Abstract
The mechanisms governing the efficient tumor spheroid penetration and transport by poly(amidoamine) (PAMAM) dendrimers displaying varying numbers of cyclic RGD targeting peptides (2, 3, 7, or 10) were evaluated in this work. The cell-free binding affinities and cellular internalization kinetics of PAMAM-RGD conjugates to malignant glioma cells were determined experimentally, and the results were incorporated into a mathematical model to predict the transport of these materials through a multicellular tumor spheroid. The theoretical analysis demonstrated that greater RGD crosslinking may improve transport through tumor spheroids due to their decreased integrin-binding affinity. This study provides evidence that altering the density of tumor-targeting ligands from a drug delivery platform is a feasible way to optimize the tumor-penetration efficiency of an anticancer agent, and provides insight into the physicochemical mechanisms governing the relative effectiveness of these conjugates.
Keywords: tumor spheroids, PAMAM dendrimers, siRNA delivery, tumor drug penetration, RGD peptides
INTRODUCTION
The inability of anticancer drug molecules to penetrate through solid tumors presents a significant barrier to their efficacy. It is important that a drug molecule reaches all of the malignant cells in a tumor at a concentration sufficient to elicit a therapeutic effect. The distribution of both small molecule chemotherapy drugs and larger antibodies within tumors in vivo is confined to regions adjacent to blood vessels, leaving some regions untouched by the therapeutic compounds (Minchinton and Tannock 2006; Thurber et al. 2008).
Because of their tendency to accumulate in tumor beds due to the enhanced permeability and retention effect, the development of nanoscale anticancer therapeutics has yielded a wealth of promising results. Yet, it is important to improve the intratumoral distribution of these particles, as their relatively large size considerably slows their diffusion within tumors. Several studies have found that nanoscale drug and gene delivery systems comprised of polymers (Han et al. 2007; Mellor et al. 2006; Oishi et al. 2007), peptides (Saleh et al. 2010), or liposomes (Kostarelos et al. 2004; Kostarelos et al. 2005) exhibit poor diffusion into multicellular tumor spheroids, an in vitro solid tumor model. The penetration of these macromolecules into tumors can be improved, however, by tuning their properties such as size and charge (Kostarelos et al. 2004; Kostarelos et al. 2005), or by the incorporation of certain targeting ligands including glucosamine (Dhanikula et al. 2008), lactose (Oishi et al. 2007), or RGD peptides (Sugahara et al. 2009; Waite and Roth 2009).
Several studies have addressed the mechanisms governing the distribution of small molecule drugs (Tzafriri et al. 2009) or antibody therapeutics (Graff and Wittrup 2003; Saga et al. 1995; Thurber et al. 2008; Thurber and Wittrup 2008) in solid tumors, but significantly less work has been done to extend this understanding to nanoscale drug delivery systems. Some mechanisms governing nanoparticle transport through solid tumors are depicted in Figure 1. Reaction transport modeling has identified several important parameters that control the distribution of antibodies into solid tumors including binding affinity, cell internalization kinetics, and rate of free diffusion (Thurber et al. 2008). A similar approach has been applied to describe the penetration of nanoparticles into tumor spheroids (Goodman et al. 2008).
Figure 1.
Transport mechanisms governing nanoparticle penetration through solid tumors. Nanoparticles are transported through tumors by (A) free diffusion in extracellular space, and can be inhibited by (B) cell binding and/or by (C) cell internalization. The structure of nanoparticles can be tuned to alter their interactions with cells and the tumor bed, thus optimizing their transport through solid tumors.
One benefit of utilizing synthetic drug delivery vehicles is the ability to chemically tune their structure to control properties such as particle size, charge, and the presentation of targeting groups. As these physical properties of a nanoparticle can alter their interactions with tumor tissue by changing their effective diffusion coefficient, cellular affinity, or rate of internalization, it is possible to exploit these properties to impart favorable penetration and distribution throughout solid tumors. Our previous work showed that conjugating cyclic RGD to a poly(amidoamine) (PAMAM) dendrimer enhances the penetration and delivery of short-interfering RNA (siRNA) through tumors in a manner that depends on the targeting ligand density (Waite and Roth 2009). In this work, we derive insights into how the density of RGD targeting ligands affects penetration into tumor spheroids via a biophysical approach. The effects of targeting ligand density on integrin binding affinity and cell internalization kinetics were measured, and the experimentally determined parameters were used in a reaction-transport model to estimate the distribution of these materials through a solid tumor spheroid model, which is compared with experimental data.
MATERIALS AND METHODS
Materials
All reagents, buffers, and sensor chips used in surface plasmon resonance experiments were purchased from GE Healthcare (Piscataway, NJ). Unless otherwise stated, all chemicals were purchased from Sigma, and all cell culture products were obtained from Invitrogen (Carlsbad, CA).
Mathematical Model of Tumor Transport
A mathematical model similar to one previously developed (Goodman et al. 2008) to describe the cell binding, internalization, and diffusion of nanoparticles in a three-dimensional tumor spheroid model was utilized in this work. A key distinction for this work is that PAMAM-RGD conjugates were assumed to enter cells additively via two pathways: (1) non-specific electrostatic interactions with cells, and (2) by receptor-mediated interactions between RGD targeting peptides and cell-surface integrin proteins. The subscripts 1 and 2 indicate these respective pathways. Entry from both pathways is by endocytosis but with distinct rates and numbers of binding sites. A system of partial differential equations (Equations 1-4) was utilized to describe the diffusion of PAMAM-RGD/siRNA complexes into a tumor spheroid model of malignant glioma:
| (1) |
| (2) |
| (3) |
| (4) |
In these equations, t is the time variable, r is the radial coordinate, C(t,r) is the concentration of free particles in the spheroid volume, D is the effective diffusion coefficient of particles through the spheroid, ε is the average spheroid porosity, Cbj(t,r) is the concentration of particles bound to pathway j (where j=1 denotes non-specific sites and j= 2 denotes integrins), Csj(t,r) is the concentration of available sites of type j per cell, Ci(t,r) is the concentration of internalized particles (summed over both pathways), and ka1, kd1, and ki1 are the non-specific and ka2, kd2, and ki2 are the integrin-specific association, dissociation, and internalization rate constants, respectively.
The following initial and boundary conditions were applied:
The effective diffusion coefficient of particles through the spheroid, D, was estimated by comparing the model output to experimental data, an approach that has been taken by others to estimate the effective diffusion coefficients of particles through biological tissues(Goodman et al. 2008; Saltzman 2001). The value was fixed at one-sixth of the free diffusivity, with the latter calculated using the Stokes-Einstein relationship. The average spheroid porosity, ε, was estimated from image analysis of the spheroid architecture, and was set to ε = 0.09 (See Supplementary Data). The porosity estimation was used to account for the limited space available for free, unbound polymer, C, to exist within spheroids due to their complex architecture. The concentration of integrin proteins per cell (Cintegrins) was estimated from a study reporting that ~1.1×104 β3 integrin receptors per cell are expressed on the U87 cell line (Benedetto et al. 2006). The number of non-specific binding sites per cell was assumed to be similar to one previously reported(Goodman et al. 2008).
Equations 1-4 were solved numerically using the pdepe function within the MATLAB R2007b software package (The Math Works, Natick, MA).
Cell Culture and Spheroid Formation
U-87 MG cells were maintained in D-MEM medium supplemented with 10% FBS, L-glutamine, and penicillin-streptomycin. Cells were cultivated in an environmental chamber at 37°C and 5% CO2. Multicellular tumor spheroids were generated using the U87 cell line by the hanging drop method as described previously (Waite and Roth 2009).
Surface Plasmon Resonance Spectroscopy
The binding kinetics between PAMAM-RGD conjugates and the purified human integrin protein αvβ3 (Millipore, Billerica, MA) were determined using a Biacore 3000 surface plasmon resonance (SPR) instrument (GE Healthcare Life Sciences, Piscataway, NJ). The running buffer for all SPR experiments was HBS-EP (150 mM NaCl, 3 mM EDTA, 10 mM HEPES, 0.005% P20 surfactant, pH 7.4). Prior to immobilization onto a sensor chip, the integrin protein was desalted into Hepes buffered saline (HBS) using Amicon Ultra 30K MWCO centrifugal filters (Millipore, Billerica, MA) to remove the Tris-containing salts in the formulation provided by the manufacturer. Integrin protein was immobilized onto a channel of the sensor chip surface (CM4 sensor chip) by amine coupling using a similar method as described previously (Shukla et al. 2005). The carboxyl groups on the sensor chip surface were activated by injecting 70 μl of a 1:1 v/v mixture of 0.2 M EDC and 0.05 M NHS. Protein coupling was achieved by injection of 70 μl of integrin protein (44.9 μg/mL in 10 mM acetate buffer at pH 4) across the activated surface. Ethylenediamine at 1 M in water was then injected to deactivate residual NHS-esters on the sensor chip. The process yielded ~1.1 ng/mm2 (~1100 RU) of integrin protein immobilized to the sensor chip surface. A reference surface was created by activating the carboxyl groups of a blank surface with EDC/NHS followed by blocking with ethylenediamine. PAMAM-RGD conjugates were diluted in running buffer at concentrations ranging from 20-200 nM. Carboxymethyl dextran sodium salt, 10 mg/mL in 0.015 M NaCl containing 0.02% NaN3, was added at a concentration of 1 mg/mL to minimize non-specific binding of cationic dendrimers to the anionic dextran substrate on the sensor chip surface. The dendrimer conjugates were injected at a flowrate of 10 μL/min for 3 minutes across the integrin-conjugated active surface and the reference surface. Dissociation was monitored for 90 seconds following dendrimer injection.
Due to differences in size and affinity as compared to the dendrimers, it was necessary to acquire binding sensorgrams for the free cyclic RGD peptide (cRGDfC) using a protein surface with a higher level of integrin protein immobilization (~5 ng/mm2) on a CM5 sensor chip, which contains a higher density of dextran substrate than the CM4 chip. The cyclic RGD peptide was dissolved in HBS-EP running buffer at concentrations of 0.125 mM and 0.25 mM and was injected over the protein and reference surfaces at 10 μL/min for 3 minutes.
The final sensorgrams were obtained by subtracting the reference surface signal from the protein-conjugated surface signal. Regeneration of the surfaces was achieved by injection of 10 mM glycine-HCl, pH 2.5 at 100 μL/min for 30 seconds after each analyte injection. The data for all analytes were analyzed by simultaneously fitting all concentrations of analyte to a 1:1 Langmuir binding model using the BIAevaluation 4.1 software package.
Fluorescent Labeling of PAMAM and PAMAM-RGD Conjugates
Generation 5 PAMAM dendrimers, and the PAMAM-RGD conjugates (G5-2RGD and G5-10RGD) previously synthesized(Waite and Roth 2009) were fluorescently labeled with a green dye, DyLight488 using the manufacturer's protocol (Pierce Rockford, IL). Approximately 1 dye molecule was conjugated per dendrimers, as quantified by absorbance at 493 nm.
Cellular Internalization Kinetic Measurements and Rate Constant Determination
The dynamics of cellular internalization by PAMAM dendrimers and by PAMAM- RGD conjugates were studied in a cellular internalization assay using a protocol similar to one described previously (Dhanikula et al. 2008). U87 cells were seeded at a density of 105 cells/mL (400 μL/well) onto 24-well tissue culture plates and were allowed to adhere overnight. Fluorescently labeled dendrimers (500 nM in culture media) were added to the cells and were harvested at various time points (4, 10, 24, or 48 hours). the cells were washed three times with PBS prior to cell lysis with 200 μL of Triton-X in 0.1N NaOH. Cell lysates were stored frozen at -20°C until analysis of lysates from all time points was performed simultaneously. The dendrimer concentrations detected at each time point were determined by measuring the DyLight fluorescence of each sample with a DTX800 Multimode Detector instrument (Beckman Coulter, CA, USA). The fluorescence values were converted to polymer concentrations using a standard curve of fluorescently-labeled dendrimers.
Equations describing a single cell that binds and internalizes nanoparticles were utilized in this work to parameterize the rate of cellular internalization of PAMAM-RGD conjugates (Equations 5-10) (Lauffenburger and Linderman 1993; Wilhelm et al. 2002). Experimental cell internalization data were fit to Equation 8 to determine the rates of internalization of PAMAM-RGDs.
| (5) |
| (6) |
| (7) |
The solution of Equations 5-7 yields:
| (8) |
| (9) |
| (10) |
where Nbj(t) is the number of bound particles, C0 is the initial concentration of particles, Nsj(t) is the number of available binding sites on the cells, Rj0 is the initial number of available binding sites on the cells, and Nij(t) is the number of internalized particles for pathway j.
The magnitude of non-specific cell interactions by PAMAM-RGDs was assumed to be equal to that of unmodified PAMAM dendrimers (ka1, kd1, and ki1), and was determined by fitting the cell internalization data of unmodified PAMAM to Equation 8 with α2 = 0. PAMAM-RGDs were then assigned additional integrin-specific rate constants (ka2, kd2, and ki2). The integrin-specific association and dissociation rate constants for PAMAM-RGDs (ka2 and kd2) were determined from surface plasmon resonance (SPR) biosensor experiments which measured the binding kinetics between PAMAM-RGDs and αvβ3 integrin proteins immobilized on a sensor chip. The rate of cellular internalization (ki2) of PAMAM-RGD conjugates was determined by fitting cellular internalization data to Equation 8 using the curve fitting toolbox in the MATLAB R2007b software package (The Math Works, Natick, MA).
Statistics
Statistical comparisons within the SPR biosensor data, for the cellular internalization kinetics, and for the experimental spheroid uptake data were performed using a one-way ANOVA test with Tukey's all-pairs post hoc comparison test.
Model Assumptions and Limitations
The following assumptions were made when using the theoretical transport model employed in this work:
The transport model was employed approximating the shape of the tumor spheroids as being strictly spherical. While the spheroids generated in this work were not perfect spheres, spherical coordinates have been utilized in several previous studies describing drug transport in tumor spheroids (Goodman et al. 2008; Thurber and Wittrup 2008), and have provided a reasonable shape estimate of the spheroids to describe drug penetration in this model.
The modes of cell association and internalization were characterized as strictly integrin-mediated and non-specific; furthermore, they were assumed to be additive and independent. While additional distinctions might also be made (e.g. according to internalization pathway as clathrin mediated vs. caveolin-mediated), only the given terms were used to avoid overfitting.
The mathematical expressions utilized to evaluate the cell internalization and SPR kinetic data assumed saturable binding between RGD peptides and integrin receptors based on constant total number of receptors (Mammen et al. 1998). The turnover rates of cellular receptors, especially adhesion receptors such as integrins, are typically low enough to support this assumption.
Constant spheroid porosity was assumed. While it is well known that tumors and tumor spheroids are complex, heterogeneous structures, our measurements did not reveal a consistent trend in porosity as a function of radial distance for the spheroids utilized in this work (Supplementary Data). Hence, we elected to assume a constant, average porosity value (ε = 0.09) rather than to utilize a complex mathematical function which may confound or skew the model predictions.
RESULTS
Binding affinity and kinetic measurements
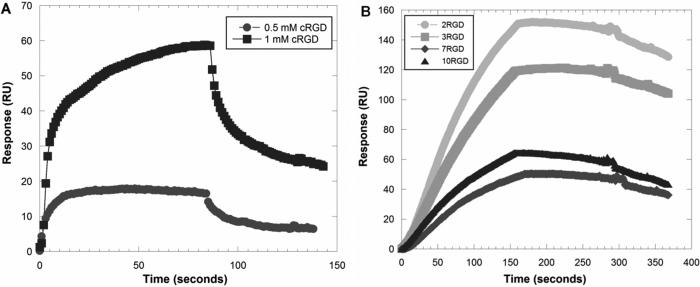
The binding interactions between PAMAM-RGD conjugates and αvβ3 human integrin proteins were studied using a surface plasmon resonance (SPR) biosensor instrument. Sensorgrams were generated to compare the monovalent interactions between the free cyclic RGD peptide and the human integrin protein to the multivalent interactions observed when displaying multiple copies of the RGD peptide from a PAMAM dendrimer scaffold. As expected, PAMAM-RGD conjugates exhibited a greater extent of binding and a slower rate of dissociation from the integrin protein surface as compared to the monovalent RGD peptide (Figure 2). Similar trends have been observed by others studying multivalent presentation of small ligands from PAMAM dendrimer scaffolds (Hong et al. 2007; Shukla et al. 2005).
Figure 2.
Representative SPR sensorgrams of binding interactions between monovalent cyclic RGD peptide (A) and multivalent PAMAM-RGD conjugates (B) with immobilized human integrin protein. The free RGD peptide was used at concentrations of 0.5 mM and 1 mM (A), and the PAMAM-RGD conjugates are shown at a concentration of 50 nM. The sensorgrams acquired for PAMAM-RGD and free RGD peptide were done on separate chip surfaces with differing amounts of immobilized integrin protein.
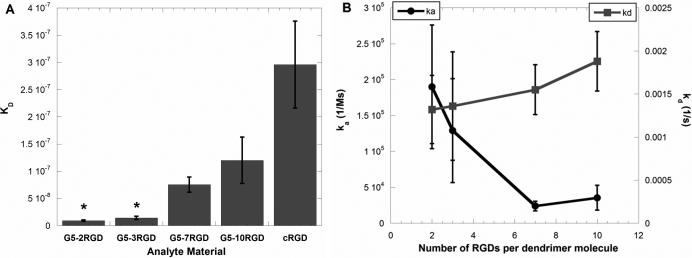
Further, the differences in binding affinities and kinetics between the multivalent PAMAM-RGD constructs displaying various numbers of the cRGD peptide (2, 3, 7, or 10) were compared. Kinetic association and dissociation rate constants (ka and kd) and the ratio of these constants, KD, were determined for each polymer-peptide conjugate by fitting the SPR sensorgram data to a Langmuir binding model (Figure 3). PAMAM-RGD conjugates displaying a low number of RGD peptides (2 or 3) were found to have a significantly higher binding affinity (lower KD value) to the integrin protein than those displaying a high number of peptides (7 or 10) (Figure 3A). This trend is reflected in the more rapid rate of association (ka) to the integrin protein surface for dendrimers with a low number of RGD peptides (2 or 3) than for dendrimers with a high number of peptides (7 or 10), which exhibit a slower rate of association to the protein surface (Figure 3B). Further, the KD measured for PAMAM-RGD conjugates was lower than was measured for the free cRGD peptide, indicating a multivalent enhancement in affinity when conjugating cRGD onto a dendrimer scaffold (Figure 3A). This is consistent with previous reports demonstrating multivalent enhancement in the affinity of small molecules when they are displayed from the surface of PAMAM dendrimers (Hong et al. 2007; Shukla et al. 2005). On the other hand, the decreased rate of association for PAMAM with higher numbers of RGD functionalized (7 or 10) suggests an interplay between avidity enhancement via multivalent binding and steric hindrance. Others have observed decreased association rates for multivalent ligands presented on nanoparticles(Tassa et al. 2010) in SPR studies or decreased affinities in other binding assays (Garanger et al. 2006; Montet et al. 2006).
Figure 3.
Dissociation constants (KD) (A) and kinetic rate constants (ka and kd) (B) for PAMAM-RGD conjugates as determined from fitting SPR sensorgram data. Data represent average fitted values from 3 independent SPR experiments for 3 concentrations of each analyte, and error bars represent SEM. Statistically similar KD values within the PAMAM-RGD conjugates are denoted with asterisks (p<0.05).
Unmodified G5 PAMAM dendrimers were used as a control material to evaluate non-specific binding interactions between dendrimers and the SPR sensor chip. Electrostatic interactions were observed between cationic PAMAM and the anionic dextran on the sensor chip surface. However, we were able to distinguish this non-specific response from the desired integrin-RGD interaction present with PAMAM-RGD materials. A discussion of the studies performed to understand the electrostatic binding interactions between PAMAM and the sensor ship is provided in Supplementary Data.
Cellular Internalization Dynamics
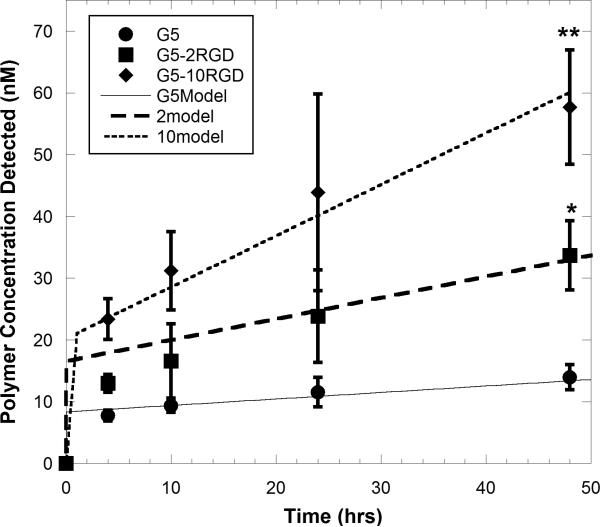
The dynamics of cellular internalization of PAMAM and PAMAM-RGDs were evaluated by measuring the concentration of fluorescently labeled polymers internalized into U87 cells at various times over a 48 hour period. The uptake dynamics of unmodified PAMAM (G5) were compared to those of dendrimers displaying a low number (2) and high number (10) of RGD peptides. For all dendrimers studied, increasing concentration was detected within U87 cells over time, indicating cellular internalization of the polymers (Figure 4). Polymers displaying RGD targeting peptides (either 2 or 10) had significantly higher concentrations internalized into U87 cells (p < 0.05) than the unmodified parent polymer, with the polymer displaying 10 copies of the RGD peptide having the highest amount internalized after 48 hours (Figure 4).
Figure 4.
Polymer concentration detected within U87 cells after constant polymer exposure for up to 48 hours, and corresponding model fits for cell internalization. U87 cells were treated with 500 nM of fluorescently labeled dendrimers in culture medium, and cells were lysed at various time points to detect the concentration of polymers associated with the cells. Asterisks denote that all polymers were significantly different from one another by 48 h (p < 0.05). Data represent mean±SD (n=6).
The cellular internalization rates (ki1 and ki2) for each polymer were determined by fitting internalization data to Equation 8 (Figure 4). The integrin-mediated rates of cellular internalization were about 2.5 (PAMAM-2RGD) to 5-fold (PAMAM-10RGD) more rapid than the non-specific rate (Table I).
Table I.
Kinetic binding and internalization rate constants.
| Non-Specific Terms | Specific integrin-mediated terms | |||||
|---|---|---|---|---|---|---|
| ka1 ((nM*hr)-1) | kd1 (hr-1) | ki1 (hr-1) | ka2 ((nM*hr)-1) | kd2 (hr-1) | ki2 (hr-1) | |
| G5 | 0.184 | 8.93 | 0.0126 | N/A | N/A | N/A |
| G5-2RGD | 0.184 | 8.93 | 0.0126 | 0.684±0.31 (SPR) | 4.77±1.42 (SPR) | 0.0297±0.019 |
| G5-3RGD | 0.184 | 8.93 | 0.0126 | 0.465±0.26 (SPR) | 4.91±2.27 (SPR) | 0.0297±0.019 |
| G5-7RGD | 0.184 | 8.93 | 0.0126 | 0.086±0.024 (SPR) | 5.57±1.04 (SPR) | 0.0608±0.011 |
| G5-10RGD | 0.184 | 8.93 | 0.0126 | 0.127±0.062 (SPR) | 9.13±1.11 (SPR) | 0.0608±0.011 |
Values for PAMAM and PAMAM-RGD conjugates determined from fitting either SPR sensorgram curves or cell internalization data. Non-specific terms (ka1, kd1, and ki1) are from fitting the U87 internalization data for unmodified G5 dendrimer, and integrin-specific ka2 and kd2 terms are from SPR experiments (error is SEM). The integrin-specific internalization terms (ki2) are from the U87 cell internalization data of PAMAM-RGDs (error is the 95% confidence interval ).
Mathematical Model Results
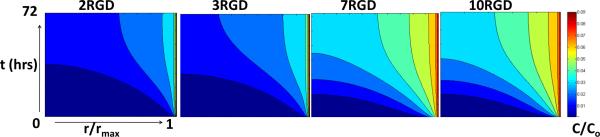
The experimentally determined parameters that represent the interactions between PAMAM-RGDs and U87 cells (ka, kd, and ki) as well as those representing spheroid architecture (ε) were incorporated into a mathematical model of tumor transport and cell interaction to estimate the concentration profile of PAMAM-RGD particles throughout a 3-dimensional tumor spheroid. Concentration profiles calculated from the transport model predict that increasing the number of RGD peptides per PAMAM dendrimer (from 2 to 10) will result in a more homogeneous concentration profile of polymer particles across the cross-section of a spheroid (Figure 5). Polymers containing a low number of RGD peptides (2 or 3) show limited penetration throughout the spheroid according to the model, with a high concentration of material confined to the peripheral region of the spheroid. By increasing the number of RGD peptides to 7 or 10, a more homogeneous profile may be achieved, with a higher concentration of material being delivered to regions farther from the spheroid surface.
Figure 5.
Model concentration profiles of PAMAM-RGD polymers as a function of distance from the spheroid center (r/rmax) and time (hours). Concentration profiles were calculated for PAMAM-RGD conjugates displaying 2, 3, 7, and 10 RGD peptides and are reported as unbound concentrations, C, normalized by the initial polymer concentration exposed to the spheroids, Cmax, of 100 nM.
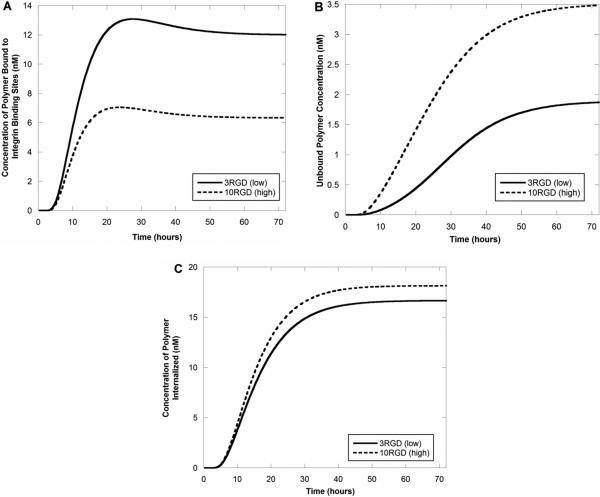
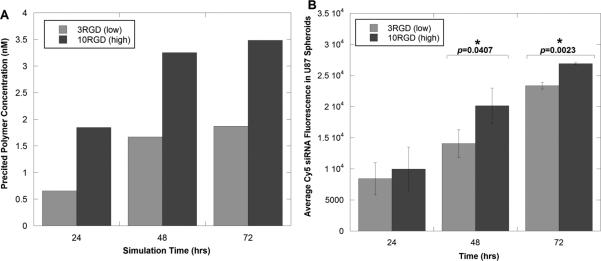
Upon comparing the predicted concentrations of bound polymers to internalized ones, it is the differential binding to integrin receptors that likely causes the overall differences in polymer diffusion through the spheroid (Figure 6). As polymers with a low degree of RGD crosslinking (2RGD and 3RGD) exhibit a significantly higher integrin binding affinity than those with a high degree (7RGD and 10RGD), a higher concentration of the low crosslinking, high-affinity polymers binds to integrins, leaving less polymer free to diffuse through the spheroid (Figure 6A and B). As a result, free polymers with high RGD crosslinking are present at higher concentrations near the spheroid center (Figure 6B), and a higher concentration is internalized into tumor cells (Figure 6C). Further, in all simulations run, the predicted concentration of material increased in all regions of the spheroid as a function of time, as expected. These theoretical results are in good agreement with our previous experimental results that showed a significantly higher concentration of siRNA present in tumor spheroids when delivered by a PAMAM dendrimer displaying a high number (10) of RGD targeting peptides compared to one with a low number (3) of peptides (Figure 7).
Figure 6.
Model concentration profiles of PAMAM-RGD polymers as a function of time at the spheroid center. Concentration profiles showing the predicted concentrations of integrin-bound polymers (A), free polymers (B), and internalized polymers (C) are shown.
Figure 7.
Comparison of time-dependent concentration profiles of PAMAM-RGD conjugates (A) predicted in the theoretical diffusion model with (B) average overall siRNA fluorescence intensity observed experimentally in U87 tumor spheroids (experimental data adapted from (Waite and Roth 2009)). Asterisks in (B) indicate time points at which there was a statistically significant difference in fluorescence intensity between the 3RGD and 10RGD materials (p<0.05).
DISCUSSION
The inability of anticancer drugs and drug delivery systems to penetrate and distribute throughout solid tumors represents a major barrier to drug delivery. Mathematical modeling and scaling analyses have proven to be useful tools in describing the distribution of drugs or antibodies throughout solid tumors. Multicellular tumor spheroids (MCTS) have been a particularly useful experimental model for drug penetration studies, and their simple geometry lends itself well to use in theoretical models. Several previous studies have developed accurate and descriptive mathematical models describing the penetration of antibodies or nanoparticles through tumor spheroids (Goodman et al. 2008; Graff and Wittrup 2003). In this work, we employed a similar methodology, and extended it to evaluate the effect of targeting ligand density on distribution of PAMAM-RGD conjugates through MCTS of malignant glioma cells.
As the binding affinity of molecules is known to be an important parameter dictating their extent of tumor penetration, the binding affinities of a panel of PAMAM-RGD conjugates displaying between 2 and 10 RGD peptides to the immobilized human integrin αvβ3 protein were measured using a surface plasmon resonance (SPR) biosensor. Interestingly, dendrimers displaying a low number of RGD peptides (2 or 3) were found to have a significantly higher binding affinity to the immobilized integrin protein than dendrimers displaying a high number of peptides (7 or 10). This is in contrast to some previous studies where using a PAMAM dendrimer scaffold was found to promote multivalent interactions between folic acid and the folate receptor, resulting in an increased binding avidity when numerous copies of folic acid were displayed (Hong et al. 2007). These contrasting trends are likely due to the inherent differences between the ligand-receptor systems studied. The spacing between RGD peptides has proven to be a critically important parameter in achieving or hindering multivalent binding to integrin proteins or integrin-expressing cells (Koo et al. 2002; Liu 2009; Maheshwari et al. 2000; Wang et al. 2009). In some other studies, an optimal presentation of RGD peptides from a multivalent array was observed, where having too high a density of RGD peptides decreased the affinity to integrin-expressing cells, presumably due to steric hindrance (Garanger et al. 2006; Montet et al. 2006). Taken together, the results suggest that an optimal presentation is strongly context-dependent and hence that new ligand-bearing conjugates should be evaluated experimentally and in a setting that corresponds to the desired application.
In addition to binding affinity, rate of cellular internalization is another important parameter governing tumor penetration. Here, cellular internalization rates for fluorescently labeled PAMAM-RGD conjugates were evaluated quantitatively. The addition of RGD peptides to PAMAM dendrimers significantly increased the amount of polymer that was internalized into U87 cells over a period of 48 hours, as expected. Presumably, this is due to the additional mode of cellular internalization (receptor-mediated) afforded by the introduction of RGD targeting peptides in addition to the non-specific electrostatic interactions known to exist between cationic PAMAM dendrimers and cells (Perumal et al. 2008). Increasing the extent of RGD crosslinking also increased the rate of cellular internalization somewhat, with dendrimers displaying 10 peptides internalizing twice as rapidly than those displaying 2 peptides.
It is notable that the effect of increasing RGD functionalization is different between the cell internalization and SPR binding measurements. There are several fundamental differences between the experimental conditions that may explain this apparent discrepancy. It is important to note the difference in time scales between the binding kinetics and cell internalization experiments; the binding kinetic SPR studies were evaluated on the order of ~5 minutes, whereas the cell internalization experiments were performed over a period of 48 hours. It is possible that steric hindrance might impede the initial attachment of dendrimers displaying many RGD peptides (which is detected by the SPR instrument) and that a longer period of time (on the order of hours) may be required for multivalency to influence binding and cellular accumulation of these materials. Furthermore, SPR measurements utilize integrins rigidly immobilized on a sensor chip, which may restrict multivalent cooperativity. Cellular membranes, in contrast, are fluid, allowing dynamic clustering of receptors and facilitating multivalent interactions.
Previously, we developed a competitive binding cell adhesion assay to compare the abilities of PAMAM-RGD conjugates to inhibit the adhesion of U87 cells to the ECM protein, fibrinogen (Waite and Roth 2009). The results of this assay showed the same trends observed in the SPR experiments in this work, where dendrimers displaying low numbers of RGD peptides were more efficient at inhibiting adhesion of U87 cells to fibrinogen protein than were dendrimers displaying a high number of RGDs. This complementary assay provides further support for the trends observed in the SPR experiments in this work. Taken together, the combination of the binding kinetic and cell internalization experiments demonstrate that altering RGD ligand density has a pronounced impact on integrin binding kinetics on malignant glioma cells and impacts cell internalization dynamics to a lesser extent.
These experimentally determined binding affinity and cell internalization kinetic constants were incorporated into a reaction/transport model to describe polymer distribution through a tumor spheroid. The predicted concentration profiles for PAMAM-RGD conjugates showed that dendrimers with a high level of RGD modification (7 or 10) had a more homogeneous distribution throughout tumor spheroids than did polymers with a low level of RGD modification (2 or 3). These results are in good agreement with our previous study showing that PAMAM dendrimers with 10 RGD peptides were able to deliver significantly higher concentrations of siRNA to malignant glioma spheroids than were PAMAM dendrimers displaying just 3 RGD peptides(Waite and Roth 2009). Our previous work also indicated that PAMAM-RGD conjugates can interfere with the interactions between tumor cells and ECM proteins(Waite and Roth 2009). While not directly incorporated into the mathematical model developed here, the ability of PAMAM-RGD to disrupt cell-ECM contacts may be another important mechanism involved in the efficient tumoral transport of these bioconjugates.
The present study provides insight into the mechanisms governing the efficient tumor penetration by PAMAM-RGD conjugates. Somewhat surprisingly, this work demonstrated that the addition of more than 2-3 RGD peptides to a dendrimer platform enhanced penetration through a tumor spheroid in spite of, or possibly due to, decreasing the binding affinity to cell-surface integrin proteins. It has been reported that low affinity antibodies have more homogeneous distributions through solid tumors than their high-affinity counterparts (Graff and Wittrup 2003), and this study provides complementary evidence for similar behavior with tumor-targeted nanoscale materials. These results are promising for researchers working with synthetic drug delivery systems, demonstrating that a relatively minor architectural change such as varying targeting ligand density can significantly impact tumor penetration in a predictable manner. The methodology presented here can be applied by researchers working with a diverse set of targeting systems to predict and optimize the penetration of anticancer agents through solid tumors. This sort of quantitative approach can guide the rational design of effective nanotherapeutics for the treatment of human cancer.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Yu Chen and Dr. Longqin Hu for access to and assistance with the surface plasmon resonance biosensor. Dominik Naczynski, Joseph Kim, and Dr. Prabhas Moghe are acknowledged for access to the confocal microscope and for technical assistance with acquiring images. We are grateful for financial support from the NIH (R01 EB008278-07), and for a NSF IGERT fellowship (DGE-0504497) to CW.
REFERENCES
- Benedetto S, Pulito R, Crich SG, Tarone G, Aime S, Silengo L, Hamm J. Quantification of the expression level of integrin receptor alpha(v)beta3 in cell lines and MR imaging with antibody-coated iron oxide particles. Magn Reson Med. 2006;56(4):711–6. doi: 10.1002/mrm.21023. [DOI] [PubMed] [Google Scholar]
- Dhanikula RS, Argaw A, Bouchard JF, Hildgen P. Methotrexate loaded polyether-copolyester dendrimers for the treatment of gliomas: enhanced efficacy and intratumoral transport capability. Mol Pharm. 2008;5(1):105–16. doi: 10.1021/mp700086j. [DOI] [PubMed] [Google Scholar]
- Garanger E, Boturyn D, Coll JL, Favrot MC, Dumy P. Multivalent RGD synthetic peptides as potent alphaVbeta3 integrin ligands. Org Biomol Chem. 2006;4(10):1958–65. doi: 10.1039/b517706e. [DOI] [PubMed] [Google Scholar]
- Goodman TT, Chen J, Matveev K, Pun SH. Spatio-temporal modeling of nanoparticle delivery to multicellular tumor spheroids. Biotechnol Bioeng. 2008;101(2):388–99. doi: 10.1002/bit.21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff CP, Wittrup KD. Theoretical analysis of antibody targeting of tumor spheroids: importance of dosage for penetration, and affinity for retention. Cancer Res. 2003;63(6):1288–96. [PubMed] [Google Scholar]
- Han M, Bae Y, Nishiyama N, Miyata K, Oba M, Kataoka K. Transfection study using multicellular tumor spheroids for screening non-viral polymeric gene vectors with low cytotoxicity and high transfection efficiencies. J Control Release. 2007;121(1-2):38–48. doi: 10.1016/j.jconrel.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr., Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14(1):107–15. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci. 2002;115(Pt 7):1423–33. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- Kostarelos K, Emfietzoglou D, Papakostas A, Yang WH, Ballangrud A, Sgouros G. Binding and interstitial penetration of liposomes within avascular tumor spheroids. Int J Cancer. 2004;112(4):713–21. doi: 10.1002/ijc.20457. [DOI] [PubMed] [Google Scholar]
- Kostarelos K, Emfietzoglou D, Papakostas A, Yang WH, Ballangrud AM, Sgouros G. Engineering lipid vesicles of enhanced intratumoral transport capabilities: correlating liposome characteristics with penetration into human prostate tumor spheroids. J Liposome Res. 2005;15(1-2):15–27. doi: 10.1081/lpr-64953. [DOI] [PubMed] [Google Scholar]
- Lauffenburger D, Linderman J. Receptors: Models for Binding, Trafficking, and Signaling. Oxford University Press; Oxford: 1993. [Google Scholar]
- Liu S. Radiolabeled cyclic RGD peptides as integrin alpha(v)beta(3)-targeted radiotracers: maximizing binding affinity via bivalency. Bioconjug Chem. 2009;20(12):2199–213. doi: 10.1021/bc900167c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113(Pt 10):1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- Mammen M, Choi S-K, Whitesides GM. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angewandte Chemie International Edition. 1998;37(20):2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Mellor HR, Davies LA, Caspar H, Pringle CR, Hyde SC, Gill DR, Callaghan R. Optimising nonviral gene delivery in a tumour spheroid model. J Gene Med. 2006;8(9):1160–70. doi: 10.1002/jgm.947. [DOI] [PubMed] [Google Scholar]
- Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6(8):583–92. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. Multivalent effects of RGD peptides obtained by nanoparticle display. J Med Chem. 2006;49(20):6087–93. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- Oishi M, Nagasaki Y, Nishiyama N, Itaka K, Takagi M, Shimamoto A, Furuichi Y, Kataoka K. Enhanced growth inhibition of hepatic multicellular tumor spheroids by lactosylated poly(ethylene glycol)-siRNA conjugate formulated in PEGylated polyplexes. ChemMedChem. 2007;2(9):1290–7. doi: 10.1002/cmdc.200700076. [DOI] [PubMed] [Google Scholar]
- Perumal OP, Inapagolla R, Kannan S, Kannan RM. The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials. 2008;29(24-25):3469–76. doi: 10.1016/j.biomaterials.2008.04.038. [DOI] [PubMed] [Google Scholar]
- Saga T, Neumann RD, Heya T, Sato J, Kinuya S, Le N, Paik CH, Weinstein JN. Targeting cancer micrometastases with monoclonal antibodies: a binding-site barrier. Proc Natl Acad Sci U S A. 1995;92(19):8999–9003. doi: 10.1073/pnas.92.19.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh AF, Aojula H, Arthanari Y, Offerman S, Alkotaji M, Pluen A. Improved Tat-mediated plasmid DNA transfer by fusion to LK15 peptide. J Control Release. 2010;143(2):233–42. doi: 10.1016/j.jconrel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Saltzman WM. Drug Delivery: Engineering Principles for Drug Therapy. Oxford University Press; 2001. [Google Scholar]
- Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, Baker JR., Jr Tumor angiogenic vasculature targeting with PAMAM dendrimer-RGD conjugates. Chem Commun (Camb) 2005;(46):5739–41. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]
- Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;16(6):510–20. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassa C, Duffner JL, Lewis TA, Weissleder R, Schreiber SL, Koehler AN, Shaw SY. Binding affinity and kinetic analysis of targeted small molecule-modified nanoparticles. Bioconjug Chem. 2010;21(1):14–9. doi: 10.1021/bc900438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber GM, Schmidt MM, Wittrup KD. Factors determining antibody distribution in tumors. Trends Pharmacol Sci. 2008;29(2):57–61. doi: 10.1016/j.tips.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber GM, Wittrup KD. Quantitative spatiotemporal analysis of antibody fragment diffusion and endocytic consumption in tumor spheroids. Cancer Res. 2008;68(9):3334–41. doi: 10.1158/0008-5472.CAN-07-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafriri AR, Levin AD, Edelman ER. Diffusion-limited binding explains binary dose response for local arterial and tumour drug delivery. Cell Prolif. 2009;42(3):348–63. doi: 10.1111/j.1365-2184.2009.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite CL, Roth CM. PAMAM-RGD Conjugates Enhance siRNA Delivery Through a Multicellular Spheroid Model of Malignant Glioma. Bioconjugate Chemistry. 2009;20(10):1908–1916. doi: 10.1021/bc900228m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shi J, Kim YS, Zhai S, Jia B, Zhao H, Liu Z, Wang F, Chen X, Liu S. Improving tumor-targeting capability and pharmacokinetics of (99m)Tc-labeled cyclic RGD dimers with PEG(4) linkers. Mol Pharm. 2009;6(1):231–45. doi: 10.1021/mp800150r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm C, Gazeau F, Roger J, Pons JN, Bacri JC. Interaction of anionic superparamagnetic nanoparticles with cells: Kinetic analyses of membrane adsorption and subsequent internalization. Langmuir. 2002;18(21):8148–8155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.