GC content of 35.02%, which is 10 396 bp larger than that of strain 56601. The small chromosome (CII) consists of 359 372 bp with an average GC content of 35.14%, identical to that of strain 56601 (Table 1, GenBank accession number CP001221 for CI and CP001222 for CII). The replication origins of both CI and CII were located by referring to those of strain 56601, originally identified by GC skew analysis 7. In total, 3 726 protein coding sequences (CDSs) were predicted, with biological function assigned for 2 320 CDSs. In addition, 1 918 CDSs could be placed into clusters of orthologous groups (COGs) 15 via homology search. We identified approximately 56 intact insertion sequence (IS) elements scattered throughout the strain IPAV genome. Meanwhile, we identified 55 IS elements in the rectified genome of strain 56601, almost twice as many as were reported in the previous study (30 ISs) 7. Most of the ISs are identical between the two strains, except for three in strain IPAV (IS1500, 1908963-1910197; ISlin1, 3392331-3393734; ISlin1, 3525068-3526455) and two in strain 56601 (ISlin1, 3225556-3227302; ISlin1, 3227341-3228607). Compared with Leptospira biflexa, the existence of a large number of ISs suggests that IS integration events happened continuously throughout the history of these strains and that the five different ISs between the two strains might have happened after the two strains diverged.
Figure 2.
Circular representation of IPAV chromosomes with predicted CDSs. Left, large chromosome (CI); right, small chromosome (CII). The outer scales are numbered in megabases from the origins of replication (Ori). Circles range from 1 (outer circle) to 7 (inner circle) for CI and from I (outer circle) to V (inner circle) for CII. Circle 1, representation of genome structure when compared with L. interrogans strain Fiocruz L1-130 (yellow, the collinear region; blue, the large inverse region). Circle 2/I and 3/II, genes on plus and minus strands (colors representing functional categories according to COGs). Circle 4, tRNA genes (red) and rRNA genes (blue). Circle 5/III, IS elements (both intact and remnant are included; green, ISlin1; black, other types of ISs). Circle 6/IV, GC skew (calculated using a 2 500 bp (CI)/500 bp (CII) window sliding 200 bp (CI)/100 bp (CII) at a time; red, values > 0; green, values < 0). Circle 7/V, GC content.
Table 1. General features of Leptospira spp. genomes.
Rectification of the strain 56601 genomic sequence followed by reannotation
Some sequencing errors that existed in the originally published strain 56601 genome were resolved. First, the sequence of an approximately 6.7-kb DNA fragment that was missing in the previous genome was identified and added at revised CI coordinates of 3 181 009 to 3 187 723, between the originally annotated CDSs of LA_3200 and LA_3201. This region encodes a 23S ribosomal RNA (rrl), a putative lipoprotein that was validated by mass spectrometry (MS), and the N-terminal 89 aa of LA_3201, which increased its CDS to 250 aa. Second, the sequence of an approximately 6.5-kb DNA region spanning the revised CI coordinates 3 248 961 to 3 255 503 was erroneously reversed in the originally published Lai 56601 genome map. Four other large fragments were rectified, including sequence rectification from 414 967 to 415 363 (LA_0417), a sequence addition from 2 051 058 to 2 051 577 (LA_2081a and LA_2083) with respect to the revised CI coordinates, a sequence addition from 332 934 to 333 358 (LB_340 and LB_340a) with respect to the revised CII coordinates, and a deletion of an 831-bp direct repeat flanking the Genome Island I (GI I) 16. Finally, 43 single nucleotides were rectified according to the resequencing of the corresponding sites. The updated nucleotide sequence for strain 56601 contains 4 698 134 bp, of which CI contains 4 338 762 bp and CII contains 359 372 bp.
The completely reannotated genome of the virulent L. interrogans serovar Lai strain 56601 was deposited in the NCBI database with the original accession numbers (AE010300 for CI and AE010301 for CII). Among the predicted 4 727 CDSs annotated previously 7, one-fifth of them were shorter than 70 codons, mainly due to the short (30 codons) cutoff value. These “hypothetical proteins” shared no homology with known proteins/domains and are not expressed in the cell, as we discovered through proteomic profiling. In this reannotation, the length of coding sequences was reevaluated according to our new proteomic knowledge 17. With the length of 60 codons as the cutoff value, only 3 653 CDSs were predicted for strain 56601. Then, 65 CDSs shorter than 60 codons were added manually based on the relatively high confidence predictions. This reannotated genome of strain 56601 contains 3 718 CDSs, with 1 088 being removed from the previous version and 79 being newly included. Although 18 of the newly added CDSs have assigned functions, 6 are truncated transposases, and the remaining proteins are “hypothetical”. The original LA designations for the CDSs annotated in the preceding genome database were maintained. The additional or revised CDSs annotated by this work were designated by appending their 5′ CDSs with a lowercase alphabetic letter (e.g., LA_0395a following LA_0395, and LA_0727a and LA_0727b following LA_0727). All 3 718 CDSs were checked for their probable functions, which resulted in 2 318 CDSs with assigned biological functions and 148 conserved hypothetical proteins similar to the proteins in the NCBI's CDD database. The remaining 1 252 hypothetical proteins do not have enough homology with proteins of other genera and are unique to the Leptospira genome.
Proteomics-assisted genome annotation
To improve the annotation of the genome of L. interrogans, MS-based proteomic analysis was performed for both strains grown under routine experimental conditions. Using MS/MS spectra obtained from strains 56601 and IPAV, sequences were searched against 22 216 (strain 56601) and 22 269 (strain IPAV) six-frame translated proteins that served as theoretical databases. We found additional 13 genes that were missed in the previous gene prediction process employing computational software. A detailed analysis indicated that among the 13 missed genes, 8 overlapped with their adjacent genes on one end, 2 were located completely inside other genes, and the remaining 3 are in the intergenic regions. Of these 13 CDSs, the smallest one is a hypothetical protein (67 aa) in IPAV. For the two missed genes located completely inside other genes (LIF_A2691 (3 323 647-3 323 850) inside LIF_A2690 (3 323 351-3 324 025); LA_0853a (852 272-852 679) inside both LA_0853 (851 443-852 315)and LA_0854 (852 312-853 562)), we did not remove the corresponding CDSs that conflicted with the two proteomic-profiling-verified genes in terms of translational frame because we indeed detected proteomic expression of both frames of overlapping genes in the two strains.
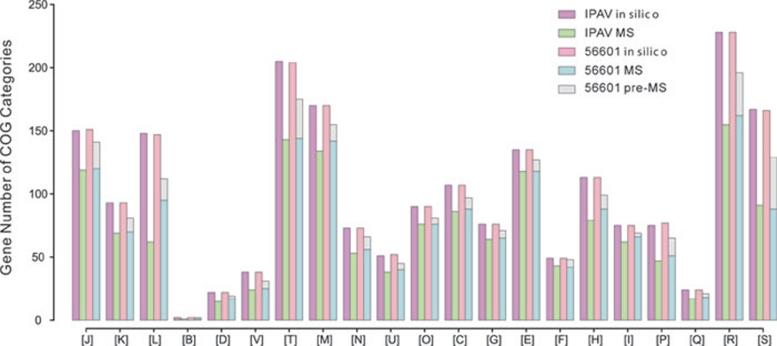
Using MS/MS spectra to search the CDS databases, a total of 2 608 and 2 673 CDSs were detected in strains IPAV and 56601, respectively. Because the predicted proteins aligned to at least two unique peptides from the MS data, they were considered reliably validated. Further functional analysis focused on the 2 078 (55.8%) and 2 225 (59.8%) CDSs confirmed with at least two unique peptide hits from IPAV and 56601, respectively (Figure 3). We evaluated these proteins based on their COG functional category classification. Of the 1 918 proteins from IPAV with assigned COG categories, 70.8% were detected in proteomic profiling, whereas only 39.8% of the remaining 1 808 CDSs were confirmed. Similarly, in strain 56601, 74.8% of the 1 918 proteins with assigned COG categories were detected and only 43.9% of the remaining 1 800 CDSs were observed. Similar statistics were also observed in the proteomic profiling of Mycoplasma pneumoniae and Mycoplasma mobile 18, 19. We also compared our data for both IPAV and 56601 with the recently published proteome-wide cellular protein data for the L. interrogans serovar Copenhageni 20. A total of 1 703 orthologous proteins (72.1% of all proteins identified in IPAV and 56601) were shared by the two studies, including 1 233 functional proteins and 366 hypothetical proteins.
Figure 3.
Functional category of in silico predicted proteins and MS-validated proteins. The colors used in the bars represent different objects: purple, IPAV all predicted proteins; green, IPAV MS-validated proteins; red, 56601 all predicted proteins; blue, 56601 MS-validated proteins; gray, 56601 proteins that were not detected in this work but confirmed by our previous MS data. Proteins were clustered by COG assignment: (J) translation, ribosomal structure and biogenesis; (K) transcription; (L) replication, recombination and repair; (B) chromatin structure and dynamics; (D) cell-cycle control, cell division and chromosome partitioning; (V) defense mechanisms; (T) signal transduction mechanisms; (M) cell wall/membrane/envelope biogenesis; (N) cell motility; (U) intracellular trafficking, secretion and vesicular transport; (O) posttranslational modification, protein turnover and chaperones; (C) energy production and conversion; (G) carbohydrate transport and metabolism; (E) amino-acid transport and metabolism; (F) nucleotide transport and metabolism; (H) coenzyme transport and metabolism; (I) lipid transport and metabolism; (P) inorganic ion transport and metabolism; (Q) secondary metabolites biosynthesis, transport and catabolism; (R) general function prediction only; (S) function unknown.
IS elements as markers for phylogenetic trace of Leptospira species and subspecies
IS elements usually contribute significantly to the plasticity of genomic structure. We examined the differences of the IS elements by their types and numbers among the seven sequenced leptospiral genomes and noticed that some of them might serve as molecular markers to trace the divergence of distinct strains from their common progenitor (Supplementary information, Table S1).
Despite a wide range of differences in the structural arrangement of the genome, all strains of L. interrogans shared a 90-bp ISlin1 remnant at the same site of their CI chromosomes (coordinates 1 388 225 to 1 388 314 as for L. interrogans IPAV) with that of Leptospira borgpetersenii (coordinates 2 651 714 to 2 651 804 as for L. borgpetersenii JB109, 85% identical to that of IPAV) (Figure 4, Supplementary information, Table S1). This is the only IS marker that is shared by all pathogenic Leptospira species known so far, which obviously indicates that L. interrogans and L. borgpetersenii evolved from a common ancestor after the integration event of ISlin1 at this site. Similarly, various types of IS elements at common loci can be observed within strains of the same species, for example, the two strains of L. biflexa, the two strains of L. borgpetersenii, the three strains of L. interrogans and the two strains of L. interrogans serovar Lai (Figure 4, Supplementary information, Table S1). However, there is no single common IS element that is shared among all seven strains, which seems mainly due to the fact that few IS elements reside in the two saprophytic species L. biflexa (six or nine copies each, respectively). This result suggests that IS-related variation in the process of evolution of Leptospira occurred mainly in the pathogenic species that diverged from a common progenitor with the saprophytic Leptospira.
Figure 4.
Distribution of IS elements in large chromosomes of leptospires. The scale is numbered in megabases from the origins of replication. The bars represent the chromosomes. The short vertical lines in each bar indicate the IS elements at different coordinates (red, intact ISs, >1 000 bp; blue, remnant ISs, 100-1 000 bp). The symbols below the genomes are selected IS markers (pathogenic Leptospira specific, sky blue circles; saprophytic Leptospira specific, orange circles; L. borgpetersenii specific, green triangles; L. interrogans specific, magenta triangles; L. interrogans serovar Lai specific, navy blue rhombuses; strain Fiocruz L1-130 specific, brown asterisk; strain 56601 specific, purple asterisk; strain IPAV specific, red asterisk; strain JB197 specific, light yellow asterisk; strain L550 specific, gray asterisk; strain Ames specific, blue asterisk). For the IS markers in each evolutionary level, only one was selected to illustrate in this figure. The colors of the branches in the phylogenic tree consistent with the colors used on the IS markers and the dashed lines indicate that no IS marker is identified at the corresponding levels.
The distribution of different IS elements differs dramatically among Leptospira species 6, 21. We constructed the phylogenetic tree for ISlin1, the dominant and most polymorphic mobile element in L. interrogans (Supplementary information, Figure S1). ISlin1 on the large chromosome was selected when there was greater than 80% sequence identity and reciprocal length coverage in both. In IPAV, the ISlin1 can be grouped into two clusters, groups I and II. Group I includes nine ISs, six of which have cognates in serovar Copenhageni. The ISlin1 F29 in IPAV is the cognate of ISlin1 C3 in Copenhageni, and both are located at the bottom of the trees and have low similarity with other ISlin1 in IPAV and Copenhageni, indicating a distinct history in the ancestor of serovars Lai and Copenhageni. In group II, 30 ISs are clustered together in a large clade. Because they are highly similar to each other, this group is not well separated by bootstrap analysis. Most members of this group have no cognate in Copenhageni and are intact from the beginning to the end in each copy. This result strongly suggests that ISs in this group still have mobile activities that could lead to frequent insertion and recombination events such as the large inversion observed between serovars Lai and Copenhageni, which is suggested to be caused by ISlin1 F33 and ISlin1 F6 21. In conclusion, although Copenhageni harbors 12 seemingly intact ISlin1 elements, at least 10 of them have lost their transposon function due to truncation, and according to Johan Malmstron 20, no ISlin1 transposase was detected by MS in Copenhageni. Most of the ISlin1 elements in IPAV or 56601 may still retain their mobile activity, especially those of group II, and many members seem to have been inserted into the host chromosome after serovar divergence.
Comparative genomic analysis of L. interrogans strains IPAV and 56601
A comparative genomic study of the two serovar Lai strains, the avirulent IPAV and the highly virulent type 56601 strain, was performed with the genome sequence of serovar Copenhageni as an outlier to distinguish strain-specific variations. Comprehensive genetic conservation was observed from chromosome-by-chromosome alignments between the two serovar Lai strains, with a total of 33 insertions and 53 deletions detected in IPAV relative to 56601 (Supplementary information, Table S2). However, only nine (27.3%) insertions are > 1-bp variations, whereas 33 (62.3%) deletions are > 1-bp variations. In addition, 301 SNVs and 3 large-sequence polymorphisms (LSPs) were identified in either the CDSs or the intergenic regions (Supplementary information, Table S3). Among these variations, a total of 101 genes, including 44 functional genes (Table 2), 29 hypothetical genes and 28 transposases, were affected either in the coding region or in the 5′ upstream region. Furthermore, by comparing the genetic variations in the 44 functional genes with their corresponding loci in L. interrogans serovar Copenhageni, 18 gene variations were designated “56601-specific” because these sequences in IPAV are identical to that of serovar Copenhageni. Variations in the remaining 26 genes were consequently termed “IPAV-specific”.
Table 2. Genes that have genetic variations in the two strains.
| IPAVa | Variationb | 56601a | Specc | Gene product | Prot Quant Alterd |
|---|---|---|---|---|---|
| LIF_A0265 | snv (n) | LA_0312 | IPAV | Cell wall hydrolase | * |
| LIF_A1206 | snv (s) | LA_1499 | IPAV | Cytoplasmic membrane protein | * |
| LIF_A1400 | del | LA_1731 | IPAV | Biotin/lipoate protein ligase | * |
| LIF_A2549 | del (up) | LA_3166 | IPAV | Metallophosphoesterase | * |
| LIF_B181 | snv (s) | LB_225 | IPAV | Lipoprotein | * |
| LIF_B239 | snv (n) | LB_298 | IPAV | L-lysine 2, 3-aminomutase | * |
| LIF_A0087 | snv (n) | LA_0098 | 56601 | tRNA (Uracil-5-)-methyltransferase TrmA | * |
| LIF_A2574 | insert | LA_3199 | 56601 | Anticodon nuclease prrC | * |
| LIF_A2853 | insert | — | 56601 | Putative lipoprotein | * |
| LIF_A0252 | del | LA_0299 | IPAV | Carbon-starvation protein A, CstA | N |
| LIF_A1136 | snv (n) | LA_1422 | IPAV | Serine/threonine kinase with GAF and PP2C domains | N |
| LIF_A1290 | snv (n) | LA_1603 | IPAV | Glycosyl transferase | N |
| LIF_A1058 | snv (n) | LA_1313 | IPAV | Glutamine synthetase GlnA | — |
| LIF_A1137 | del | LA_1422 | IPAV | Serine/threonine kinase with GAF and PP2C domains | — |
| LIF_A2090 | snv (n) | LA_2554 | IPAV | Phosphate sodium symporter | — |
| LIF_A3093 | snv (n) | LA_3871 | IPAV | Acriflavine resistance protein | — |
| LIF_A1928 | snv (n) | LA_2360 | 56601 | Ribonucleotide reductase NrdA | — |
| LIF_A0023 | snv (n) | LA_0025 | IPAV | Flagellar switch complex protein, FliG | = |
| LIF_A0264 | snv (n) | LA_0311 | IPAV | Metal-dependent hydrolase | = |
| LIF_A0333 | snv (n) | LA_0391 | IPAV | ATP-dependent protease ClpA | = |
| LIF_A0845 | snv (n) | LA_1045 | IPAV | Membrane carboxypeptidase | = |
| LIF_A0876 | snv (n) | LA_1082 | IPAV | ADP-ribose pyrophosphatase | = |
| LIF_A1181 | snv (n) | LA_1471 | IPAV | Memb-bound H+ translocate pyrophosphatase (H+-PPase) | = |
| LIF_A2430 | snv (s) | LA_2989 | IPAV | NH(3)-dependent NAD(+) synthetase | = |
| LIF_A2439 | snv (s) | LA_3002 | IPAV | tRNA nucleotidyltransferase/poly A polymerase | = |
| LIF_A2491 | snv (n) | LA_3085 | IPAV | Guanosine polyPi pyrophosphohydrolase/synthetase SpoT | = |
| LIF_A3424 | snv (n) | LA_4291 | IPAV | Putative lipoprotein | = |
| LIF_A0210 | snv (n) | LA_0243 | 56601 | Cytochrome c oxidase polypeptide I CyoB | = |
| LIF_A0392 | del | LA_0459 | 56601 | N-acetylmuramoyl-L-alanine amidase | = |
| LIF_A1716 | snv (s) | LA_2114 | 56601 | ATP-binding protein - ABC transporter complex | = |
| LIF_A2120 | insert | LA_2591 | 56601 | Flagellar MS-ring protein FliF | = |
| LIF_A3034 | snv (up) | LA_3793 | 56601 | Putative hemolysin | = |
| LIF_A3075 | del | LA_3845 | 56601 | AcrR family transcriptional regulator | = |
| LIF_A0628 | snv (n) | LA_0765 | IPAV | RNA polymerase alpha subunit | + |
| LIF_A1885 | snv (n) | LA_2309 | IPAV | Long-chain-fatty-acid CoA ligase | ++ |
| LIF_A1107 | snv (n) | LA_1378 | 56601 | GTP-binding protein BipA | ++ |
| LIF_A2500 | snv (n) | LA_3096 | 56601 | Anti-sigma factor antagonist | ++ |
| LIF_A2698 | snv (n) | LA_3365 | 56601 | Hydroxyethylthiazole kinase ThiM | ++ |
| LIF_A0838 | snv (n) | LA_1036 | IPAV | His kinase and response regulator hybrid protein | Y |
| LIF_A0515 | del | LA_0632 | 56601 | PEP-dependent enzyme IIA component of the PTS system PtsN | Y |
| LIF_A1585 | del | LA_1963a | 56601 | Multidrug efflux pump | Y |
| LIF_A2575 | insert | LA_3200 | 56601 | Type I restriction enzyme hsdR | Y |
| LIF_A2854 | insert | — | 56601 | RNA polymerase ECF-type sigma factor | Y |
| LIF_A2857 | insert | — | 56601 | Signal transduct protein containing EAL domain | Y |
The transposases and hypothetical proteins in both strains are not included.
snv (n), nonsynonymous SNV in coding region; snv (s), synonymous SNV in coding region; snv (up), SNV in gene upstream; del, nucleotide deletion in coding region; del (up), nucleotide deletion in gene upstream; insert, nucleotide insertion in coding region.
Specificity of each variation is designed by checking its corresponding site on L. interrogans serovar Copenhageni.
++, the protein abundance of IPAV is significantly higher than that of strain 56601 (P < 0.01); +, higher (P < 0.05); Y, the protein is only expressed in IPAV; —, the protein abundance of IPAV is significantly lower than that of strain 56601 (P < 0.01); —, lower (P < 0.05); N, the protein is only expressed in strain 56601; =, the protein abundance in each strain has no significant difference.
, the protein is not expressed in either strain.
The largest insertion (14 025 bp) in the genome of IPAV was found between LIF_A2852a and LIF_A2862, from 3 516 796 to 3 530 821 (Figure 5A). It introduced nine CDSs, including genes encoding a putative lipoprotein (LIF_A2853), an RNA polymerase ECF-type sigma factor (LIF_A2854), an EAL domain-containing protein (LIF_A2857, similar to diguanylate phosphodiesterase), and five hypothetical proteins (LIF_A2855, LIF_A2856, LIF_A2858, LIF_A2860 and LIF_A2861) as well as an ISlin1 transposase (LIF_A2859). Although our proteomic study identified four proteins (LIF_A2854, LIF_A2855, LIF_A2857 and LIF_A2858), only LIF_A2858 is unique in the genome. The other three have their paralogous genes elsewhere. This 14-kb DNA fragment was also present with a high degree of similarity in the pathogenic serovar Copenhageni strain, only missing the ISlin1. In reference to our analysis of the IS markers, this intact mobile element (ISlin1 F31) is a member of the group II ISs (Supplementary information, Figure S1A). We infer that the 14-kb DNA fragment should exist in the ancestral of L. interrogans serovar Lai IPAV and 56601 and that the ISlin1 F31 was integrated before the ancestor diverged into different lineages. In one of these lineages, this region was deleted via ISlin1-induced DNA translocation, as was observed in 56601.
Figure 5.
Large Indels in the genomes of L. interrogans strains IPAV or 56601. (A) Loss of 14-kb DNA sequence in strain 56601. (B) Type I R/M gene cluster. Genes are color coded according to their functional categories. A red asterisk is labeled above a gene if its product can be detected by MS.
Another insertion in IPAV occurred in the Type I restriction modification (R/M) gene cluster (Figure 5B). This defense mechanism system consists of HsdM, HsdS, HsdR and sometimes a tRNALys-specific anticodon nuclease (ACNase), which, in combination, carries out DNA methylation and restriction functions 22. In the IPAV strain, we identified the intact Type I R/M gene cluster with an anticodon nuclease gene incorporated (LIF_A2572-LIF_A2575). In strain 56601, however, the anticodon gene LA_3199 and its downstream gene hsdR (LA_3200) are truncated as a result of a 2 796-bp deletion. In fact, MS data revealed that HsdM and HsdS had peptide hits in both strains, but HsdR, responsible for the endonuclease activity, was detected only in IPAV. Because the homologous region in Copenhageni exhibits high similarity to that of the IPAV strain, the corresponding nucleotides in strain 56601 were likely lost after the segregation of the two strains. One may conclude that the DNA cleavage and tRNA recognition mechanisms in strain 56601 are impaired.
Other large (> 40 bp) insertions/deletions in strain IPAV include tandem repeats and gains or losses of nucleotides in either CDSs or intergenic regions. Insertions in CDSs include two hypothetical proteins (LIF_A1368 and LIF_A3272) and four transposases (LIF_A1558, LIF_A1559, LIF_A2750 and LIF_A2751). It is worthwhile to mention that LIF_A1368 is a gene located at the end of the lipopolysaccharide (LPS) biosynthetic locus. We did not detect its translation product by MS analysis. If it is involved in LPS biosynthesis, further study is necessary because mutations in this locus have been demonstrated to strongly affect the virulence 23. Deletions in IPAV are equivalent to insertions in strain 56601, which include six hypothetical proteins (LA_0509, LA_1665, LA_1761, LA_1762, LA_2773 and LA_3161) and four transposases (LA_3249, LA_3250, LA_3252 and LA_3253).
Protein quantification and comparative proteomic analysis of the IPAV and 56601 strains
Individual protein abundances and the statistical significance of expression differences in orthologs identified from strains of IPAV and 56601 cultured in EMJH medium are listed in Supplementary information, Table S4. Among the selected 1 627 pairs of orthologs, 149 proteins were upregulated in IPAV (Z > 1.96, corresponding to 95% confidence), and 25 proteins were not detected in strain 56601; conversely, 187 proteins were upregulated (Z < −1.96, P < 0.05) in strain 56601, and 41 proteins were not observed in IPAV (Supplementary information, Table S4).
The upregulated proteins in 56601 were functionally enriched in the COG categories of translation and DNA replication/repair, whereas upregulated proteins in IPAV were enriched for the categories of energy production/conversion, posttranslational processing and lipid metabolism (chi-square test, P < 0.05) (Figure 6). Among these genes, only 15 of these differentially expressed orthologs were found to have mutations in their upstream intergenic regions, while another 16 genes had mutations that occurred in their CDSs. Of these, the six in-frame deletions/insertions and five nonsynonymous SNVs might influence the stability of the protein. All of the remaining genes were free of mutations in their upstream intergenic or coding regions, and apparent variations in the expression level may be caused by mutations in transcriptional regulators or signal transduction systems controlling these genes.
Figure 6.
Functional classification of upregulated orthologs in IPAV and 56601. Green: upregulated genes in IPAV. Blue: upregulated genes in 56601. The upregulated genes include those orthologs that are only expressed in one of the strains. Proteins were clustered according to COG assignment. If a protein had more than one COG category, all were used. A purple/red asterisk is situated in the functional category where the enrichment in IPAV/56601 was discovered by chi-square test.
In strain 56601, 17 proteins involved in translation were upregulated. These proteins included many ribosomal proteins, tRNA synthetases and the ribosome-binding factor, RbfA. Interestingly, we found that the BipA protein displayed a significant bias in expression between the two strains (Z = 7.74, P = 9.77 × 10−15). A total of 171 peptides were mapped on BipA (LIF_A1107) in strain IPAV, but only 17 peptides hit its ortholog in strain 56601, LA_1378. The BipA protein shares sequence similarity with EF-G and can interact with the 70S ribosome, acting as a regulatory factor for normal protein synthesis. However, as was reported, overexpression of BipA will inevitably disrupt the normal functions of the ribosome and impair protein synthesis 24. Because a dramatic difference in protein expression was observed, we postulated that the overexpression of BipA in IPAV might have a negative impact on protein translation and ribosomal biogenesis (shown by the comparative proteomic data). It is noteworthy that BipA in strain 56601 has a 56601-specific R66I nonsynonymous variation in the N-terminal P_loop_NTPase domain. Multiple sequence alignment of BipA from 50 different bacteria indicated that this mutated site is always a basic residue, such as Lys (25), Arg (4) or His (20). Therefore, this R66I mutation might be the cause of the lower level of BipA detected in the virulent strain 56601, which could ultimately lead to more active protein translation in this highly virulent strain.
Proteomic data indicated that purine/pyrimidine metabolism and DNA replication/repair in strain 56601 were much more active than in strain IPAV. Several enzymes involved in these pathways were upregulated in strain 56601, including DNA polymerase III, polymerase I, DNA helicase, DNA ligase, DNA gyrase and the mismatch repair protein MutL/MutS. In addition, the ribonucleotide-diphosphate reductase, nrdA (LA_2360), which is responsible for the conversion of ribonucleotides into deoxyribonucleotides, was expressed approximately three times more in strain 56601 than its ortholog in IPAV. Interestingly, this rate-limiting DNA biosynthesis enzyme has a 56601-specific missense mutation causing an A703T substitution within a barrel domain. It was previously reported that, compared with other metabolic pathways, inactivation of purine/pyrimidine pathway genes in Escherichia coli and some other bacteria was the key factor for limiting their growth in human serum 25. Therefore, these variations are likely, to a certain extent, to contribute to the avirulence property of strain IPAV.
Although not statistically enriched, another functional category exhibiting a notable bias in protein expression between the two strains was the cell envelope and outer membrane biogenesis category. In this class, proteins such as glycosyltransferase, N-acetylneuraminic acid synthetase, lauroyl/myristoyl acyltransferase, UDP-glucose 6-dehydrogenase, 3-deoxy-D-manno-octulosonic-acid transferase and CapA were upregulated in strain 56601. In addition, expression variation for many lipoproteins not classified in this category was also noticed (Table 3). For example, both the exclusively outer membrane lipoproteins, LipL48 (Z = −15.33, P = 0) and LipL36 (P = 3.63× 10−2), maintained high expression levels in strain 56601 compared with those in IPAV. In contrast, significant increases in abundance were detected in IPAV compared with strain 56601 for LipL41 (Z = 10.23, P = 0), OmpL1 (Z = 17.61, P = 0) and the peripheral membrane protein, LipL45 (Z = 20.31, P = 0).
Table 3. Unequally expressed orthologs of OMPs and lipoproteins in IPAV and 56601.
|
L. interrogans strain IPAV |
L. interrogans strain 56601 |
|||||
|---|---|---|---|---|---|---|
| Locus | Ave. abundance | Locus | Ave. abundance | Z-score | P-value | Gene product |
| LIF_A0412 | 2.72 | LA_0492 | 3.52 | −2.094 | 3.63E−02 | LipL36 |
| LIF_A1110 | 0.30 | LA_1384 | 0.58 | −1.996 | 4.60E−02 | Putative lipoprotein |
| LIF_A1578 | 0.81 | LA_1957 | 1.54 | −2.229 | 2.58E−02 | Putative lipoprotein |
| LIF_A1809 | 0.48 | LA_2219 | 1.00 | −2.010 | 4.45E−02 | Putative lipoprotein |
| LIF_A2046 | 0.44 | LA_2497 | 0.81 | −2.442 | 1.46E−02 | Putative lipoprotein |
| LIF_A2603 | 16.59 | LA_3240 | 36.76 | −15.332 | 0 | LipL48 |
| LIF_A2614 | 2.24 | LA_3262 | 4.32 | −2.589 | 9.63E−03 | Putative lipoprotein |
| LIF_A2871 | 0.17 | LA_3571 | 1.05 | −2.888 | 3.88E−03 | Conserved hypothetical lipoprotein |
| LIF_A2902 | 0.04 | LA_3611 | 0.24 | −2.578 | 9.93E−03 | Putative lipoprotein |
| LIF_B116 | 0 | LB_143 | 0.50 | — | — | LipL45-related protein |
| LIF_A0011 | 35.85 | LA_0011 | 30.28 | 4.591 | 4.42E−06 | Outer membrane lipoprotein LipL21 |
| LIF_A0054 | 0.71 | LA_0061 | 0.39 | 2.428 | 1.52E−02 | RlpA-like lipoprotein |
| LIF_A0124 | 4.84 | LA_0136 | 1.55 | 11.055 | 0 | LipL45-like lipoprotein |
| LIF_A0125 | 1.73 | LA_0137 | 1.41 | 2.183 | 2.91E−02 | Hypothetical lipoprotein |
| LIF_A0506 | 39.84 | LA_0616 | 27.43 | 10.232 | 0 | LipL41 |
| LIF_A1179 | 0.45 | LA_1468a | 0 | — | — | Putative lipoprotein |
| LIF_A1561 | 3.73 | LA_1939 | 2.59 | 3.567 | 3.61E−04 | Putative lipoprotein |
| LIF_A1635 | 2.07 | LA_2023 | 1.04 | 2.958 | 3.10E−03 | Putative lipoprotein |
| LIF_A1636 | 5.09 | LA_2024 | 4.07 | 3.745 | 1.81E−04 | Conserved hypothetical lipoprotein |
| LIF_A1825 | 1.73 | LA_2240 | 0.25 | 2.464 | 1.37E−02 | Hypothetical lipoprotein |
| LIF_A1873 | 13.31 | LA_2295 | 3.94 | 20.305 | 0 | LipL45 |
| LIF_A2387 | 1.65 | LA_2936 | 0.51 | 4.768 | 1.86E−06 | LipL45-related lipoprotein |
| LIF_A2417 | 5.70 | LA_2973 | 4.56 | 2.212 | 2.70E−02 | Putative lipoprotein |
| LIF_A3350 | 1.56 | LA_4202 | 0.72 | 2.634 | 8.44E−03 | Putative lipoprotein |
| LIF_B155 | 9.96 | LB_194 | 6.40 | 5.949 | 2.70E−09 | Putative lipoprotein |
| LIF_B174 | 1.05 | LB_216 | 0.27 | 3.840 | 1.23E−04 | Lipoprotein |
| LIF_B198 | 0.80 | LB_250 | 0 | — | — | Hypothetical lipoprotein |
Proteins involved in energy production, lipid metabolism and posttranslational processing were overrepresented in strain IPAV. They included a series of key enzymes in the citric-acid cycle including citrate synthase (LIF_A0645), isocitrate dehydrogenase (LIF_A3240), the succinyl-CoA synthetase alpha/beta subunits (LIF_A0892/ LIF_A0893), fumarate hydratase (LIF_A0159) and malate dehydrogenase (LIF_A1738) as well as genes responsible for fatty acid/lipid metabolism, which generates acetyl-CoA for the initial step of the citric-acid cycle. However, there was no genetic mutation found in these genes with the exception of one coding for a long-chain fatty acid CoA ligase. Therefore, we assume that mutations in trans-regulatory proteins might account for this phenomenon, as discussed in previous sections.
Mutations affecting genes related to signal-induced regulation
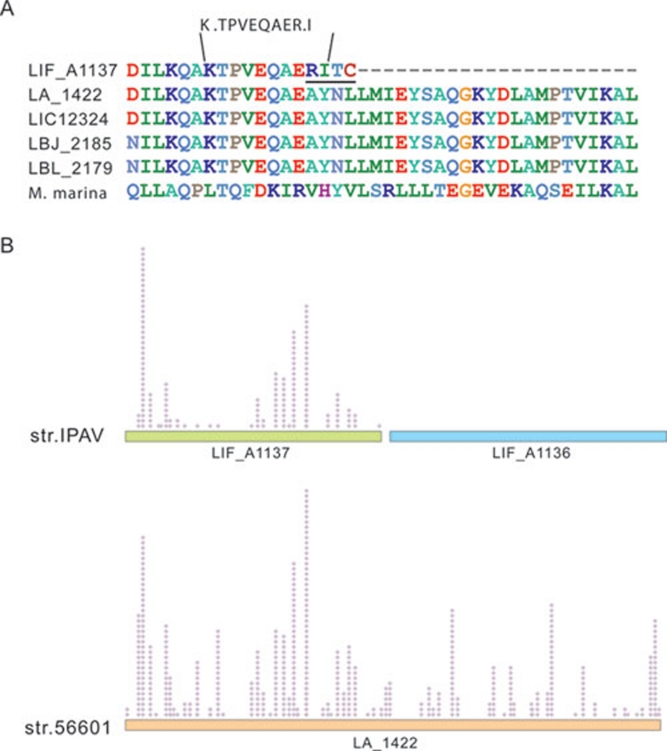
There is growing evidence that Ser/Thr protein kinases (STPKs) also exist in the genomes of microbes in addition to their ubiquitous homologs in eukaryotes 26. Prokaryotes use STPKs and phosphatases not only to regulate many intracellular metabolic processes but also to control stress responses, cell density, cell segregation, carbon and nitrogen assimilation and virulence 27, 28. In certain species, functionally related kinases and phosphatases are located in a single operon and are co-transcribed to maintain balanced expression and cooperative signal transduction activities for certain pathways 29. Leptospira engages this module in one enzyme by fusing the kinase domain with the phosphatase domain. Although three copies of this kind of hybrid genes exist in the genome of strain 56601, only one (LA_1422, 1 780 aa) is highly expressed (520 peptides identified), whereas the other two gene products (LA_1164 and LA_3113) are almost undetectable in our MS data. In fact, similar MS results can also be found in the work for Copenhageni 20, 30. Compared with the genes from strain 56601 and Copenhageni, a single nucleotide in the coding region is deleted in IPAV, leading to a frameshift and yielding two truncated CDSs (LIF_A1136, 930 aa and LIF_A1137, 851 aa). Moreover, we identified one peptide (K.TPVEQAER.I) matching the changed amino-acid residues in IPAV, LIF_A1137 (Figure 7A). Although a total of 160 peptides that contained a kinase domain mapped to LIF_A1137, no peptides were identified in LIF_A1136, which contains a PP2C phosphatase domain (Figure 7B). Given that the other two paralogs (LIF_A0944 and LIF_A2517) are minimally expressed, it can be inferred that as the mutation occurred in strain IPAV the 3′-proximal phosphatase was no longer expressed, and its regulatory function was obviously impaired.
Figure 7.
Comparison of the mutation-affected STPK in IPAV to its orthologs in other species. (A) Multiple sequence alignment of STPK sequences retrieved from NCBI (strain 56601, NP_711603.1; Fiocruz L1-130, YP_002258.1; JB197, YP_801432.1; L550, YP_798524.1; Microscilla marina, ZP_01687505.1). (B) MS-detected peptides mapping on the STPKs in IPAV and 56601. The bars with different colors represent the genes, and the purple dots above the genes represent the detected peptides at their corresponding sites.
In prokaryotes, the two-component system (TCS) plays an essential role in signal transduction. When compared with four other pathogenic leptospires, the strain IPAV histidine kinase LIF_A0838 has a point mutation at amino-acid residue 426, which results in replacement of Gly by Arg in a highly conserved ATP-binding site. Considering that autophosphorylation of the histidine kinase is dependent on this well-conserved ATP-binding domain 31 and that TCS not only regulates basic housekeeping functions but also governs toxins and other proteins important for pathogenesis 32, the mutation in LIF_A0838 may partially account for the avirulence of IPAV.
Mutations affecting genes related to stress response
The cst genes, a group of cyclic AMP (cAMP)-dependent carbon-starvation response genes, are involved in response to nutrient-source deficiency 33, 34. The results from previous studies indicated that the virulence factor CstA is involved in peptide transport, which could expand the range of substrates that a bacterium can utilize and can assist the cell in escaping carbon starvation 35, 36. Protein topology analysis on Leptospira CstA showed that it contained at least 15 transmembrane helices. This hydrophobicity profile agrees well with its function in peptide transport. However in IPAV, the cstA gene LIF_A0252 has an in-frame deletion of 21 nucleotides between helixes 10 and 11, which leads to a 7-aa shorter product compared with the orthologous CstA in pathogenic strains 56601, Copenhageni and L. borgpetersenii. Notably, IPAV CstA was not identified by our MS data, whereas 16 peptides were found for the CstA in strain 56601. In light of this finding, we hypothesize that this critical mutation may influence the stability of the CstA protein. Additionally, the absence of the cstA gene in saprophytic L. biflexa suggests that it could be involved in the pathogenesis of L. interrogans.
Enzymes of the Rel/Spo family enable bacteria to survive during periods of nutrient limitation by producing an intracellular signaling molecule, (p)ppGpp, which triggers the so-called stringent response 37. Previous studies demonstrated that the ability of bacteria to synthesize (p)ppGpp and mount a stringent response is an essential physiological adaptation required for stages of pathogenesis, such as initial growth of adherent bacteria 38, toxin production 39, motility 40 and expression of other virulence regulators 41. Comparative analysis revealed that the IPAV spoT gene (LIF_A2491) has two missense mutations at the amino-acid residues, Val293 and Lys407, resulting in replacement by Ala and Glu, respectively. Apparently, the V293A mutation in the hydrolysis domain may have little impact on its function, as Val and Ala are both aliphatic amino acids. However, the K407E mutation occurred at a conserved site within the TGS motif and may impair the enzymatic function by decreasing the regulation efficiency of TGS toward both the N-terminal synthesis domain and the hydrolysis domains 42, eventually weakening the capacity for in vivo persistence and long-term survival of IPAV under starvation.
Mutations affecting transmembrane channels
The H+-translocating pyrophosphatase (H+-PPase), which exists in a wide variety of organisms, is a membrane-bound proton pump that hydrolyzes inorganic pyrophosphate to generate a proton gradient across the plasma membrane 43, 44. In prokaryotes, the proton gradient is used mainly for intermediate energy storage, transportation of nutrients into the cell and flagellar rotation. According to the literature, H+-PPase always contains 14-16 transmembrane domains as well as several large cytoplasmic loops with important functional motifs 43. Sequence alignment of H+-PPases from different organisms showed high similarity at the amino-acid level, especially in the carboxyl terminus. In IPAV, the H+-PPase homolog, LIF_A1181, had a point mutation at the highly conserved residue 644 with a His substituted by a Gln in the C-terminus (Figure 8). This site is just three residues away from Ser641, which has been demonstrated to be essential for PPi hydrolysis by site-directed mutagenesis studies in Streptomyces coelicolor 43. This change in IPAV may imply that the function of the H+-PPase as an energy source for secondary active transport systems and a scavenger of cytoplasmic PPi may be impaired.
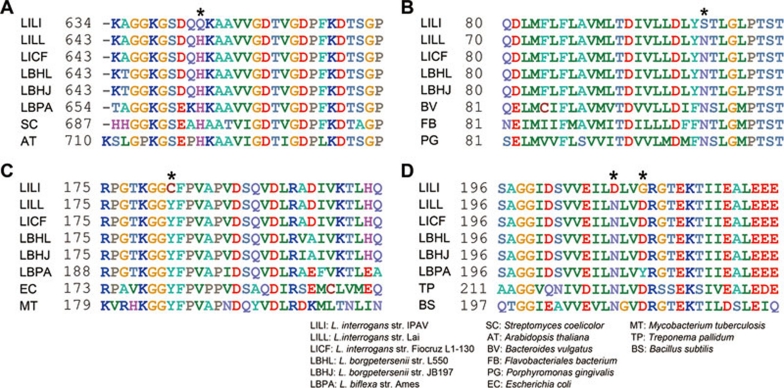
Figure 8.
Multiple sequence alignments of proton-PPase (A), phosphate sodium symporter (B), GlnA (C) and FliG (D). Each site of substitution is indicated by an asterisk above the sequence. The numbers refer to the sites of amino-acid residues. The GenBank accession numbers of the used sequences are as follows: Streptomyces coelicolor (SC, NP_627745.1), Arabidopsis thaliana (AT, NP_173021.1), Bacteroides vulgatus (BV, YP_001299118.1), Flavobacteriales bacterium (FB, ZP_01105510.1), Porphyromonas gingivalis (PG, NP_906040.1), Escherichia coli (EC, NP_418306.1), Mycobacterium tuberculosis (MT, YP_001283561.1), Treponema pallidum (TP, NP_218840.1) and Bacillus subtilis (BS, NP_389504.1).
In L. interrogans, a sodium-dependent phosphate transporter gene was identified by similarity search. Homologous proteins found in Saccharomyces cerevisiae and Neurospora crassa were designated high-affinity phosphate sodium symporters and were induced under phosphate-limiting conditions to assist the organisms in survival during phosphate starvation 45, 46. In IPAV, this phosphate sodium symporter (LIF_A2090) has an N101S mutation located in a highly conserved transmembrane helix in the PHO4 domain (Figure 8). Significantly, a sharp decrease in protein abundance of this symporter was observed in IPAV when compared to strain 56601. A total of 216 peptides mapped to this protein in 56601, but only 52 peptides were detected for LIF_A2090 in IPAV. No mutation occurred in its predicted promoter region, and whether the alteration in the CDS caused the difference in expression remains to be determined. Nonetheless, this high-affinity symporter does not exist in the non-pathogenic L. biflexa, indicating its essential role in phosphorus uptake when pathogenic Leptospira encounter a phosphate-limiting condition in vivo.
Mutations affecting genes related to metabolism, cell division and motility
Glutamine, a source of nitrogen in the biosynthesis of some amino acids and many other metabolites, is produced by glutamine synthetase encoded by glnA, which catalyzes the ATP-dependent condensation reaction of ammonium and glutamate. Comparison of multiple sequences between Leptospira and other bacteria revealed that the glutamine synthetase of IPAV (LIF_A1058) had a point mutation at amino-acid residue 182 that led to replacement of a highly conserved Tyr with a Cys (Figure 8). According to a study of the glutamine synthetases of S. typhimurium and M. tuberculosis, this Tyr is involved in forming a pocket and directly functions as one of the ammonium binding sites 47, 48. Therefore, we might infer that the ability to synthesize glutamine in IPAV may be impaired due to a decrease in the ammonium substrate-binding efficiency. In addition, the expression of glnA in strain 56601 and IPAV exhibited a distinctive feature revealed by our proteomic study: a total of 3 341 peptides mapped to the 56601 GlnA, but only 702 peptides mapped to its ortholog in IPAV. This phenomenon is consistent with a previous report, which showed that the expression levels of glnA are significantly greater in the pathogenic serovars 49.
Bacteria employ different types of genes to complete cell division for their continuous survival. In L. interrogans, we identified four peptidoglycan (PG) hydrolases containing peptidase M23 family domains, as well as one or two N-terminal LysM motifs, which have been demonstrated to have the ability to bind various types of peptidoglycans 50. There is evidence that in Staphylococcus aureus, a PG hydrolase acts as a virulence factor that digests peptidoglycans during the cell division separation and assists in the dissemination of daughter cells in the host 51. It was also reported that a PG hydrolase in Chlamydia pneumoniae was upregulated during long-term chronic infection 52. In IPAV, the smallest PG hydrolase (LIF_A0265) has a G146V mutation located in the conserved peptidase domain. None of the four gene products was identified by MS in strains 56601 or IPAV cultured in the laboratory, and whether the PG hydrolase is expressed in L. interrogans and the LIF_A0265 mutation affects its hydrolase activity are worthwhile subjects for further analysis.
The turning of bacterial flagella is controlled by a rotary motor switch, a complex apparatus that is composed of three proteins: FliG, FliM and FliN 53. Previous studies have demonstrated that the middle domain of FliG is important for flagellar assembly, and that mutations in or around a well-conserved hydrophobic patch from Ile198 to Ile231 directly weaken binding to FliM 54. In IPAV, FliG (LIF_A0023) has two neighboring mutations, N208D and D211G, located in the conserved hydrophobic patch in the middle third of this protein (Figure 8). Active motility of strain 56601 was observed under a dark-field microscope as compared to avirulent IPAV and L. biflexa in vitro. Interestingly, although a conserved Asp residue dominated among the proteins from different genera, the corresponding second mutation site in L. biflexa is a Tyr.
Mutations affecting genes related to degradation
ADP-ribose is an intermediate that is produced during the metabolism of NAD+, mono- or poly-ADP-ribosylated proteins and cyclic ADP-ribose. A high concentration of ADP-ribose in the cell causes nonenzymatic ADP-ribosylation, which inactivates various proteins and interferes with metabolic regulation via enzymatic ADP-ribosylation 55, 56. The ADP-ribose pyrophosphatases (ADPRases), which constitute a subfamily of the Nudix hydrolases, hydrolyze ADP-ribose to AMP and ribose-5-phosphate. In IPAV, the ADPRase (LIF_A0876) has a point mutation in the conserved amino-acid residue 148 that leads to a substitution of acidic Asp by nonpolar Gly in the C-terminal domain. According to the structural study of the ADPRase of Mycobacterium tuberculosis (its corresponding amino acid is also an acidic residue), this site forms a conserved loop that contacts the substrate, triggers a conformation change of the enzyme and finally accomplishes catalysis 57. Therefore, the alteration in IPAV ADPRase is likely to be deleterious, pending experimental confirmation.
ClpA, a Hsp100/Clp chaperone and an integral component of the ATP-dependent ClpAP protease, participates in regulatory protein degradation and the dissolution of protein aggregates 58. In Leptospira, ClpA contains an N-terminal domain consisting of two short-tandem Clp_N motifs and two AAA+ modules (ATPase) head-to-tail from the middle to the C-terminal end. In the literature, the two ATPase domains of E. coli ClpA play different roles: the first ATPase domain (D1 in ClpA) mainly contributes to complex assembly/stability, while the second one (D2 in ClpA) possesses higher ATPase activity and contacts ClpP 59. The IPAV clpA (LIF_A0333) has a missense A303T mutation in the D1 domain, where a functionally important loop related to conformational variability resides 59. If Leptospira clpA has a function similar to that of its ortholog in E. coli, one may presume that the mutation in IPAV might affect ClpAP structure and consequently impair its activity.
56601-specific mutations
Among all of the mutated genes found between IPAV and strain 56601, 13 genes with biological functions (not including previously described genes located in the large sequence deletions) contain 56601-specific mutations when their corresponding sites are compared to Copenhageni (Table 2), including trmA, cyoB, ptsN, bipA, nrdA, fliF, thiM and acrR. Because Copenhageni is also a pathogenic strain, these 56601-specific mutations may not contribute directly to its virulence. However, because many of the mutated genes do not have a paralog in the genome of 56601, mutations that occur in conserved sites may influence the function of their gene products. We indeed found altered expression for some of these genes in the proteomic analysis between IPAV and strain 56601.
Discussion
The virulence-attenuated L. interrogans strain IPAV, assigned to serovar Lai by repeated serological tests, was derived from a virulent strain isolated from a Chinese patient whose clinical record has been unrecoverable thus far. Its characteristic avirulence was described in leptospirosis animal models followed by histological confirmation. In contrast to strain 56601, strain IPAV could not cause conspicuous pathological changes in the liver, kidney and lung tissues, and no clinical manifestations could be detected in any animal inoculated with a lethal dose intraperitoneally. Thus, a comparative study between stain IPAV and, the previously sequenced, highly virulent Lai-type strain 56601 was performed with the aim of elucidating the genetic basis for virulence.
Because we determined the entire genome sequence of strain IPAV using a high-density pyrosequencing method 11, all of the single-nucleotide indels residing in homopolymers were reevaluated against the sequence of the 56601 genome. Ambiguous bases were resequenced by conventional Sanger method using the corresponding PCR products as the templates. Meanwhile, some sequencing errors that existed in the published genome of strain 56601 were resolved in this study, and the genome was completely reannotated.
Comparative genomic analysis indicated that the two strains shared highly similar genome structures and gene orders; 33 insertions, 53 deletions, 301 SNVs and 3 LSPs were observed when IPAV was compared with strain 56601. It was particularly interesting to identify a 14-kb DNA fragment, containing nine genes, in IPAV that was absent in strain 56601 and thus obviously not involved in virulence determination. This fragment also existed in the L. interrogans serovar Copenhageni but was absent in the parasitic L. borgpetersenii and the saprophytic L. biflexa. Further molecular epidemiological investigation is needed to trace its phylogeny in different species of L. interrogans. This result implied that the ancestral strain of IPAV was unlikely to be directly derived from 56601. Other supporting evidence includes the partial deletion of a Type I R/M gene cluster uniquely found in 56601 among all of the Leptospira species that have been sequenced and the presence of five ISs (one IS1500 and two ISlin1 in IPAV; two ISlin1 in strain 56601) that are different between the two strains.
In this study, proteogenomics – a comprehensive strategy combining whole genome sequencing with proteomic studies 19, 60, 61, 62 – was employed for genome annotation in both strains. Quantitative proteomic studies for the two strains indicated that approximately half of the predicted proteins were validated (55.8% for strain IPAV and 59.8% for strain 56601), slightly more than that in recent studies for serovar Pomona 63 and Copenhageni 30. Moreover, by searching the MS spectra against the six-frame translations of the genome sequence, 13 novel genes were detected. Many of these genes overlapped with their flanking neighbors, and as a result, were missed by the gene-calling programs. Hypothetical proteins are either real products that are unique to each organism or false hits that are predicted by the greedy gene-calling algorithms. From MS-based proteomic data, 651 hypothetical proteins (46.3% of all hypothetical proteins) were detected with at least two unique peptides mapped in the L. interrogans strains. These results point to the importance of applying MS data in the validation of gene annotations, which is often lacking from genome studies alone. The lower confirmation rate of hypothetical proteins compared with those in COG categories could be explained by one or more of the following reasons: (1) hypothetical proteins are often expressed at lower concentrations and thus are less likely to be detected by MS, (2) hypothetical proteins have “uncommon” functions that might be induced under specialized conditions different from our studies, and (3) the gene-calling programs are biased in their statistics models toward predicting COG genes, which needs to be addressed by statistical analysis.
A comparative proteogenomic strategy was employed to further analyze the probable genetic factors that may contribute to the virulence-attenuation of strain IPAV. First, the probable effect of transposable elements related to the avirulence of IPAV was excluded because all of the indels caused by ISs that differed between the two strains were focused in intergenic regions.
Although we observed a significant proportion (more than 20% of all functional orthologs) of changes in the proteomic profiles of the two strains under laboratory growth conditions, only a few of these changes could be directly accounted for by nonsynonymous mutations identified in the corresponding genes. Therefore, genetic variation in regulatory proteins is likely the major cause of the global change in expression profiles, which in turn may eventually affect the virulence attenuation of IPAV in combination with other mechanisms, such as direct mutations in some virulence-related genes.
Possible genetic variations affecting the vitality of the cells may play a significant role in virulence determination. According to the MS data, both protein translation and DNA replication/repair were more active in strain 56601. Considering the 56601-specific mutations and distinct abundance of orthologous proteins observed for BipA, regulating the protein synthesis, and NrdA, the rate-limiting enzyme for DNA biosynthesis, we assumed that these variations may alter the primary physiology of the two strains leading to the higher activity and vitality of strain 56601 and making it favorable for pathogenesis during infection. In addition, owing to the mutation at the conserved substrate binding site and the sharp decrease of protein abundance of glutamine synthetase GlnA, nitrogen metabolism or glutamine biosynthesis in IPAV must be largely impaired compared with 56601. Meanwhile, mutations in two starvation response genes, cstA and spoT, may also directly decrease resistance ability to the stress imposed by an unfavorable host environment during infection.
Several genes involved in signal transduction and transcription regulation were found to have genetic variations in IPAV or 56601. These included a signal transduction-related STKP, a TCS gene, an anti-sigma factor antagonist and an AcrR trans-acting factor. According to this study, a nucleotide deletion in the STKP may severely influence the cellular process in IPAV, due to its truncation and the loss of the phosphatase function. Three other trans-regulators were affected with nonsynonymous SNV in coding regions. As revealed by the previous work of Yin-yang MDLC-MS/MS, the anti-sigma factor antagonist (LA_3096) can be phosphorylated in vivo 17, indicating an essential role in transcription regulation. These signal transduction pathways regulate a wide variety of intracellular processes in bacteria and sometimes are essential for coordinating responses to environmental changes. Given the fundamental importance of these genes, it is likely that the genetic mutations in these genes seriously impact the ability for strain IPAV to adapt inside host bodies (e.g., evading immune response or responding to environmental changes in host), thus resulting in virulence attenuation in strain IPAV.
Finally, we observed that some cell-surface proteins were either mutated or had alterations in the protein profiles. Two transmembrane channels, H+-PPase and phosphate sodium symporter, both contained IPAV-specific SNV mutations, but the latter had a dramatic decrease in its protein abundance in IPAV. We also observed high levels of expression of virulence-related proteins belonging to the TolC family and some ABC transporters, which interact in combination to facilitate the efflux of cytoplasmic components 64. In addition, a TonB-like protein and a TonB-dependent outer membrane receptor were upregulated in strain 56601, indicating the intensified ability to uptake certain iron sources 65 from the environment to help its growth. LigB was reported to bind fibronectin during infection 66, 67. However, mutagenesis of the gene by allelic exchange did not affect the virulence of the species 68. We detected its expression under the laboratory culture conditions, but only in the pathogenic strain of 56601 (LA_3778). Whether it contributes to the virulence requires in-depth investigation. Notably, some outer-membrane proteins and lipoproteins were expressed unequally in the two strains, such as OmpL1, LipL45, LipL48, LipL41 and LipL36. These membrane constituents, the predominant leptospiral surface-exposed proteins, play pivotal roles in host-cell interaction, and some of them have been verified as being essential for in vivo infection of pathogenic leptospires 69. These differences in expression ought to account for the avirulence of IPAV to a large extent and are likely caused by alterations in signal transduction and/or transcription regulation, as mentioned above.
Materials and Methods
Bacteria
L. interrogans serovar Lai strain IPAV was a gift from Pro. I. Saint-Girons and M. Picardeau. L interrogans serovar Lai strain 56601 was obtained by the Institute for Infectious Disease Control and Prevention, Beijing, China, and was maintained by serial passages in guinea pigs for the preservation of virulence. Both strains were grown in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium at 28 °C with shaking under aerobic conditions and simultaneously collected at mid-log phase at the density of 3 × 108bacteria per ml (OD600 ≈ 0.075) for further genome sequencing and MS analysis.
Histopathology studies
The animal study was approved by the animal research committee of the Shanghai Jiao Tong University School of Medicine. For each strain, 25 guinea pigs of either sex were assigned to five groups. All animals were injected intraperitoneally with 1 ml of 56601 or IPAV culture (3 × 108) and then euthanized after defined time periods. Tissues from lung, liver and kidney were collected from guinea pigs for histology studies. After fixing in neutral-buffered 4% formaldehyde, the processed tissues were stained by H&E according to routine histological procedures.
Genome sequencing
For genome sequencing, 5 μg of genomic DNA of strain IPAV was extracted from cells. The large-scale IPAV genome sequencing was performed on a GS 20 system 11. A total of 1.8 million reads were generated by 454 GS systems in three runs, and 872 719 high-quality reads were obtained using the system filter criteria. The high-quality reads (813 924, 93%) were assembled with the 454 assembly tools, which had an average depth of 17-fold coverage of the genome and yielded 996 contigs. Among these, 448 large contigs, which ranged from 610 bp to 72 277 bp with an average length of 10 114 bp, represented 98% of the draft sequence. In the finishing process, the order of the selected large contigs was determined using the Blast program with the original published genome sequence of strain 56601. Physical gaps were filled through sequencing PCR products that spanned these regions using ABI 3730 xl DNA sequencers. Sequence assembly was accomplished by the Phred/Phrap/Consed software package 70, 71. A gap that exceeded 6 kb and five gaps that contained multiple copies of tandem repeats were directly filled using a shotgun strategy. To ensure final accuracy, errors in the homopolymer sites that arose from the pyrosequencing method were resolved via comparison with the corresponding sites on 56601, and the ambiguous bases were resequenced using the ABI 3730 xl DNA sequencer. This traditional sequencing method was also used to find and revise sequence errors in strain 56601 when the chromosome-by-chromosome alignment generated inconsistent matches between the two strains.
Sequence annotation and analysis
Open reading frames (ORFs) were identified by GLIMMER 72 and GeneMark 73. The exact predicted CDSs and all six-frame CDSs were translated into proteins and used as the database for mass spectra searching, respectively. After incorporating the two search results, functional annotation was done by comparison with the in-house NCBI NR protein database using BLASTP, followed by a manual check. COG functional classification for each gene was assigned using RPS-BLAST against the CDD database 74. Transfer RNA genes were identified with tRNAscan-SE 75. Considering that the original sequence of 56601 was revised in this work, we reannotated this genome in parallel with IPAV (reannotation for strain 56601 has been done before 6, 17, both of which were based on the old version of genome sequence). Comparison of the chromosome sequences was performed using ACT 76. A phylogenic tree based on the ISlin1 sequences was constructed using NJ methods of the MEGA package 77, and the reliability of each branch was tested by 1 000 bootstrap replications.
Gel electrophoresis separation and in-gel proteolysis
The protein samples from strains 56601 and IPAV were prepared as follows. The harvested cells were suspended in lysis buffer (2% SDS, 50 mM Tris-HCL, 2 mM PMSF, 2 mM sodium fluoride, 2 mM sodium orthovanadate) and sonicated. After centrifuging, the concentration of protein extract was determined by the bicinchoninic acid assay. Samples of 200 μg were subjected to SDS-PAGE on 12.5% gels. The gels were entirely stained with Coomassie blue and excised into 22 sections, which were subjected to in-gel digestion with trypsin as described previously 78.
LC-MS/MS
Liquid chromatography was performed on a Surveyor liquid chromatography system coupled to a LTQ linear ion trap mass spectrometer (Thermo Fisher). The HPLC solvents used were 0.1% formic acid (v/v) aqueous (A) and 0.1% formic acid (v/v) acetonitrile (B). The proteolytic peptide mixtures were eluted using a gradient of 2%- 40% B for 130 min. The temperature of the heated capillary was set at 170 °C. A voltage of 3.0 kV was applied to the ESI needle. The normalized collision energy was 35.0%. The mass spectrometer was set as one full MS scan (400-1800 m/z) followed by MS/MS of the 10 most intense ions. A dynamic exclusion of 120 s was applied.
MS/MS data analysis
After two independent biological replicates were performed, the acquired MS/MS spectra were searched against the databases consisting of forward and reverse sequences of all six-frame CDSs and the final confirmed CDSs using the TurboSEQUEST program in the BioWorks 3.2 software package. The following search criteria were employed: full tryptic specificity was required and two missed cleavages were allowed, carbamidomethylation was set as a fixed modification and oxidation (M) was set as a modification variable, and precursor and fragment ion mass tolerances were 3.0 Da and 1.0 Da (default), respectively. A cutoff of 1.0% false-positive ratio (FPR) 79 and at least 0.1 DeltaCn were set to filter the identified peptides (for raw MS results refer to Supplementary information, Dataset S1-S4). To evaluate the concordance between the two replicates in IPAV and 56601, Pearson correlation tests were performed by calculating identified peptides of the whole proteins in the two parallel datasets. The Pearson correlations are 0.87754 and 0.8685888 for IPAV and strain 56601, respectively.
Protein quantification and data comparisons between the two strains
We chose the proteins assigned by at least two unique peptides to perform the quantification. Protein abundance was calculated mainly based on the simplified APEX-index 80 with slight modification. We utilized the in silico predicted proteotypic peptides to substitute the previously used theoretical tryptic peptides. The protein abundance was defined as:
 |
where PEPI and PEPMIX are the number of detected peptides of an individual protein and the sample of protein mixtures, respectively. PTI and PTMIX represent the number of in silico predicted proteotypic peptides of an individual protein and the sample of protein mixtures, respectively. The proteotypic peptides were predicted using PeptideSieve software 81. The parameters were set as follows: experimental design (PAGE_ESI), peptide length (5-50), peptide mass (400-6000), allowed mis-cleavage (< 2) and threshold score (60).
To ensure the high confidence of the comparisons of the proteomic data, we chose orthologous proteins that both had more than two unique peptides, and at least one of the two orthologs had more than 10 total peptides to perform the comparison. A statistical formulation, assuming protein expression profiling that is well approximated by a normal distribution described previously 82, 83, was introduced to measure which pairs of orthologs were significantly differentially expressed in the two strains:
 |
Briefly, Pi = PEPI/PEPMIX, so the numerator is the difference in proportions of orthologous proteins in IPAV and Lai 56601 (abbreviated as “Lai” in the equation), and Pi 0 = (PEPI IPAV + PEPI Lai)/(PEPMIX IPAV + PEPMIX Lai), so the denominator is the standard error of the difference if the null hypothesis is true. Using the normally distributed Z statistic, different expression of orthologous proteins can be decided by Z > 1.96 or < −1.96 (P < 0.05). Additionally, proteins that had fewer than two unique peptides but were observed in the other strain with more than two unique peptides and 10 total peptides were defined as non-expressed proteins in one strain but expressed in the other.
Functional distribution of the strain-specific upregulated proteins was classified according to COG functional categories. The enrichment of certain categories was evaluated using the chi-square test, and significant classes with P < 0.05 were selected as overrepresented.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgments
We thank Professor M Picardeau for the gift of L. interrogans strain IPAV and Bao-Yu Hu and Yang Yang for the support in performing bacteria culture. We also thank Dr Junwei Yang for suggestions in revising the manuscript. We thank the genome sequencing team at Chinese Human Genome Center at Shanghai for their assistance. This work was supported in part by grants from the National Natural Science Foundation of China (30770111, 30900051 and 30970125), the National Key Program for Infectious Diseases of China (2008ZX10004 and 2009ZX10004), the Program of Shanghai Subject Chief Scientist (09XD1402700) and the Program of Shanghai Research and Development (10JC1408200).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website)
Supplementary Information
Distribution of the IS elements discovered in Leptospira spp.
Insertions and deletions detected in IPAV genome.
SNVs detected in IPAV genome.
Orthologous proteins of strains IPAV and 56601 identified by MS.
Phylogenic analysis of ISlin1 in L. interrogans.
References
- Paster BJ, Dewhirst FE, Weisburg WG, et al. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA. Spirochaetal lipoproteins and pathogenesis. Microbiology. 2000;146 (Pt 7):1491–1504. doi: 10.1099/00221287-146-7-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti AR, Nally JE, Ricaldi JN, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009;7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faine S, Adler B, Bolin C, Perolat P.Leptospira and LeptospirosisMelbourne: Medisci, 1999
- Bulach DM, Zuerner RL, Wilson P, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci USA. 2006;103:14560–14565. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren SX, Fu G, Jiang XG, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- Murray GL, Srikram A, Henry R, et al. Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. 2009;11:311–314. doi: 10.1016/j.micinf.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Murray GL, Morel V, Cerqueira GM, et al. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect Immun. 2009;77:810–816. doi: 10.1128/IAI.01293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata S, Trueba G, Bulach DM, et al. Characterization of a lipopolysaccharide mutant of Leptospira derived by growth in the presence of an anti-lipopolysaccharide monoclonal antibody. FEMS Microbiol Lett. 2010;309:144–150. doi: 10.1111/j.1574-6968.2010.02029.x. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow P, Bourhy P, da Cruz McBride FW, et al. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 2007;3:e97. doi: 10.1371/journal.ppat.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HL, Jiang XC, Zhang XY, et al. Thrombocytopenia in the experimental leptospirosis of guinea pig is not related to disseminated intravascular coagulation. BMC Infect Dis. 2006;6:19. doi: 10.1186/1471-2334-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y, Xu J, et al. Serum activity of platelet-activating factor acetylhydrolase is a potential clinical marker for leptospirosis pulmonary hemorrhage. PLoS ONE. 2009;4:e4181. doi: 10.1371/journal.pone.0004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Bourhy P, Salaun L, Lajus A, et al. A genomic island of the pathogen Leptospira interrogans serovar Lai can excise from its chromosome. Infect Immun. 2007;75:677–683. doi: 10.1128/IAI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XJ, Dai J, Xu H, et al. High-coverage proteome analysis reveals the first insight of protein modification systems in the pathogenic spirochete Leptospira interrogans. Cell Res. 2010;20:197–210. doi: 10.1038/cr.2009.127. [DOI] [PubMed] [Google Scholar]
- Jaffe JD, Berg HC, Church GM. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- Jaffe JD, Stange-Thomann N, Smith C, et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom J, Beck M, Schmidt A, et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AL, Ko AI, Martins EA, et al. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarowicz A, Klyz A, Kwiatek A, Stein DC. Analysis of type I restriction modification systems in the Neisseriaceae: genetic organization and properties of the gene products. Mol Microbiol. 2001;41:1199–1210. doi: 10.1046/j.1365-2958.2001.02587.x. [DOI] [PubMed] [Google Scholar]
- Murray GL, Srikram A, Henry R, et al. Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol Microbiol. 2010;78:701–709. doi: 10.1111/j.1365-2958.2010.07360.x. [DOI] [PubMed] [Google Scholar]
- Owens RM, Pritchard G, Skipp P, et al. A dedicated translation factor controls the synthesis of the global regulator Fis. Embo J. 2004;23:3375–3385. doi: 10.1038/sj.emboj.7600343. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Samant S, Lee H, Ghassemi M, et al. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog. 2008;4:e37. doi: 10.1371/journal.ppat.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L. Manganese-dependent protein O-phosphatases in prokaryotes and their biological functions. Front Biosci. 2004;9:1382–1397. doi: 10.2741/1318. [DOI] [PubMed] [Google Scholar]
- Wehenkel A, Bellinzoni M, Grana M, et al. Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim Biophys Acta. 2008;1784:193–202. doi: 10.1016/j.bbapap.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Hussain H, Branny P, Allan E. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J Bacteriol. 2006;188:1628–1632. doi: 10.1128/JB.188.4.1628-1632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshghi A, Cullen PA, Cowen L, Zuerner RL, Cameron CE. Global proteome analysis of Leptospira interrogans. J Proteome Res. 2009;8:4564–4578. doi: 10.1021/pr9004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanin PM, Thomason PA, Stock JB.Histidine protein kinases: key signal transducers outside the animal kingdom Genome Biol 20023REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche MG, Wanner BL, Crepin S, Harel J. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev. 2008;32:461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- Matin A. Role of alternate sigma factors in starvation protein synthesis--novel mechanisms of catabolite repression. Res Microbiol. 1996;147:494–505. doi: 10.1016/s0923-2508(96)90151-5. [DOI] [PubMed] [Google Scholar]
- Schultz JE, Matin A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J Mol Biol. 1991;218:129–140. doi: 10.1016/0022-2836(91)90879-b. [DOI] [PubMed] [Google Scholar]
- Tenor JL, McCormick BA, Ausubel FM, Aballay A. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr Biol. 2004;14:1018–1024. doi: 10.1016/j.cub.2004.05.050. [DOI] [PubMed] [Google Scholar]
- Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Beresford M, Epton HA, et al. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol. 2002;184:621–628. doi: 10.1128/JB.184.3.621-628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S, Nandi S, Bhadra RK. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J Bacteriol. 2003;185:4672–4682. doi: 10.1128/JB.185.16.4672-4682.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson DL, Lines JL, Pesci EC, Venturi V, Storey DG. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect Immun. 2004;72:5638–5645. doi: 10.1128/IAI.72.10.5638-5645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Tedin K. The bacterial signal molecule, ppGpp, regulates Salmonella virulence gene expression. Mol Microbiol. 2004;52:1827–1844. doi: 10.1111/j.1365-2958.2004.04122.x. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical. Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Mimura H, Nakanishi Y, Hirono M, Maeshima M. Membrane topology of the H+-pyrophosphatase of Streptomyces coelicolor determined by cysteine-scanning mutagenesis. J Biol Chem. 2004;279:35106–35112. doi: 10.1074/jbc.M406264200. [DOI] [PubMed] [Google Scholar]
- Lin HH, Pan YJ, Hsu SH, et al. Deletion mutation analysis on C-terminal domain of plant vacuolar H(+)-pyrophosphatase. Arch Biochem Biophys. 2005;442:206–213. doi: 10.1016/j.abb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Persson BL, Petersson J, Fristedt U, et al. Phosphate permeases of Saccharomyces cerevisiae: structure, function and regulation. Biochim Biophys Acta. 1999;1422:255–272. doi: 10.1016/s0304-4157(99)00010-6. [DOI] [PubMed] [Google Scholar]
- Versaw WK, Metzenberg RL. Repressible cation-phosphate symporters in Neurospora crassa. Proc Natl Acad Sci USA. 1995;92:3884–3887. doi: 10.1073/pnas.92.9.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill HS, Pfluegl GM, Eisenberg D. Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation. Biochemistry. 2002;41:9863–9872. doi: 10.1021/bi020254s. [DOI] [PubMed] [Google Scholar]
- Liaw SH, Kuo I, Eisenberg D. Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site. Protein Sci. 1995;4:2358–2365. doi: 10.1002/pro.5560041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongboonkerd V, Chiangjong W, Saetun P, et al. Analysis of differential proteomes in pathogenic and non-pathogenic Leptospira: potential pathogenic and virulence factors. Proteomics. 2009;9:3522–3534. doi: 10.1002/pmic.200700855. [DOI] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Kajimura J, Fujiwara T, Yamada S, et al. Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol. 2005;58:1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- Hogan RJ, Mathews SA, Kutlin A, Hammerschlag MR, Timms P. Differential expression of genes encoding membrane proteins between acute and continuous Chlamydia pneumoniae infections. Microb Pathog. 2003;34:11–16. doi: 10.1016/s0882-4010(02)00187-0. [DOI] [PubMed] [Google Scholar]
- Lowder BJ, Duyvesteyn MD, Blair DF. FliG subunit arrangement in the flagellar rotor probed by targeted cross-linking. J Bacteriol. 2005;187:5640–5647. doi: 10.1128/JB.187.16.5640-5647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Hill CP, Blair DF. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson EL, Cervantes-Laurean D, Jacobson MK. Glycation of proteins by ADP-ribose. Mol Cell Biochem. 1994;138:207–212. doi: 10.1007/BF00928463. [DOI] [PubMed] [Google Scholar]
- Okuda K, Hayashi H, Nishiyama Y. Systematic characterization of the ADP-ribose pyrophosphatase family in the Cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2005;187:4984–4991. doi: 10.1128/JB.187.14.4984-4991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang LW, Gabelli SB, Cunningham JE, O'Handley SF, Amzel LM. Structure and mechanism of MT-ADPRase, a nudix hydrolase from Mycobacterium tuberculosis. Structure. 2003;11:1015–1023. doi: 10.1016/s0969-2126(03)00154-0. [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Ansong C, Purvine SO, Adkins JN, Lipton MS, Smith RD. Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief Funct Genomic Proteomic. 2008;7:50–62. doi: 10.1093/bfgp/eln010. [DOI] [PubMed] [Google Scholar]
- Tanner S, Shen Z, Ng J, et al. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Prince JT, Marcotte EM. Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005;15:1118–1126. doi: 10.1101/gr.3994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira ML, Pimenta DC, de Morais ZM, Vasconcellos SA, Nascimento AL. Proteome analysis of Leptospira interrogans virulent strain. Open Microbiol J. 2009;3:69–74. doi: 10.2174/1874285800903010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AL, Verjovski-Almeida S, Van Sluys MA, et al. Genome features of Leptospira interrogans serovar Copenhageni. Braz J Med Biol Res. 2004;37:459–477. doi: 10.1590/s0100-879x2004000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvel H, Bommezzadri S, Zidane N, et al. Comparative and functional genomic analyses of iron transport and regulation in Leptospira spp. J Bacteriol. 2006;188:7893–7904. doi: 10.1128/JB.00711-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J, Barocchi MA, Croda J, et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol. 2003;49:929–945. doi: 10.1046/j.1365-2958.2003.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Raman R, Sharma Y, Chang YF. Calcium binds to leptospiral immunoglobulin-like protein, LigB, and modulates fibronectin binding. J Biol Chem. 2008;283:25140–25149. doi: 10.1074/jbc.M801350200. [DOI] [PubMed] [Google Scholar]
- Croda J, Figueira CP, Wunder EA, Jr, et al. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun. 2008;76:5826–5833. doi: 10.1128/IAI.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Matsunaga J. Leptospira: a spirochaete with a hybrid outer membrane. Mol Microbiol. 2010;77:805–814. doi: 10.1111/j.1365-2958.2010.07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, et al. ACT: the artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Kumar A, Tyagi NK, Arevalo E, Miller KW, Kinne RK. A proteomic study of sodium/D-glucose cotransporter 1 (SGLT1): topology of loop 13 and coverage of other functionally important domains. Biochim Biophys Acta. 2007;1774:968–974. doi: 10.1016/j.bbapap.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, et al. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- Mallick P, Schirle M, Chen SS, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- Kal AJ, van Zonneveld AJ, Benes V, et al. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10:1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of the IS elements discovered in Leptospira spp.
Insertions and deletions detected in IPAV genome.
SNVs detected in IPAV genome.
Orthologous proteins of strains IPAV and 56601 identified by MS.
Phylogenic analysis of ISlin1 in L. interrogans.