Abstract
Germline stem (GS) cells were established from gonocytes and spermatogonia of postnatal mouse testes. GS cells proliferate in the presence of several kinds of cytokines, and a small percentage of GS cells also show spermatogonial stem cell (SSC) activity, i.e., they differentiate into sperm after being transplanted into infertile mouse testes without endogenous spermatogenesis. Interestingly, in GS cell culture, we also found that pluripotent stem cells (multipotent germline stem cells (mGS cells)) could be derived and these mGS cells do not have normal androgenetic genomic imprinting marks that are shown in GS cells, e.g., H19 hypermethylation. A new culture system for fetal male germ cells (embryonic GS (eGS) cells) has also been recently developed. Although these cells exhibited SSC potential, the offspring from cultured cells showed heritable imprinting defects in their DNA methylation patterns. In an attempt to understand the self-renewal machinery in SSCs, we transfected H-Ras and cylin D2 into GS cells, and successfully reconstructed the SSC self-renewal ability without using exogenous cytokines. Although these cells showed SSC activity in germ cell transplantation assays, we also found development of seminomatous tumors, possibly induced by excessive self-renewing signal. These stem cell culture systems are useful tools not only for understanding the mechanisms of self-renewal or epigenetic reprogramming but also for clarifying the mechanism of germ cell tumor development.
Keywords: germline stem cells, spermatogonial stem cells, spermatogenesis, self-renewal, genomic imprinting
Introduction
Spermatogonial stem cells (SSCs) are the tissue-specific stem cell population of the postnatal testes and they are responsible for the high productivity of spermatogenesis 1. Postnatally, SSCs arise from more undifferentiated precursors termed as gonocytes, which are derived from primordial germ cells (PGCs) that migrate to the genital ridge and participate in the formation of the embryonic gonad 2. On formation of seminiferous tubules during embryogenesis, PGCs turn into gonocytes. Transformation of gonocytes into SSCs occurs between 0 and 6 days postpartum (dpp) in male mice 3, 4. A single SSC can produce two stem cells or two differentiated cells by self-renewal or differentiating division 4, 5. In the absence of known unique biochemical or phenotypic markers to distinguish SSCs from their initial daughters and without a culture system in vitro, it is difficult to study the biology of SSCs 6. However, in 1994, a germ-cell transplantation technique was reported in mice: this technique could identify SSCs by their ability to generate a colony through spermatogenesis after being transplanted into the seminiferous tubules of mice that were previously treated with a cytostatic agent (busulfan) or of mutant W/Wv (W) mice lacking a functional seminiferous epithelium because of defective spermatogenesis 7.
Glial cell line-derived neurotrophic factor (GDNF), a distantly related member of the transforming growth factor-β superfamily, was originally identified as a survival factor of the embryonic dopaminergic neurons of the midbrain 8. GDNF also plays critical roles outside the nervous system – in the regulation of kidney morphogenesis and spermatogenesis 9. GDNF is expressed in Sertoli cells in the testis and is reported to be a critical factor in vivo for the replication of spermatogonia 10. Heterozygous GDNF-knockout mice gradually lost their spermatogenic ability due to SSC depletion, whereas overexpression of GDNF produced clusters of undifferentiated spermatogonia that could not differentiate 10. In the presence of GDNF, gonocytes and spermatogonia proliferated on feeder cells as islands or clumps, and these cultured cells were designated as GS cells 11, 12, 13. Further, GS cells could be cultured either without serum or without a feeder layer 13. Similar cultures were subsequently established from SSCs of not only pups 14 but also adult mice 15 by other groups. GS cells expressed several markers that are expressed in SSCs, for example, β1 integrin, α6 integrin, and CD9, and these GS cells were able to differentiate into sperm when they were transplanted into seminiferous tubules of infertile W mice. GS cells have a stable karyotype and androgenetic DNA methylation patterns like neonatal gonocytes and sperm, and are tractable for gene targeting 12, 16. Interestingly, a small portion of GS cells can convert into pluripotent stem cells (mGS cells) 12. These mGS cells could differentiate into various types of cells in vitro when ES cell differentiation protocols were applied, and they formed germline chimeras when microinjected into blastocysts. Generation of knockout mice from mGS cells by homologous recombination has also been reported 17. With respect to the origin of mGS cells (whether from residual pluripotent cells that remained from the fetal stage 18 or from de-differentiated GS cells in vitro), it is likely that mGS cells (ES-like cells) can be derived from normal cultured GS cells as shown by the establishment of a clonal line of mGS cells from genetically modified GS cells integrated with a targeting vector 19. Although this study indicates that each GS cell (or SSC) has only a small chance to convert into a pluripotent stem cell in vitro accompanied by the loss of spermatogenic potential, the mechanism of conversion or reprogramming is unknown. In addition, other studies on the generation of pluripotent stem cells from mouse testes have also been reported 20, 21, 22, 23; however, the reprogramming mechanism remains unclear. In addition to the acquisition of pluripotency by SSCs, self-renewal and epigenetic modifications in SSCs are also important issues in tissue stem cell studies. Development of a spermatogonia culture system that provides an opportunity to enrich the number of SSCs in vitro for biochemical and molecular analyses will help to enhance our understanding of SSC biology.
Genomic imprinting and epigenetic reprogramming
Genomic imprinting is an epigenetic mechanism that causes functional differences between paternal and maternal genomes and plays an essential role in mammalian development, growth, and behavior 24, 25. DNA methylation is an important epigenetic mechanism that regulates transcription of many kinds of genes, such as those involved in tumorigenesis and genomic imprinting. Imprinted genes are expressed monoallelically and are divided into two groups of genes: maternally expressed gene Meg and paternally expressed gene Peg. To date, 143 imprinted genes have been identified (see http://www.har.mrc.ac.uk/research/genomicimprinting/) in mice, and many of these genes form a cluster (e.g., Dlk1-Dio3 domain on chromosome 12) 26. Moreover, there is at least one differentially methylated region (DMR) in a cluster of imprinted genes, and such DMRs regulate the expression of imprinted genes. Genomic imprinting memory is erased in PGCs 27, 28 and re-established during gametogenesis. There are three paternally imprinted regions, namely, H19, Meg3IG, and Rasgrf1, in which DNA are methylated during spermatogenesis 29, 30. Other regions are maternally imprinted at oocyte maturation and the process also involves DNA methylation 31, 32. While global DNA demethylation occurs after fertilization in non-imprinted genes, the methylation established during gametogenesis persists in the imprinted genes. Therefore, imprinted regions in somatic cells are differentially methylated between paternal and maternal alleles 33.
Characteristics of germline stem cells
GS cells have normal androgenetic DNA methylation patterns with hypermethylation in DMRs of paternally imprinted regions such as H19 and Meg3IG, and hypomethylation in DMRs of maternally imprinted regions such as Igf2r, Snrpn, Peg5, and Peg10 12. Interestingly, mGS cells established during GS cell culture have methylation patterns that differ from those of GS cells, and DNA methylation analysis of mGS cells showed demethylation in the DMRs of paternally imprinted genes including H19 and Meg3 12. This result suggests that changes in epigenetic modifications such as DNA methylation patterns may play pivotal roles in the reprogramming mechanism to pluripotency (conversion from SSCs to mGS cells).
GS cells could be cultured for more than 2 years and achieve ∼1085-fold expansion. Unlike other germline cells that often acquire genetic and epigenetic changes in vitro, GS cells retained the stable euploid karyotype and androgenetic imprinting during this period and produced not only normal spermatogenesis but also fertile offspring with normal somatic imprinting patterns. Although the telomere length in GS cells decreased progressively at a constant rate (∼0.7 kb per month), the remarkable genetic and epigenetic stability and proliferative potential of GS cells may prevent abnormal germline transmission of genetic and epigenetic damages 34.
Heritable imprinting defects in embryonic GS-derived offspring
Following germ cell transplantation, GS cells can recolonize seminiferous tubules and produce spermatozoa, which are used to produce offspring. In vitro microinsemination is particularly useful for offspring production because it does not require a large number of spermatozoa for fertilization. Therefore, these transplantation and microinsemination techniques are very powerful tools for the functional analysis of the developmental potential of GS cells. F1 offspring from various GS cell lines have normal somatic cell methylation patterns, e.g., half methylated and half unmethylated DMRs in their somatic cells. However, a new cell line derived from fetal gonocytes unexpectedly showed a heritable imprinting defect 35. In an attempt to clarify the effect of altered DNA methylation patterns in the reprogramming mechanism for pluripotency, we developed a new culture system of fetal germ cells. These established male germ cells from embryos at 12.5–18.5 days postcoitum (dpc) were designated as eGS cells. These cells were morphologically indistinguishable from GS cells from postnatal mouse testes and proliferated actively. The eGS cells also showed the typical phenotype of GS cells, such as, high expression of β1 integrin, α6 integrin, and CD9, which are also expressed in SSCs. Unlike embryonic germ (EG) cells that were established by culturing PGCs with leukemia inhibitory factor (LIF), basic fibroblast growth factor (bFGF), and stem cell factor 36, 37, eGS cells did not form teratomas but, rather, induced spermatogenesis and exhibited spermatogonia potential. Furthermore, there were no differences in the DNA methylation patterns in either imprinted genes or repetitive genes such as intracisternal A particles (IAP) and long interspersed nuclear element 1 (Line1) between GS and eGS cells. Methylated DMRs of two paternally imprinted regions, H19 and Meg3IG, and unmethylated DMRs of three maternally imprinted regions, namely, Igf2r, Peg10, and Snrpn, were detected, and this methylation pattern did not change after additional culture; this result implies that eGS cells have stable DNA methylation. However, through a chromatin immunoprecipitation (ChIP) assay, site-specific changes of histone modifications in DMRs of imprinted regions were detected, e.g., reduced H3K27 monomethylation and increased H3K27 trimethylation in H19 DMR in eGS cells. Further, real-time polymerase chain reaction (PCR) analysis also revealed some differences in the gene expression of potential histone epigenetic modifiers such as Chaf1a, Enhancer of Zeste homolog 2 (Ezh2), and Suppressor of Zeste 12 (Suz12). Because Ezh2, a component of the polycomb repressive complex 2 (PRC2), mediates H3K27 trimethylation 38, our results suggest that abnormal histone modification patterns in eGS cells may be caused by aberrant expression of epigenetic modifiers such as Ezh2 and Suz12. Although eGS cells had stem cell activity and produced F1 offspring by transplantation and subsequent microinsemination, the offspring from eGS cells (from embryos at 13.5 dpc) had abnormal body weight and aberrant hypermethylation in both H19 and Snrpn DMRs, suggesting a functional difference between eGS cells and GS cells (which were from postnatal mice). This defect is probably due to in vitro culture, considering the normal DNA methylation in offspring produced by transplantation of fresh gonocytes from embryos at 13.5 dpc, which confirms our previous observation that fetal germ cell transplantation produces offspring with normal imprinting 39. Interestingly, the abnormal DNA methylation of H19 DMR in eGS-cell-derived F1 offspring was not erased after germline transmission and persisted in both male and female germlines for at least four generations (Figure 1). These results suggest that erasing or re-establishing imprinting memory in offspring of eGS cells may be disrupted by abnormal histone modifications. Taken together, this work demonstrates the first long-term culture system for male fetal germ cells and that culturing fetal germ cells induces epigenetic abnormalities that could be transmitted to subsequent generations. Because the mechanism of this heritable imprinting defect in eGS cells remains unclear, further studies will be necessary, e.g., to examine the roles of epigenetic modifiers in GS and eGS cells, the rescue of epigenetic abnormalities by artificial epigenetic reprogramming, and the genomic instability of eGS-cell-derived offspring.
Figure 1.
Characteristics of GS and eGS cells. (A) GS cells are cultured in the presence of growth factors, e.g., GDNF, EGF, and bFGF. Cultured GS cells have a self-renewing property, androgenetic genomic imprinting, and normal karyotype that is stable during a 2-year-long culture. GS cells also have stem cell activity because GS cells form germ cell colonies by germ cell transplantation into seminiferous tubules of infertile W mouse testes. The mGS cells are derived from GS cells acquiring pluripotency accompanied by loss of spermatogenic potential during GS cell culture. (B) eGS cells from fetal male germ cells from embryos at 13.5 dpc. Although eGS cells are morphologically indistinguishable from GS cells established from postnatal mouse testes, express markers identical to that expressed in SSCs, normal DNA methylation patterns, and spermatogenic potential, these cells showed abnormal histone modification patterns in DMRs of imprinted genes. Moreover, eGS-cells-derived F1 offspring and subsequent generations showed abnormal DNA methylation patterns in H19 DMR. This heritable imprinting defect may be caused by residual aberrant histone modifications that could not be reprogrammed. Bars, 100 μm.
Self-renewal in spermatogonial stem cells
SSCs divide continuously and produce sperm throughout a male's life. Therefore, self-renewal and differentiation of SSCs are very important mechanisms for continual spermatogenesis. Because self-renewing SSCs are present in small numbers in mouse testes (0.02%-0.03% of the total testes cell population) 1, 5, it was difficult to analyze the molecular mechanisms of SSC self-renewal. However, the development of this GS cell culture system allowed us to study the self-renewal ability of SSCs through molecular and biochemical analyses.
GDNF-regulated genes, for example, B cell CLL/lymphoma 6, member B (Bcl6b); Bazf, Ets variant gene 5 (Etv5), and Lim homeobox protein 1 (Lhx1), were identified in cultures of self-renewing germ cells. Moreover, it was revealed that Bcl6b is crucial for SSC maintenance in vitro and is also important for spermatogenesis in vivo 40. GDNF-independent SSC self-renewal factors such as promyelocytic leukemia zinc-finger protein (Plzf) and TATA box-binding protein (TBP)-associated factor 4b (Taf4b) were also identified 41, 42. It was reported that the classical mouse mutant luxoid, contains a nonsense mutation in the Plzf-encoding gene, which affects adult germline stem cell self-renewal 41. The gonad-specific TAF4b, a component of TFIID is a transcriptional regulator enriched in the mouse testis, and this factor is important for the maintenance of spermatogenesis by regulating the precise expression of gene products essential for germ cell proliferation such as c-Ret, Plzf, and Stra8 42.
Akt, which is activated by phosphoinositide-3 kinase (PI3K), maintains the pluripotency and growth of ES cells 43, which is another cell type that has germline potential. In our previous study, we elucidated the role of the PI3K-Akt pathway using GS cells and demonstrated that activation of the PI3K-Akt pathway plays an important role in the self-renewal division of SSCs 44. When Akt (the myristoylated form of Akt-Mer; myr-Akt-Mer) was transfected into GS cells and activated by 4-hydroxy-tamoxifen (4OHT), the Akt-transfected GS cells (Akt-GS cells) could proliferate actively in the absence of GDNF for at least 5 months. These cells showed similar expression of markers that are expressed in SSCs to that of wild-type (WT) GS cells and retained normal androgenetic DNA methylation patterns. Through use of the germ-cell transplantation technique, it was revealed that Akt-GS cells supported stem cell self-renewal, but differentiation was stopped at the round spermatid stage. However, offspring were successfully obtained through microinsemination using round spermatid derived from Akt-GS cells, and these offspring also showed normal epigenetic properties such as proper somatic-cell DNA methylation patterns in DMRs of imprinted regions. Because inhibition of the PI3K-Akt pathway stops the proliferation of GS cells and induces apoptosis, this pathway is likely to be involved in SSC self-renewal and survival. However, Akt-GS cells require bFGF supplementation for continuous proliferation. This result suggests that the activation of Akt alone is not sufficient to trigger SSC self-renewal and that other molecules are necessary to provide additional signals equivalent to bFGF signals.
With regard to the role of Akt in SSC self-renewal, another study suggests that Akt is responsible for SSC survival because germ cell clumps did not form in cultures maintained with an Akt inhibitor; apoptotic cells increased in the Akt-inhibitor cultures, and the number of colonies was significantly reduced in recipient testes transplanted with cells from Akt-inhibitor cultures 45. This study also suggests that Akt signaling plays a role in the promotion of SSC survival but not in self-renewal. For instance, while Akt inhibition blocked GDNF-mediated upregulation of Bcl6b, Erm, and Lhx1, which are essential for SSC self-renewal, the expression of Plzf and Pou5f1 was also significantly reduced by Akt inhibition. As Plzf and Pou5f1 are not GDNF-regulated, these results suggest that Akt signaling likely plays a general role to promote SSC survival, but not specifically SSC self-renewal. Nevertheless, it is difficult to exclude the possibility that Akt is involved in SSC self-renewal because the expression of GDNF-upregulated Bcl6b, Erm, and Lhx1 was blocked by Akt inhibitor. In general, the precise signaling mechanisms governing SSC self-renewal remain to be investigated.
Reconstruction of SSC self-renewal in vitro by Ras-cyclin activation
Since SSC self-renewal depends on several cytokines, studies of the molecular machinery for SSC self-renewal have been hampered. Although activated Akt-GS cells could proliferate in the absence of GDNF, activation of Akt alone was not sufficient to drive SSC self-renewal; the cells also required co-stimulation with bFGF because epidermal growth factor (EGF) could not replace bFGF 44. Despite the increasing number of identified genes involved in SSC self-renewal 40, 41, 42, 46, how the exogenous cytokine signals are converted to drive the SSC self-renewal machinery remains unclear.
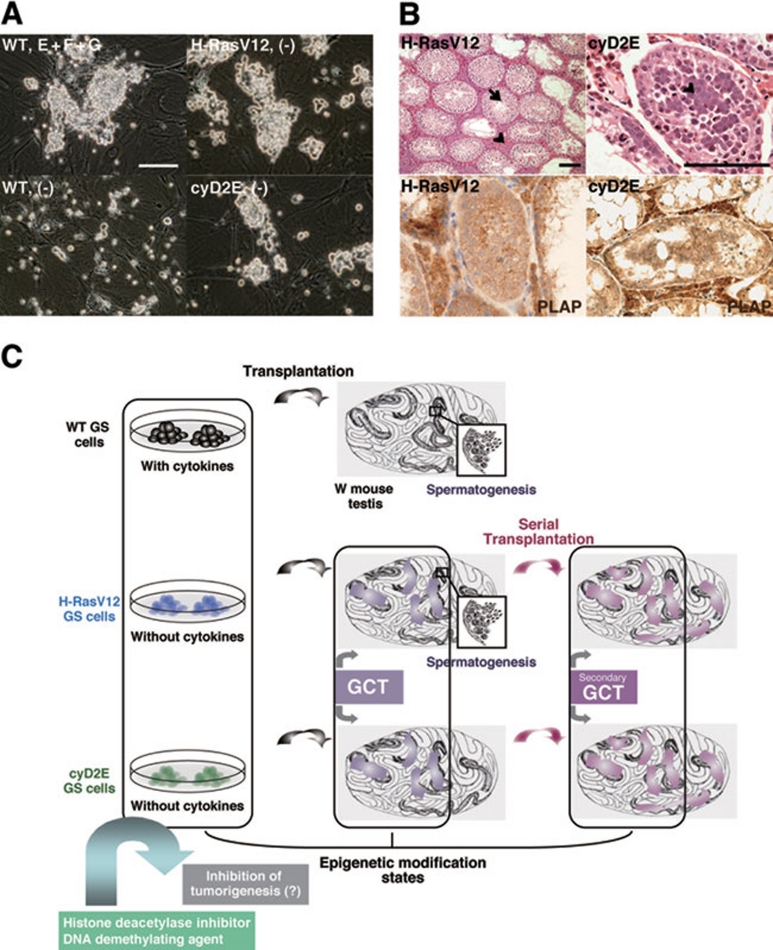
In our recent study, we successfully reconstructed SSC self-renewal abilities in vitro without exogenous cytokines and investigated the molecular mechanism of self-renewal and tumorigenesis in these SSCs 47. We analyzed the involvement of the Ras proto-oncogene 48, 49, 50 in SSC self-renewal. Ras was activated through addition of EGF, bFGF, or GDNF. This activation was inhibited by an inhibitor (PP2) of Src, which was previously shown to be involved in SSC and spermatogonia proliferation 45, 51. Therefore, it was suggested that Ras was activated downstream of Src. Based on this finding, lentivirus expressing the activated form of Ras, H-RasV12, was introduced into GS cells, and these H-RasV12-transfected GS (H-RasV12-GS) cells proliferated in the absence of cytokines (Figure 2A, top right). On the other hand, when H-RasN17, a dominant-negative form of Ras, was transfected into GS cells, proliferation of GS cells was inhibited and these H-RasN17-GS cells died by culture day 12. These results indicate that Ras is not only sufficient but also necessary for GS cell proliferation.
Figure 2.
SSC self-renewal in vitro and tumorigenesis by Ras-cyclin D2 activation. (A) H-RasV12- and cyclin D2/E (cyD2E)-transfected GS cells can proliferate in the absence of growth factors, while WT GS cells cannot be cultured in the absence of growth factors. (B) Development of germ cell tumors (GCTs) in recipient testes of H-RasV12- and cyD2E-transfected GS cells (top). Although abnormal undifferentiated spermatogonia proliferation was detected in both recipients, the observation of both spermatogenesis (arrow) and interstitial tumor infiltration (arrowhead) was made only in H-RasV12-GS cell recipients. Secondary GCTs were developed by serial transplantation for both H-RasV12-GS and cyD2E-GS cells and these GCTs expressed PLAP, the marker of seminomatous tumors (bottom). (C) Schemes of an antitumor strategy in mouse. Based on our findings, H-RasV12-GS and cyD2E-GS cells may show different epigenetic modifications (DNA methylations or histone modifications) compared to WT GS cells. Further, it might be possible to detect some differences in testicular cells between WT GS recipients and H-RasV12-GS and cyD2E-GS recipients containing GCTs and secondary GCTs. Altering epigenetic marks (states) through the addition of histone deacetylase inhibitor and DNA demethylating agent into H-RasV12-GS and cyD2E-GS cells might be able to inhibit tumorigenesis. Bars, 100 μm.
To understand how Ras drives GS cell proliferation, we analyzed the roles of G1 cyclins such as cyclin D and cyclin E. In particular, cyclin D genes are upregulated in response to cytokine stimulation, and it is possible that they are the targets of Ras signaling. Indeed, these genes are upregulated by cytokine treatment in WT GS cells, and analysis of the H-RasV12-GS cells also showed strong expression of cyclin D and E genes without cytokines. Although cyclin E1 or cyclin D alone did not induce GS cell proliferation, co-transfection of cyclin D and E1 restored the proliferation of GS cells, indicating a synergistic effect of cyclin D and E in driving the cell cycle. The cyclin D2 + E1 double transfectants (cyD2E-GS) were expanded without cytokines (Figure 2A, bottom right). It was also suggested that cyclins D1 or D3 may partly compensate for GDNF, because cyclin D1 + E1 (cyD1E-GS) or cyclin D3 + E1 (cyD3E-GS) double transfectants grew when they were cultured only with EGF and bFGF.
Flow-cytometry analysis showed that cyclin-transfected GS cells expressed markers such as EpCAM, α6 integrin, and β1 integrin, all of which are expressed in SSCs, without significant changes in expression levels. However, cell-cycle analysis by Hoechst 33342 staining revealed more cells in the G2/M phase in H-RasV12-GS and cyD2E-GS cells cultured without cytokines than in the WT GS cells.
A germ-cell transplantation technique was used to determine whether H-RasV12-GS and cyD2E-GS cells cultured without self-renewal factors have stem cell activity. After 3 months of transplantation, H-RasV12-GS and cyD2E-GS cells produced germ cell colonies, indicating their SSC activity. Moreover, by transplanting the cells at different time points during culture, we also found that SSCs in both H-RasV12-GS and cyD2E-GS cell cultures consistently increased during long-term culture. These results demonstrated that activation of the Ras-cyclin D2 pathway drives SSC self-renewal in vitro.
Another important observation from this transplantation experiment was the development of germ cell tumors (GCTs). Both H-RasV12-GS and cyD2E-GS cells produced GCTs. Although some H-RasV12-GS cells underwent seemingly normal spermatogenesis, cyD2E-GS cells could not differentiate after being transplanted into infertile W mice (Figure 2B, top). The GCTs that developed from both types of cells expressed placental alkaline phosphatase (PLAP), a marker of seminomas 52. These results strongly suggest that excessive self-renewing stimulus induces oncogenic transformation. Moreover, it was also revealed that GCTs derived from H-RasV12-GS and cyD2E-GS cells contain cancer stem cells because GCTs could produce secondary tumors by serial transplantation, and these secondary GCTs were seminomatous tumors because they expressed PLAP (Figure 2B, bottom). They also showed increased expression of CD44, a cancer stem cell antigen 53. Importantly, our results in mice were very recently confirmed in human studies, where activating mutations in H-Ras were found in human germ cell tumors 54. In addition, cyclin D2 is known to be expressed at very early stages of human GCTs. Therefore, our GCT mouse models will be useful for understanding how GCTs develop from human germ cells.
Although the mechanism of GCT development induced by excessive self-renewing signals remains unclear, the testicular microenvironment might play an important role in GCT development because no tumorigenesis-related abnormalities were found in both types of transfected GS cells in vitro. Normal spermatogenesis in H-RasV12-GS cells and the upregulation of CD44 also suggest that in vivo testicular microenvironment provided oncogenic signals. Besides the role of microenvironment in GCT formation, it will also be interesting to know whether abnormal epigenetic modifications are involved in excessive self-renewal and GCT development of these transfected GS cells. Given that many CpG islands (including tumor suppressor gene CpG islands) have been shown to undergo de novo methylation in specific tumor types, DNA methylation undoubtedly mediates epigenetic silencing of CpG island genes in cancer 55. Furthermore, it was shown that genes methylated in cancer cells are specifically pre-marked with H3K27 trimethylation, a chromatin mark established by the polycomb complex 56. The examination of histone modification patterns in Ras- and cyclin-transfected GS cells and WT GS cells may help in deciphering the epigenetic modification patterns in early stage of tumorigenesis and cancer stem cells (Figure 2C). Thus, it might be possible to detect the early steps of GCT development not only by morphological analysis but also by epigenetic state according to tumor developmental stages. Further studies on molecular mechanisms governing self-renewal and tumorigenesis of SSCs with combined epigenetic analyses will provide unique insights that would expand the research on SSCs, contribute to the treatment of male infertility and improve our understanding of GCT etiology.
Acknowledgments
This work was supported in part by the Mochida Memorial Foundation for Medical and Pharmaceutical Research and by the JST PRESTO program in Japan.
References
- Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- Huckins C, Clermont Y. Evolution of gonocytes in the rat testis during late embryonic and early post-natal life. Arch Anat Histol Embryol. 1968;51:341–354. [PubMed] [Google Scholar]
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21:776–798. [PubMed] [Google Scholar]
- Meistrich ML, van Beek MEAB.Spermatogonial stem cellsIn: Desjardins C, Ewing LL, eds. Cell and Molecular Biology of the Testis. Oxford University Press: New York; 1993266–295. [Google Scholar]
- Brinster RL. Male germline stem cells: from mice to men. Science. 2007;316:404–405. doi: 10.1126/science.1137741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LF, Doherty DH, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci. 2003;116:3855–3862. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvönen ME, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, et al. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Miki H, Inoue K, et al. Long-term culture of mouse male germline stem cells under serum-or feeder-free conditions. Biol Reprod. 2005;72:985–991. doi: 10.1095/biolreprod.104.036400. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci USA. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Ohmura M, Tamura Y, et al. Derivation and morphological characterization of mouse spermatogonial stem cell lines. Arch Histol Cytol. 2004;67:297–306. doi: 10.1679/aohc.67.297. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ikawa M, Takehashi M, et al. Production of knockout mice by random and targeted mutagenesis in spermatogonial stem cells. Proc Natl Acad Sci USA. 2006;103:8018–8023. doi: 10.1073/pnas.0601139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehashi M, Kanatsu-Shinohara M, Miki H, et al. Production of knockout mice by gene targeting in multipotent germline stem cells. Dev Biol. 2007;312:344–352. doi: 10.1016/j.ydbio.2007.09.029. [DOI] [PubMed] [Google Scholar]
- Kubota H, Brinster RL. Technology insight: in vitro culture of spermatogonial stem cells and their potential therapeutic uses. Nat Clin Pract Endocrinol Metab. 2006;2:99–108. doi: 10.1038/ncpendmet0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Lee J, Inoue K, et al. Pluripotency of a single spermatogonial stem cell in mice. Biol Reprod. 2008;78:681–687. doi: 10.1095/biolreprod.107.066068. [DOI] [PubMed] [Google Scholar]
- Guan K, Nayernia K, Maier LS, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izadyar F, Pau F, Marh J, et al. Generation of multipotent cell lines from a distinct population of male germ line stem cells. Reproduction. 2008;135:771–784. doi: 10.1530/REP-07-0479. [DOI] [PubMed] [Google Scholar]
- Ko K, Tapia N, Wu G, et al. Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell. 2009;5:87–96. doi: 10.1016/j.stem.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Noris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. Timing of establishment of parental methylation imprints in the mouse. Genomics. 2004;84:952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem. 2002;277:5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Iwano T, et al. Genetic and epigenetic properties of mouse germline stem cells during long-term culture. Development. 2005;132:4155–4163. doi: 10.1242/dev.02004. [DOI] [PubMed] [Google Scholar]
- Lee J, Kanatsu-Shinohara M, Ogonuki N, et al. Heritable imprinting defect caused by epigenetic abnormalities in mouse spermatogonial stem cells. Biol Reprod. 2009;80:518–527. doi: 10.1095/biolreprod.108.072330. [DOI] [PubMed] [Google Scholar]
- Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Chuma S, Kanatsu-Shinohara M, Inoue K, et al. Spermatogenesis from epiblast and primordial germ cells following transplantation into postnatal mouse testis. Development. 2005;132:117–122. doi: 10.1242/dev.01555. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci USA. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Falender AE, Freiman RN, Geles KG, et al. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Umehara H, Murayama K, Okabe M, Kimura T, Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25:2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- Lee J, Kanatsu-Shinohara M, Inoue K, et al. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development. 2007;134:1853–1859. doi: 10.1242/dev.003004. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem. 2007;282:25842–25851. doi: 10.1074/jbc.M703474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kanatsu-Shinohara M, Morimoto H, et al. Genetic reconstruction of mouse spermatogonial stem cell self-renewal in vitro by Ras-cyclin D2 activation. Cell Stem Cell. 2009;5:76–86. doi: 10.1016/j.stem.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Takahashi M. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12:361–373. doi: 10.1016/s1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- Danielsen AJ, Maihle NJ. The EGF/ErbB receptor family and apoptosis. Growth Factors. 2002;20:1–15. doi: 10.1080/08977190290022185. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Braydich-Stolle L, Kostereva N, Dym M, Hofmann MC. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev Biol. 2007;304:34–45. doi: 10.1016/j.ydbio.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, de Rooij DG, Westerdahl K, Saarma M, Sariola H. Promotion of seminomatous tumors by targeted overexpression of glial cell line-derived neurotrophic factor in mouse testis. Cancer Res. 2001;61:3267–3271. [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumors: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Goriely A, Hansen RM, Taylor IB, et al. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat Genet. 2009;41:1247–1252. doi: 10.1038/ng.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]