Abstract
The free energy of unfolding of a membrane protein from lipids into water () describes its equilibrium thermodynamic stability. Knowing this parameter gives insight into a membrane protein’s sequence-structure-energy relationships. However, there are few measures of membrane protein stability because of the technical difficulties associated with unfolded and partially folded states. Here, we describe experimental process that allowed us to measure the of the outer membrane phospholipase A (OmpLA) into large unilamellar vesicles (LUVs) of 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC). To arrive at this reversible folding condition, we screened a large number of experimental variables: temperature, incubation time, salt concentration, pH, lipid composition as well as liposome morphology. The principal challenge we encountered under most conditions was hysteresis between folding and unfolding titrations. A second factor that compromised reversible folding was the observation that a fraction of the protein population tended to aggregate. We found that hysteresis could be completely eliminated on a feasible timescale by conducting experiments at acidic pH, by the slow dilution of the protein in the initial titration setup and by utilizing a low concentration of a detergent as a temporary “holdase” to solubilize the protein upon its initial dilution into folding conditions. We confirmed that the detergent did not disrupt the LUVs using fluorescence emission of lipid-sensitive dyes and light scattering. The results of our parameter search should be generally useful for efforts to measure of for other membrane proteins.
Introduction
Thermodynamic measurements of membrane protein stabilities will enable a better understanding of how membrane proteins adopt folded states that remain stably embedded in lipid bilayers and will inform on the consequences of genetic mutations that destabilize membrane protein structures. Understanding thermodynamic stability also brings insight into the formation of misfolded proteins, which can have devastating consequences in the cellular environment. In cystic fibrosis, for example, the most commonly occurring genetic mutation (Δ508) can still result in a functional cystic fibrosis transmembrane conductance regulator protein (CFTR) under some conditions [1-3]. Consequently, the human disease with this genetic variant cannot be one in which the native structure is irreversibly altered; rather, it is the folding and processing of the mutated CFTR protein that is compromised by this mutation [4] [5]. Thus, this form of cystic fibrosis is a membrane protein folding disease. Aside from the intellectual merit of knowing how sequence encodes structure, we also need a deeper understanding of the conformations and stabilities of unfolded, partially folded and misfolded membrane proteins to be able to address medical conditions like those exemplified by genetic variations in CFTR.
However, there are few reports of membrane protein stability [6-11]. Presumably these low numbers can be attributed to the technical difficulties associated with handling unfolded and partially folded conformations, which have a tendency to aggregate or precipitate [9, 12] [10]. Recently, we successfully measured the thermodynamic stability of the outer membrane phospholipase A (OmpLA) from E. Coli in 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) [11]. Using OmpLA as a scaffold for host-guest experiments, we further determined the contributions of each of the twenty natural amino acid side chains to the stability of OmpLA in a central position of a lipid membrane [11]. For these experiments to even be possible, the first task we faced was to demonstrate reversible, path-independent folding of OmpLA protein in bilayers so that we could rigorously extract from these data the equilibrium free energy difference between the folded form of OmpLA in a lipid membrane and the unfolded form in water () [11]. We made measurements of by using a well-established method of perturbing the equilibrium between the folded and unfolded forms by titrating the protein into varying amounts of a chemical denaturant [8, 10, 11, 13, 14]. Importantly, that method is only valid when the folded and unfolded forms are able to interconvert reversibly in a path independent manner [13, 14]. The reversibility must be verified in the region of the denaturant titration where equilibrium between folded and unfolded states is observed. Merely testing reversibility at the endpoints of the titrations under strongly folding or unfolding conditions is insufficient because equilibrium between the two states cannot be observed at those ends.
Verifying reversibility of membrane protein folding in the presence of large unilamellar vesicles (LUVs) was already known to be difficult. Huysmans et al. executed their own search of several experimental conditions before discovering reversible folding conditions for the protein E. Coli PagP with DLPC LUVs [10]. At low lipid-to-protein molar ratios (<800:1), PagP was shown to exhibit a path dependent hysteresis between folding and unfolding transitions when titrated in urea [10]. Likewise, Pocanschi et al. found hysteresis between the folding and unfolding paths of E. Coli OmpA with DLPC LUVs DLPC or equal-molar DLPC and 1,2-dilauroyl-sn-glycero-3-phosphoglycerol (DLPG) [15]. That hysteresis of OmpA folding was observed to persist for at least 12 days. Reversible data has been presented for a urea titration of OmpA in SUVs having a small fraction of anionic lipid headgroups [8, 16], however, even in those results there was a 10-20 percent difference in folding and unfolding paths at multiple steps in the region of the titration where both the folded and unfolded states were observed [16].
Therefore, investigating reversible folding of membrane proteins with liposomes is not a trivial exercise, however given the contributions that thermodynamic stability information can bring to an understanding of sequence-structure-energy relationships for membrane proteins, we believe it warrants further attempts. We tackled this challenge by examining the folding of OmpLA in LUVs of phospholipids because LUVs are a thermodynamically stable system and are therefore more appropriate for thermodynamic measurements as opposed to metastable small unilamellar vesicles (SUVs) [17]. In the present paper, we describe our process to find experimental conditions that promote reversible folding of OmpLA across a denaturant titration. Our results should provide a roadmap for future work with other membrane proteins (OMPs) and should also have applicability to the study of helical membrane proteins.
Results and Discussion
Considerations of membrane protein folding and unfolding
Our experimental system was simple in many respects: (1) prepare a “folding” titration by diluting unfolded protein initially in a high concentration of denaturant into various lower concentrations of denaturant; (2) prepare an “unfolding” titration by diluting folded protein initially in a low concentration of denaturant into higher concentrations of denaturant that are the same as in the samples in the folding titration; and (3) compare spectroscopic signals from samples in both titrations. If the folded and unfolded forms of OmpLA reversibly interconvert within the timescale of our experiment, then the spectroscopic signals from the two different titrations should match each other’s for each of the various denaturant concentrations. Overall, this experimental system was very similar to three previous systems used for the protein OmpA [8, 15, 16, 18] and one previous system used for the protein PagP [10]. However, there were some differences in our system compared to those previous systems below, and we describe each difference below.
Our spectroscopic signal of choice to monitor conformational changes both of OmpLA was the intrinsic fluorescence emission from OmpLA’s nine tryptophans. Intrinsic tryptophan fluorescence was useful because the spectral properties of tryptophans are sensitive to the polarity of their environment, which for membrane proteins varies largely between their folded forms in apolar membranes and their unfolded forms in water. A useful spectral parameter for following protein folding is the wavelength position of maximum emission (λmax) because it does not need to be normalized to be indicative of protein environment. Thus, λmax values are technically amenable to the examination of a large number of experimental conditions. Nevertheless, while this metric was extremely useful in screening we were always cognizant of the fact that λmax is not linearly proportional to protein conformational population [19]. For measurements of energetics, we monitored fluorescence intensity, which is linearly proportional to protein conformational population [11] [19].
The tryptophan fluorescence spectrum for folded OmpLA has a wavelength position of maximum emission (λmax) at 336 nm with our spectrofluorometer set-up. That value is consistent with the majority of OmpLA’s nine tryptophans being embedded in the apolar bilayer or protein interior in the folded state, which is the result we expected based on OmpLA’s known three-dimensional folded structure [19, 20]. The unfolded form of OmpLA demonstrates a lower quantum yield than the folded form and displays an emission spectrum with a λmax of 352 nm. That value was similar to the λmax values we previously measured for the unfolded forms of OmpW and FadL [19] and was consistent with all nine of OmpLA’s tryptophans being located in a polar environment [17, 19, 21]. Such an environment would exist if the unfolded OmpLA was not bound to the liposomes and was instead solvated by water and denaturant. Importantly, we also found that unfolded OmpLA in the absence of any lipid or detergent yields an emission spectrum with the same λmax value (see below), which we interpret to mean that it is not bound to membranes at high denaturant. A second benefit of tryptophan fluorescence spectroscopy is that OmpLA’s nine tryptophans together yield bright fluorescence even at low protein concentrations, which enabled us to dilute the protein well below levels that would be useful for other spectroscopy methods such as circular dichroism. As we describe below, working with low protein concentrations was necessary to avoid self-association of OmpLA at low guanidine HCl concentrations.
A second method we used to verify folding was enzymatic activity [11]. Activity was sound verification that OmpLA’s folded state was inserted across the lipid bilayer because it requires proper tertiary and quaternary contacts [22-26]. A third method we used to verify folding was SDS-PAGE [11]. The folded state of OmpLA exhibited the classic gel shift that is characteristic of other OMPs [8] [10] [27], whereas the unfolded state migrates according to its molecular weight.
In all previous attempts by other groups to verify the reversibility of OMP folding into lipid bilayers, urea was the chosen chemical denaturant used to perturb equilibrium between folded and unfolded populations [8] [10] [15, 16, 18]. However, both our experience and available data suggest that urea may not be powerful enough to fully denature all OMPs and remove them from LUVs. For example, Huysmans et al. found that PagP was likely still associated with LUVs of DLPC even at concentrations up to 10 M urea after several hours at 25° C and at pH 8.0 [10]. Similarly, OmpA did not appear to denature from LUVs of DLPC in about 10 M urea at 40° C and at pH 10, even after 12 days [15]. Likewise, we found that 10 M urea was not strong enough to fully denature OmpLA from LUVs of DLPC after 40 hours at 37° C and at pH 8.0 (data not shown).
Due to the solubility limits of urea, it was not practicable to work with concentrations of urea much higher than 10 M at the temperatures of our experiments. Therefore, we used the more powerful guanidine HCl as the denaturant in all of our experiments presented in this paper. We briefly tested other denaturants, but some (guanidine acetate and guanidine carbonate) were also not soluble at high enough concentrations, and others (lithium perchlorate, methyl urea, dimethyl urea, butyl urea, and thiourea) did not solubilize OmpLA.
As we describe below, we varied the lipid composition of our LUVs, but our primary lipid of choice for most experiments was DLPC. This was convenient because there is precedent for examining reversible folding of other OMPs in LUVs of DLPC to which we could compare our results with OmpLA [10] [15]. Also, DLPC matches the hydrophobic thickness of OmpLA [11, 28]. Further, lipids with longer acyl-chain lengths do not promote the efficient spontaneous folding of OmpLA [27].
Dual challenges for OmpLA: hysteresis and protein aggregation
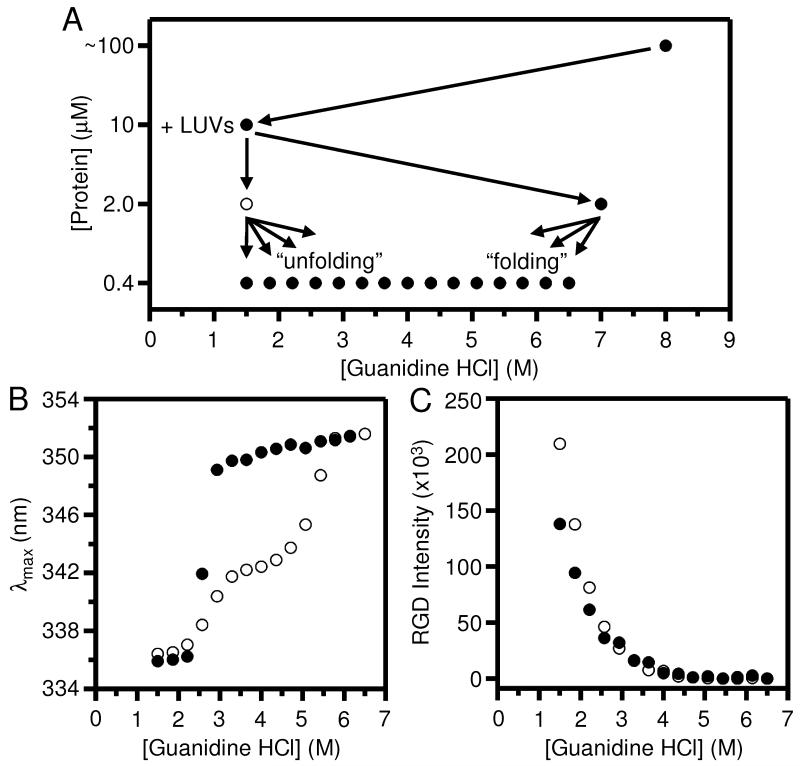
Fig. 1 shows one of our first attempts at verifying the reversibility of OmpLA folding into LUVs of DLPC in which the issue of hysteresis became quite apparent. Fig. 1A shows the dilution scheme we used to prepare the titrations. We initially dissolved a concentrated stock of protein in 8 M guanidine HCl. We then diluted a portion of that stock into folding conditions: 1.5 M guanidine HCl with the LUVs. We previously showed that a guanidine HCl concentration of 1.5 M is low enough to promote native-like folding OmpLA that is enzymatically active [11].
Fig. 1.
Aggregates of OmpLA that form in the presence of lipid vesicles contribute to the protein’s intrinsic fluorescence spectra during chemical denaturation titrations. The excitation wavelength for all protein fluorescence samples was 295 nm. (A) General dilution scheme for titrations of OmpLA with guanidine HCl. There were three dilution steps of the protein. The final dilution step produced the “folding” and “unfolding” titrations. (B) Wavelength position of maximum fluorescence intensity (λmax) for samples of OmpLA in folding (●) and unfolding (○) titrations with LUVs of DLPC at 37° C and at pH 8.0 after 40 hours. The titrations were prepared according to the scheme in (A). (C) Rayleigh-Gans-Debye (RGD) light scattering at 295 nm for the same folding and unfolding titrations shown in (B).
The protein concentration at this stage was 10.0 μM and the lipid concentration was set to where the molar ratio of lipid-to-protein was 2000-1. We preserved that same lipid-to-protein ratio in all other experiments presented here. This folding sample was incubated for five hours at 37° C and at pH 8.0. The phase transition temperature of DLPC lipids is −1° C, and all folding experiments are carried out with lipids in the liquid-disordered phase. We then split the folding sample and diluted one portion five-fold to a guanidine HCl concentration of 7.0 M in order to unfold the protein. We diluted a second portion five-fold and kept the guanidine HCl at 1.5 M in order to keep the protein folded. These unfolded and folded samples had a protein concentration of 2.0 μM. We incubated the 2.0 μM samples for another five hours at 37° C.
Finally, we prepared titrations from the unfolded and folded samples into varying final amounts of guanidine HCl by another five-fold dilution. This final dilution of the unfolded sample produced a “folding” titration. The final dilution of the folded sample produced an “unfolding” titration. The final protein concentration in the titration samples was 0.4 μM and we incubated them for at least 40 hours at 37° C before we took fluorescence measurements. We kept the samples slowly rotating during that long incubation to avoid the guanidine salt creeping out of solution.
Fig. 1B shows that the folding titration produced a single sigmoidal transition in OmpLA’s λmax from the unfolded 352 nm form to the folded 336 nm form. In contrast, the unfolded titration did not produce the same transition. Instead, there was hysteresis between the two transition curves. This hysteresis indicates that OmpLA’s folding and unfolding reactions were not reversible at all guanidine HCl concentrations on our 40 hour timescale using our experimental conditions shown in Fig. 1A.
What new experimental conditions could close the hysteresis loop? To answer that question, we sought to address two curious results shown in Fig. 1B. The first curious result was that the unfolding transition had two-steps. The first unfolding step appeared to happen around the same guanidine HCl concentrations as did the entire folding transition, but the second unfolding step occurred at a much higher range of guanidine HCl concentrations. The second curious result in Fig. 1B was that the λmax values of the folded samples in the unfolding titration (i.e., those samples below about 2.25 M guanidine HCl) were slightly higher than the λmax values for the folded samples in the folding titration at the same guanidine HCl concentrations.
We speculated that both these curious results could be caused by the same problem: aggregation of OmpLA that took some of its population off its folding pathway thereby preventing or delaying its return to a folding-competent state. Aggregation could be lessened at higher concentrations of guanidine HCl, therefore it could have been more apparent in the unfolding titration, as opposed to the folding titration, because the sample we used to prepare the unfolding titration was at a lower guanidine HCl concentration than was the sample we used to prepare the folding titration (Fig. 1A). The observation that the λmax values of the unfolding titration samples in Fig. 1B were higher than the λmax of folded protein is consistent with the conclusion that those samples actually contained a mixture of folded protein and aggregated protein since a mixture of two conformations of a protein in the same sample will produce an observed fluorescence spectrum that is a weighted average of the spectra from two forms [17, 19, 21].
To determine a λmax value of aggregated OmpLA at low guanidine HCl concentrations, we prepared an “aggregation” titration that was identical to the scheme shown in Fig. 1A except there were no LUVs added at any step (Fig. S1A). The aggregation titration revealed that the slightly higher λmax values from the low guanidine samples of the unfolding titration (Fig. 1B) could be due to a mixture of aggregated form of OmpLA (with a higher λmax) and the folded forms of OmpLA (with a lower λmax).
Rayleigh Gans Debye (RGD) light scattering data [19] was also consistent with our conclusion that aggregated OmpLA was present in the unfolding samples at low guanidine levels. Fig. 1C shows RGD scattering data from the same unfolding and folding titrations shown in Fig. 1B. RGD scattering would have come primarily from the LUVs [19], but any particle of a size on the order of our excitation light (295 nm) would also have contributed to the total scattering. Therefore, aggregated particles of OmpLA present in the unfolding titration would have boosted the observed RGD scattering compared to the scattering from samples in the folding titration. We indeed observed such a boost in Fig. 1C at low concentrations of guanidine HCl for the open circles versus closed circles. At high concentrations of guanidine HCl, the difference in scattering between the two titrations was reduced to zero. That result is consistent with the guanidine untangling the aggregates into smaller particles or even monomers that would not have contributed to RGD scattering.
Fig. S1B shows RGD scattering coming from the aggregated particles in the aggregation titration shown in Fig. S1A. Even in the absence of lipid, there were large particles in the samples below 2 M guanidine HCl. The aggregated form of OmpLA shown in Figs. S1A and S1B appears to have undergone a transition at about the same range of guanidine HCl concentrations as where the folding transition was observed (Fig. 1B, filled circles). Aggregation and folding could both reasonably occur over the same range of guanidine HCl concentrations if the protein undergoes a hydrophobic collapse when diluted into that range.
Aggregation of OMPs in the presence of liposomes had not been well studied. One exception is that Huysmans et al. also suspected that aggregation of PagP occurred at low denaturant concentrations even in the presence of LUVs [10]. They addressed the problem by keeping their experiments above a threshold level of denaturant. That strategy would not work for OmpLA because the necessary threshold of guanidine HCl to prevent its aggregation according to Fig. 1B-C and Fig. S1 would be about 3 M, which is also above the level that promotes efficient folding. Therefore, keeping the guanidine levels high would eliminate our baseline region in the folding arm of the transition curves in Fig. 1B, which would compromise extraction of thermodynamic parameters because a well-defined baseline is necessary to invoke the linear extrapolation model to analyze the data [14].
We cannot simply ignore the aggregation because it could be a source of the hysteresis in Fig. 1B. Moreover, we know that the presence of the aggregate would distort the tryptophan emission information from denaturant titrations. Therefore, we sought to prevent aggregation of OmpLA from happening in our titrations. The obvious parameter to vary is protein concentration, however, we were already diluting OmpLA to 0.4 μM, which was at the lower limit of what produced a good signal to noise ratio with our fluorometer set-up.
Therefore, we sought a different method to reduce aggregation and yet keep the same general dilution scheme for OmpLA shown in Fig. 1A. We chose to include a detergent to act as a “holdase” to keep OmpLA soluble during its initial dilution to 10.0 μM at low concentrations of guanidine HCl. We chose the detergent SB3-14 because its low CMC (see below) coupled with its low aggregation number (83) allowed us to use it at low concentrations and still have more detergent micelles than monomers of OmpLA in the initial dilution step. A second criterion for selecting this particular detergent is that it does not support folding of OmpLA (data not shown).
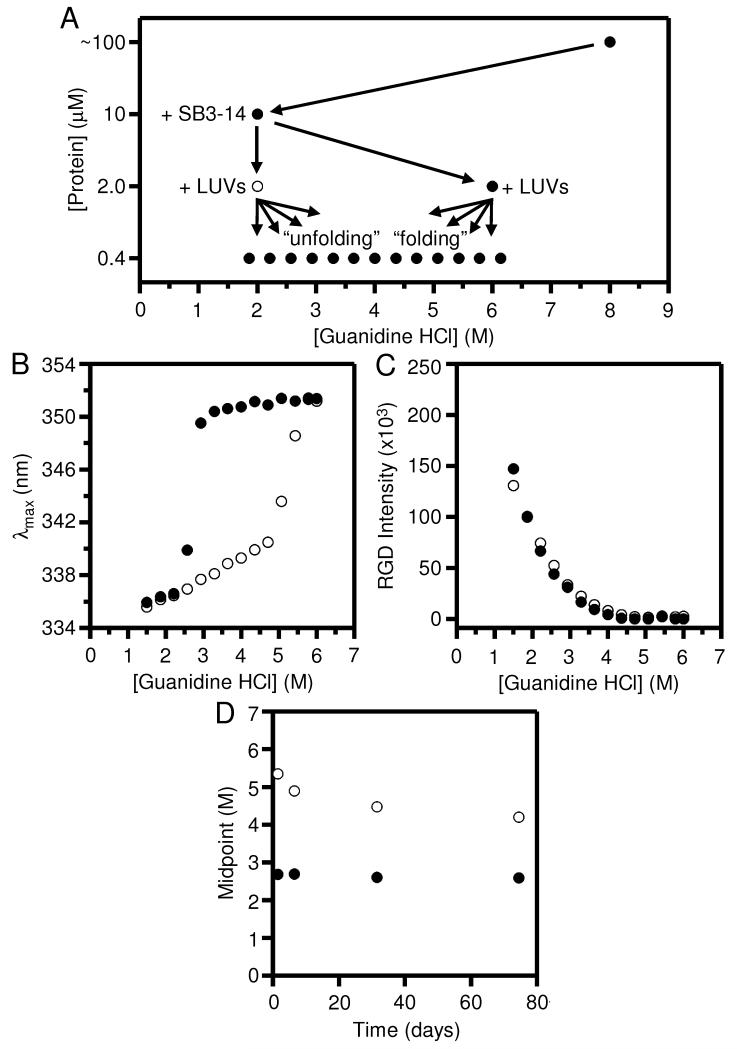
Fig. 2A summarizes our revised dilution scheme using SB3-14. We added the SB3-14 during the initial dilution of OmpLA. We set the concentration of SB3-14 just above its CMC, so there would be micelles to solubilize the protein at this stage. However, we are cognizant of the possibility that the detergent at that concentration could have disrupted the structure of the LUVs. Therefore, we did not add the LUVs in the first dilution. Instead, we diluted the SB3-14-solubilized protein five-fold into the LUVs at two different guanidine HCl concentrations. These five-fold dilutions took the SB3-14 below its CMC and left the LUVs intact (see Supporting Information and Fig. S7). There was also a further five-fold dilution prior to our fluorescence measurements (Fig. 2A).
Fig. 2.
Folding and unfolding of OmpLA remain irreversible at pH 8.0, even when SB3-14 is included as a holdase to prevent OmpLA from aggregating. The excitation wavelength for all protein fluorescence samples was 295 nm. (A) General dilution scheme for the titrations that included the SB3-14. (B) Wavelength position of maximum fluorescence intensity (λmax) for samples of OmpLA in folding (●) and unfolding (○) titrations with LUVs of DLPC at 37° C and at pH 8.0 after 40 hours. The titrations were prepared according to the scheme in (A). (C) Rayleigh-Gans-Debye (RGD) light scattering at 295 nm for the same folding and unfolding titrations shown in (B). (D) Concentration of guanidine HCl where the λmax was at its midpoint of the transition between folded and unfolded protein for the same titrations in (B) measured at various time points.
Another consideration with these experiments was that detergent CMCs are sensitive to denaturant concentration. In order to choose an appropriate concentration of SB3-14 for our dilutions, we measured its CMC at various concentrations of guanidine HCl using a Coomassie dye binding assay [29]. The peak absorbance of Coomassie shifts upon its association with detergent micelles. Fig. S2 shows results from our Coomassie measurements.
Another modification we incorporated was to increase the level of guanidine HCl to 2 M in the protein dilution steps to 10.0 μM and 2.0 μM protein to 2 M. This guanidine HCl concentration was chosen to be near the highest concentration that was still a folding condition according to Fig. 1B. By using the data shown in Fig. S2, we chose to use 1.5 mM SB3-14 so that it would be above its CMC at 2 M guanidine HCl in the initial dilution step (Fig. 2A) but would allow the five-fold dilutions into LUVs to reduce the SB3-14 below its CMC in a range of useful guanidine HCl concentrations.
Fig. 2B shows the results of folding and unfolding titrations prepared according to the scheme in Fig. 2A. A comparison of Fig. 2B to Fig. 1B shows that the SB3-14 effected a striking change to the unfolding titration without affecting the folding titration. In contrast to the unfolding titration in Fig. 1B, the unfolding data in Fig. 2B shows a single sigmoidal transition, which is centered around the same guanidine HCl concentration as the second transition in Fig. 1B (i.e., ~5.5 M), and the first transition in Fig. 1B was no longer observed. The loss of that first transition in Fig. 2B confirmed our suspicion that the first unfolding step in Fig. 1B may not have involved a conformational change to folded protein at all. Rather, we think this first transition may instead have been an unfolding of aggregated protein that formed during the initial folding stage using our original folding scheme. We therefore propose that we can explain our original two-step unfolding titration in Fig. 1B as a reflection of two processes: above 3 M guanidine HCl, when the aggregate was fully melted and presumably solubilized by the guanidine HCl and water, there was left a mixture of two populations: (1) that newly unfolded protein (that was formed by aggregate melting); and (2) folded protein in lipids that had remained resistant to denaturation at those levels of guanidine. The latter population of folded protein then unfolded during the second transition shown in Fig. 1B at guanidine levels above 5 M. With this interpretation, the single unfolding transition in Fig. 2B is consistent with the idea that the usage of SB3-14 and the slightly higher guanidine HCl concentration during the initial folding reaction suppressed the formation of aggregate populations in the folded samples. With our experimental modifications, the entire population of OmpLA would have been folded and could have again remained folded until the levels of guanidine HCl were above 5 M at which concentrations the unfolded conformation would be favored.
Corroborating these conclusions are the RGD light scattering data in Fig. 2C, which shows that there was essentially no difference in scattering between the unfolding and folding titrations in Fig. 2B, which is in contrast to the RGD light scattering data in Fig. 1C. No large particles of aggregate are apparent in the unfolding titration samples using the modified protocol. Therefore, the pre-solvation by using SB3-14 as a holdase seemed to eliminate the bulk of the aggregation challenge, although we were later forced to re-visit this problem (see below).
Even with the aggregation problem mostly solved, Fig. 2B clearly shows that the hysteresis problem was still present, i.e., the folding and unfolding transitions did not overlay. The hysteresis indicates that there is a large activation barrier to either folding or unfolding or both. Waiting for longer periods of time can sometimes close hysteresis loops. However, for lower guanidine concentrations, longer times did not solve our hysteresis problem. Fig. 2D shows the same titration samples from Fig. 2B monitored multiple times over several weeks. The plot depicts the midpoint guanidine HCl concentrations for the sigmoidal transitions in the folding and unfolding titrations at different time points, and we observed that the folding transition held steady over time. The unfolding transition did move towards the folding transition over time (i.e., the degree of hysteresis went down). However, Fig. 2D suggests the timescale to eliminate hysteresis in those experimental conditions could be very slow, and we wanted a reasonably fast solution to the hysteresis problem.
The application of heat is another strategy to lower activation barriers. Since heat is also a denaturant, we reasoned that the kinetics of unfolding should be accelerated at higher temperatures. However, when we increased the temperature of our reaction above 45° C, we observed the protein precipitating out of solution and forming visible white clumps. Therefore, searching for reversibility at higher temperatures was not practicable.
We also tried modifying the morphology of the liposomes. The protein OmpA has previously been shown to display a large degree of hysteresis with LUVs [15] and yet Hong and Tamm had success in finding less hysteresis for OmpA in SUVs of various lipid compositions [8]. In contrast, when we tested OmpLA in SUVs (Fig. S3A), we still observed hysteresis. McKibbin et al. had success monitoring folding and unfolding of opsin with bicelles of lipid and detergent [30]. We tested OmpLA in bicelles (Fig. S3B), but we again still observed hysteresis. We then tested the effects of the lipid headgroup in LUVs by substituting a fraction of the DLPC with anionic DLPG (Fig. S4A), anionic DLPS (Fig. S4B), or cationic EPC (Fig. S4B), but we also still observed hysteresis. Likewise, the addition of mono and divalent salts did not reduce hysteresis (Fig. S5).
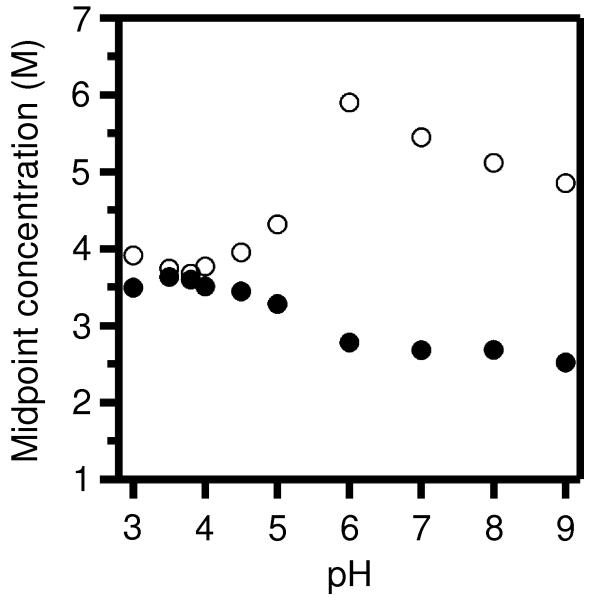
We next tested the effects of solution pH on the hysteresis, and this investigation provided much progress. Fig. 3 shows how the folding and unfolding transitions were affected by pH. The midpoint guanidine HCl concentrations of each transition at each of several pH values are shown with the same symbols as in Figs. 1 and 2. The midpoints of each of the folding and unfolding reactions aligned reasonably well at around pH 3.8 (Fig. S6A), suggesting the degree of hysteresis would be very low at that pH. At higher pH levels, however, there was still significant separation between the folding and unfolding transitions.
Fig. 3.
Folding and unfolding of OmpLA remain irreversible at a broad range of pH, however, hysteresis is lessened at acidic pH, especially at pH 3.8. The guanidine HCl concentrations of the midpoints of folding (●) and unfolding (○) titrations are plotted for titrations at various pH levels that were otheriwse prepared according to the scheme shown in Fig. 2A with incubation at 37° C for 40 hours. The excitation wavelength for all fluorescence samples was 295 nm.
Both the folding and unfolding transitions were sensitive to pH, and both transitions shifted towards each other when the pH was lowered from 6.0 to 3.8: there was a decrease in the unfolding midpoint of about 2.5 M guanidine HCl while there was an increase in the folding midpoint of about 1.0 M guanidine HCl. The unfolding process appeared to be the more sensitive reaction to pH. It is reasonable that OmpLA’s folding and unfolding reactions proceed along different coordinates, which would allow their different sensitivities to pH. The rates of the progress of OmpLA along those two reaction coordinates could be affected differently by pH depending on the local environments through which the ionizable residues of the solvent-exposed loops pass during each process.
Outer membrane proteins have an intrinsic ability to fold, which could be evolutionarily refined and part of their in vivo folding processes [8, 10, 22-27]. We speculate that the folding coordinate may involve some protection of the ionizable loop residues as they pass through the hydrocarbon core of the membrane. The protection would dampen the effects of the ionization state of those residues during folding. On the other hand, the unfolding coordinate, due to high concentrations of guanidine HCl, would have no biologically relevant analog and may occur with the ionizable residues being exposed to the acyl-chains of the lipids as the protein exits the membrane and becomes solvated. Absent such protection, the ionization state of the loop residues could be very important to the unfolding coordinate of OmpLA.
At higher pH levels, certain glutamic and aspartic acid residues would carry charge and increase the energetic barrier for the unfolding reaction coordinate, thus slowing down unfolding such that its progress at low guanidine HCl concentrations was not the same as its progress along the folding coordinate (Fig. 3). At low enough pH (e.g., 3.8), enough of those glutamic and aspartic acid residues were be neutralized such that the protein’s exit from the bilayer would have cost less energetically, thus lowering the activation barrier to exiting the membrane and allowing the protein to proceed along the unfolding coordinate fast enough to equilibrate with the folding reaction, thus closing the hysteresis loop.
Discovery of reversible folding conditions for OmpLA
At pH 3.8, the degree of hysteresis was as small as in any experiment we had yet performed (Fig. S6A). Therefore, we chose pH 3.8 to be the new default pH level for all remaining experiments. However, Fig. S6A shows that there was still some residual hysteresis present and even a small degree of hysteresis could critically alter our analysis of the transitions for energetics measurements. Therefore, we decided to explore further tweaks to our experimental protocol to completely close the hysteresis loop. We suspected that the remaining hysteresis in Fig. S6A was again due to the recurring problem of aggregation.
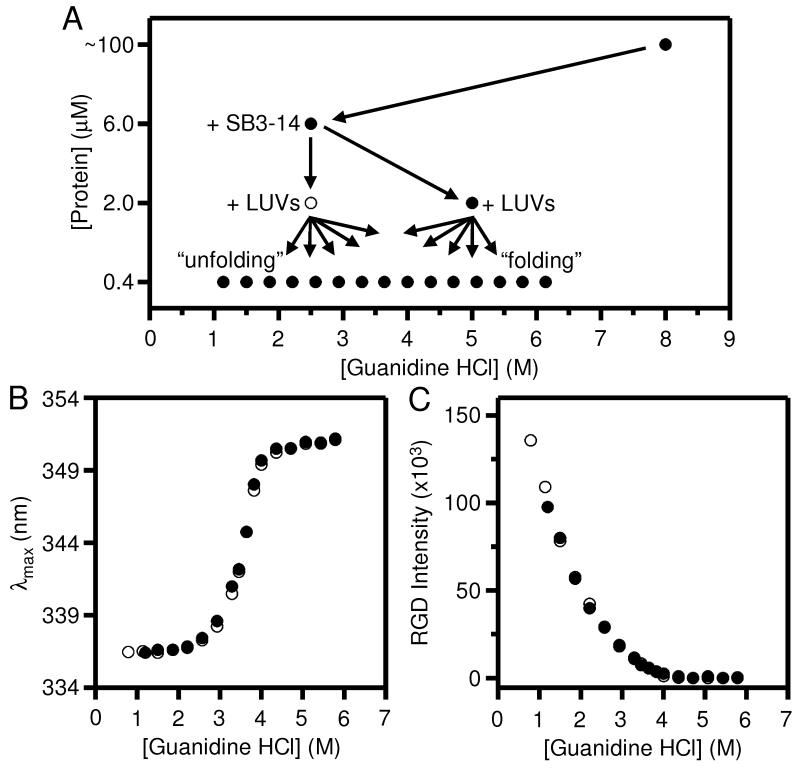
In order to be more diligent about preventing aggregation in the unfolding titration at pH 3.8, we again updated our dilution scheme to the one shown in Fig. 4A. We also made changes to the mechanics of our experiments by (a) emphasizing fast and thorough mixing of the samples when we prepared dilutions, and (b) slowing down the dilution of the 6.0 μM protein sample into the LUVs. More details about these new mechanics are in the Supporting Information.
Fig. 4.
At pH 3.8, all remaining hysteresis between the folding and unfolding titrations of OmpLA can be eliminated by avoiding the residual tendency of the protein to aggregate. The excitation wavelength for all fluorescence samples was 295 nm. (A) Updated dilution scheme for titrations of OmpLA that included changes in the protein and guanidine HCl concentrations in the first two dilution steps as compared to Fig. 2A. (B) Wavelength position of maximum fluorescence intensity (λmax) for samples of OmpLA in folding (●) and unfolding (○) titrations with LUVs of DLPC at 37° C and at pH 3.8 after 40 hours. The titrations were prepared according to the scheme in (A). (C) RGD light scattering at 295 nm for the same titrations shown in (B).
Figs. 4B and 4C show the combined results of all our updates in mixing and mechanics at pH 3.8. Fig. 4B shows that there was no remaining hysteresis between samples in the folding and unfolding titrations, which indicates that there was complete reversibility of OmpLA’s folding at all concentrations of guanidine HCl in the titrations. Fig. 4C shows that there was also complete reversibility in the degree of RGD light scattering between the two titrations. This equivalency in light scattering suggested that we finally eliminated aggregation from the unfolding samples at low guanidine HCl concentrations. It also suggested that any changes to the LUVs themselves by their exposure to the guanidine HCl solutions were also reversible. We describe the implications of LUV reversibility below.
LUVs of DLPC were reversible and remained intact throughout our titrations
Following confirmation of OmpLA’s reversible folding with LUVs, we still faced two important questions regarding the lipid bilayers in our experiments: (1) were the effects of denaturant on the LUVs also reversible? (2) did the LUVs remain intact with the addition of the denaturant and the low levels of SB3-14?
As we noted above, Fig. 4C shows that the RGD light scattering coming from the LUVs was the same for the folding and unfolding titrations at all guanidine HCl concentrations. This result implied that any change to the size of the LUVs due to the guanidine HCl was reversible when the guanidine was diluted away. Therefore, the LUVs did not dissolve into smaller structures or merge together into larger structures in the high levels of guanidine HCl because those new structures could not spontaneously reform vesicles of the original starting size. We also used guest lipids having NBD-labels to confirm the reversibility of the structure of the membrane-water interface in response to guanidine HCl by monitoring their fluorescence λmax values (Fig. S7A) and red edge excitation shifts (Fig. S7B). Further, we used those NBD-labeled lipids to verify that the low detergent concentration used in our experiments with OmpLA did not significantly compromise the structure of the lipid bilayers or the structure of the LUVs (Figs. S7C-F).
Conclusions
We have discovered experimental conditions under which OmpLA can interconvert reversibly and to equilibrium between its folded, intermediate, and unfolded states in the presence of thermodynamically stable lipid bilayers. This path independent interconversion between protein conformations was a prerequisite for measuring thermodynamic parameters for OmpLA, including its equilibrium free energy of unfolding in the absence of denaturant (). We found the reversible conditions only by first preventing OmpLA from aggregating in our samples, notably by including a detergent “holdase” to solubilize the protein before its addition to LUVs. Upon dilution, the detergent goes below its CMC and the protein folds into lipid bilayers. We propose that this use of low concentrations may be general for other OMPs as some will surely be more prone than OmpLA to aggregating in the presence of LUVs.
Other than the suppression of aggregation, the most important experimental condition that we used to help promote reversibility of OmpLA was acidic pH. We propose that any group having trouble verifying reversible folding for another OMP with LUVs also try acidic pH. However, we speculate that the particular pH that we found effective for OmpLA (3.8) may not be a “magic bullet” condition for all OMPs because of differences in the number and location of ionizable residues in their structures. Rather, each OMP may require its own round of method development along the roadmap that we provided here.
Materials and Methods
Protein preparation and purification
We expressed OmpLA to inclusion bodies, and purified it as previously described [11]. We stored inclusion body pellets at −20° C for up to one year. We always began every new titration experiment with a fresh pellet straight from −20° C storage. We prepared concentrated unfolded protein stocks in 8 M guanidine HCl (UltraPure powder, Invitrogen) in an appropriate buffer for our selected pH. Protein was never frozen once it was dissolved in guanidine HCl or buffer.
All buffers were the best available molecular biology grades from Sigma and were prepared at 100 mM and were titrated at their final concentration with either concentrated HCl or NaOH at the beginning of each titration. For pH 9.0 and above, the buffer was glycine. For pH 7.0 and pH 8.0, the buffer was glycine-glycine. This buffer was selected to be compatible with urea denaturation in case we eventually wanted to use urea. Urea forms ions that can modify several functional groups in proteins, especially at neutral and high pH [14, 31], and glycine-glycine can scavenge those ions [31]. For pH 6.0 and below, the buffer was citrate.
It is notable that at our final chosen pH of 3.8, spontaneous lipid hydrolysis would be faster than at neutral pH. However, the rate of hydrolysis would still be slow enough at the temperature of our experiments (37° C) as to be negligible on the timescale of our experiments (40 hours) [32, 33]. Indeed, if we stored our LUV suspensions in buffer at 37° C, we did not notice any change in their appearance or turbidity until more than 40 days had passed.
For all titrations, we included 2 mM EDTA in the buffers and guanidine HCl stocks to ensure that OmpLA was monomeric because the solutions had no calcium available to mediate its dimerization and thus subsequent enzymatic activity [23]. When used, alternative denaturants (guanidine acetate, guanidine carbonate, lithium perchlorate, methyl urea, dimethyl urea, butyl urea, and thiourea) were from Sigma.
Lipid and detergent preparation
We handled lipids (Avanti Polar Lipids) and prepared 100 nm LUVs as previously described [11]. When guest lipids were used, we mixed an appropriate volume of the host and guest lipids in chloroform together by vortexing and warming slightly. The lipid mixtures were then dried and prepared into LUVs according to the same procedure as for pure DLPC. We took care to flush dried lipid films with nitrogen prior to storage at −20° C because DLPC could be hygroscopic. Once hydrated, lipids were always used fresh the same day. We never stored LUVs for later use because of the concern of lipid hydrolysis. The molar lipid-to-protein ratio was always 2000-1 in all our experiments.
The SB3-14 detergent was from Sigma. We prepared it as an 8 mM stock in water from which we diluted appropriate portions into our samples.
Dilution schemes
Our dilutions and titrations of protein and lipids were prepared according to the various schemes we show in Figs. 1A, 2A, and 4A. Our initial concentrated protein stocks in 8 M guanidine HCl were usually around 80-100 μM after purification [11]. For low resolution titrations in our reversibility screen, we prepared 3.2 mL of the initial dilution step to either 6.0 or 10.0 μM protein. For the high resolution folding titrations, we prepared 6 mL of that initial step. For the next dilution, we prepared 4 mL of each of the folded and unfolded protein samples for low resolution titrations. For the high resolution titrations, we prepared 14.4 mL of that second dilution to 2.0 μM protein. The final folding and unfolding titration samples in all cases were at a final volume of 1100 μL, which was just enough volume to place the meniscus above the excitation light beam in our fluorometer.
All titration and dilution steps were incubated in a SciGene Model 400 hybridization incubator at a gentle speed of around three revolutions per minute. To prevent leakage, for large sample sizes, we used Wheaton borosilicate glass vials with PTFE-lined caps and for the 1100 μL titration samples, we used microcentrifuge tubes with rubber seals.
Fluorescence emission and light scattering
We used the tryptophan fluorescence emission protocols that we described previously in detail [19]. Briefly, our spectrofluorometer was an ISS PC1 photon-counting steady-state instrument. Except where noted, the excitation wavelength was 295 nm. The pathlength was 1 cm and we used 2.4 mm excitation slits and 2.0 mm emission slits with no other grating or filters. To reduce light scattering from the LUVs [17, 19], we used an excitation polarizer at 90° and an emission polarizer at 0°. That emission polarizer setting also eliminates the Wood’s anomaly that is an artifact of many monochromators [17].
For emission scans, we averaged at least three sets of successively recorded spectra at 1 nm resolution from 15 nm below the excitation wavelength to 400 nm, with 0.3 s of sample averaging for each reading. After transferring samples from the microcentrifuge tubes used for the 40 hour incubation into a cuvette for use in the fluorometer, we then equilibrated each sample for at least 3 minutes to the specified experimental temperature inside the instrument’s sample chamber prior to taking any measurements. Shorter equilibration times produced noisy fluorescence data.
Emission spectra from blank samples containing only relevant buffers and guanidine HCl were subtracted. We did not include LUVs in the blanks so that we could monitor the RGD light scattering [19]. We analyzed emission spectra to measure the λmax values and RGD scattering intensities by fitting the spectra to a sum of a normal distribution and a log-normal distribution as previously described [19].
Supplementary Material
Acknowledgements
This work was supported by grants from the National Science Foundation (MCB0423807, MCB0919868) and the National Institutes of Health (R01 GM079440, T32 GM008403).
Abbreviations
- 14:0 NBD PE
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl)
- 14:0-12:0 NBD PC
1-Myristoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]lauroyl]-sn-Glycero-3-Phosphocholine
- CMC
critical micelle concentration
- DLPC
1,2-dilauroyl-sn-glycero-3-phosphocholine
- DLPG
1,2-dilauroyl-sn-glycero-3-phosphoglycerol
- DLPS
1,2-dilauroyl-sn-glycero-3-phospho-L-serine
- EDTA
ethylenediaminetetraacetic acid
- EPC
ethylphosphocholine
- LUVs
large unilamellar vesicles
- OMPs
outer membrane proteins
- OmpA
outer membrane protein A
- OmpLA
outer Membrane Phospholipase A
- PagP
PhoPQ-activated gene P
- REES
red edge excitation shift
- RGD
Rayleigh-Gans-Debye
- SB3-14
3-N,N-Dimethylmyristyl-ammonio)propanesulfonate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Li C, Ramjeesingh M, Reyes E, Jensen T, Chang X, Rommens J, Bear C. The cystic fibrosis mutation (delta 508) does not influence the chloride channel activity of CFTR. Nat Genet. 1993;3:311–316. doi: 10.1038/ng0493-311. [DOI] [PubMed] [Google Scholar]
- [2].Riordan J, Rommens J, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- [3].Kerem B, Rommens J, Buchanan J, Markiewic D, Cox T, Chakravarti A, Buchwald M, Tsui L. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- [4].Clain J, Fritsch J, Lehmann-Che J, Bali M, Arous N, Goossens M, Edelman A, Fanen P. Two mild cystic fibrosis-associated mutations result in severe cystic fibrosis when combined in cis and reveal a residue important for cystic fibrosis transmembrane conductance regulator processing and function. J Biol Chem. 2001;276:9045–9049. doi: 10.1074/jbc.M008979200. [DOI] [PubMed] [Google Scholar]
- [5].Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- [6].Chen GQ, Gouaux E. Probing the folding and unfolding of wild-type and mutant forms of bacteriorhodopsin in micellar solutions: evaluation of reversible unfolding conditions. Biochemistry. 1999;38:15380–15387. doi: 10.1021/bi9909039. [DOI] [PubMed] [Google Scholar]
- [7].Faham S, Yang D, Bare E, Yohannan S, Whitelegge JP, Bowie JU. Side-chain contributions to membrane protein structure and stability. J Mol Biol. 2004;335:297–305. doi: 10.1016/j.jmb.2003.10.041. [DOI] [PubMed] [Google Scholar]
- [8].Hong H, Tamm LK. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc Natl Acad Sci U S A. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stanley AM, Fleming KG. The process of folding proteins into membranes: Progress and challenges. Archives of Biochemistry & Biophysics. 2008;469:46–66. doi: 10.1016/j.abb.2007.09.024. [DOI] [PubMed] [Google Scholar]
- [10].Huysmans GH, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. P Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ebie Tan A, Burgess NK, DeAndrade DS, Marold JD, Fleming KG. Self-association of unfolded outer membrane proteins. Macromol Biosci. 2010;10:763–767. doi: 10.1002/mabi.200900479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Santoro MM, Bolen DW. A test of the linear extrapolation of unfolding free energy changes over an extended denaturant concentration range. Biochemistry. 1992;31:4901–4907. doi: 10.1021/bi00135a022. [DOI] [PubMed] [Google Scholar]
- [14].Street TO, Courtemanche N, Barrick D. Protein folding and stability using denaturants. Methods Cell Biol. 2008;84:295–325. doi: 10.1016/S0091-679X(07)84011-8. [DOI] [PubMed] [Google Scholar]
- [15].Pocanschi CL, Patel GJ, Marsh D, Kleinschmidt JH. Curvature elasticity and refolding of OmpA in large unilamellar vesicles. Biophys J. 2006;91:L75–77. doi: 10.1529/biophysj.106.091439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hong H, Szabo G, Tamm LK. Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat Chem Biol. 2006;2:627–635. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- [17].Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Analytical biochemistry. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- [18].Sanchez KM, Gable JE, Schlamadinger DE, Kim JE. Effects of tryptophan microenvironment, soluble domain, and vesicle size on the thermodynamics of membrane protein folding: lessons from the transmembrane protein OmpA. Biochemistry. 2008;47:12844–12852. doi: 10.1021/bi800860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Moon CP, Fleming KG. Using tryptophan fluorescence to measure the stability of membrane proteins folded in liposomes. Methods Enzymol. 2011;492:189–211. doi: 10.1016/B978-0-12-381268-1.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- [21].Burstein EA, Vedenkina NS, Ivkova MN. Fluorescence and the location of tryptophan residues in protein molecules. Photochem Photobiol. 1973;18:263–279. doi: 10.1111/j.1751-1097.1973.tb06422.x. [DOI] [PubMed] [Google Scholar]
- [22].Dekker N, Tommassen J, Lustig A, Rosenbusch JP, Verheij HM. Dimerization regulates the enzymatic activity of Escherichia coli outer membrane phospholipase A. J Biol Chem. 1997;272:3179–3184. doi: 10.1074/jbc.272.6.3179. [DOI] [PubMed] [Google Scholar]
- [23].Stanley AM, Chauwang P, Hendrickson TL, Fleming KG. Energetics of Outer Membrane Phospholipase A (OMPLA) Dimerization. J Mol Biol. 2006;358:120–131. doi: 10.1016/j.jmb.2006.01.033. [DOI] [PubMed] [Google Scholar]
- [24].Stanley AM, Fleming KG. The role of a hydrogen bonding network in the transmembrane beta-barrel OMPLA. J Mol Biol. 2007;370:912–924. doi: 10.1016/j.jmb.2007.05.009. [DOI] [PubMed] [Google Scholar]
- [25].Stanley AM, Treubodt AM, Chauwang P, Hendrickson TL, Fleming KG. Lipid chain selectivity by outer membrane phospholipase A. J Mol Biol. 2007;366:461–468. doi: 10.1016/j.jmb.2006.10.055. [DOI] [PubMed] [Google Scholar]
- [26].Ubarretxena-Belandia I, Boots JW, Verheij HM, Dekker N. Role of the cofactor calcium in the activation of outer membrane phospholipase A. Biochemistry. 1998;37:16011–16018. doi: 10.1021/bi9814181. [DOI] [PubMed] [Google Scholar]
- [27].Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fleming PJ, Freites JA, Moon CP, Tobias DJ, Fleming KG. Outer membrane phospholipase A in phospholipid bilayers: A model system for concerted computational and experimental investigations of amino acid side chain partitioning into lipid bilayers. Biochim Biophys Acta, Under Review. 2011 doi: 10.1016/j.bbamem.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kleinschmidt JH, Wiener MC, Tamm LK. Outer membrane protein A of E. coli folds into detergent micelles, but not in the presence of monomeric detergent. Protein Sci. 1999;8:2065–2071. doi: 10.1110/ps.8.10.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McKibbin C, Farmer NA, Jeans C, Reeves PJ, Khorana HG, Wallace BA, Edwards PC, Villa C, Booth PJ. Opsin stability and folding: modulation by phospholipid bicelles. Journal of molecular biology. 2007;374:1319–1332. doi: 10.1016/j.jmb.2007.10.018. [DOI] [PubMed] [Google Scholar]
- [31].Lin MF, Williams C, Murray MV, Conn G, Ropp PA. Ion chromatographic quantification of cyanate in urea solutions: estimation of the efficiency of cyanate scavengers for use in recombinant protein manufacturing. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2004;803:353–362. doi: 10.1016/j.jchromb.2004.01.017. [DOI] [PubMed] [Google Scholar]
- [32].Grit M, Crommelin DJ. Chemical stability of liposomes: implications for their physical stability. Chem Phys Lipids. 1993;64:3–18. doi: 10.1016/0009-3084(93)90053-6. [DOI] [PubMed] [Google Scholar]
- [33].Zuidam NJ, Crommelin DJ. Chemical hydrolysis of phospholipids. J Pharm Sci. 1995;84:1113–1119. doi: 10.1002/jps.2600840915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.