Abstract
Mutant p53 is frequently detected in cancers lose of its ability in tumor suppression and gain of function in promoting tumor progression. Restoration of p53 functions by replacement of wild-type p53 and inhibition of its degradation or increment of its transcriptional activity has been applied in prevention and treatment of cancers. Recent evidence indicates that disrupting ceramide glycosylation can resuscitate wild-type p53 expression and p53-dependent apoptosis in mutant p53 tumors. Acting in posttranscriptional process that can turn on wild-type p53 expression and abrogate mutant p53 presents a tractable new strategy to eradicate mutant p53 cancers.
Keywords: mutant p53, resuscitation, glucosylceramide synthase, ceramide, apoptosis
Introduction
p53 protein encoded by the human gene TP53 is a key tumor suppressor in preventing tumorigenesis and cancer progression. As an essential transcription factor, p53 activates expressions of p21Waf1/Cip1, Bax, Puma, FAS and other p53-responsive genes, consequently promotes cell growth-arrest, apoptosis, DNA repair and cell differentiation. Thus, these cell processes remove damaged or transformed cells from normal tissues (1). The transcriptional activity of p53 on the p53-responsive genes is sequence-specific, and mainly relies on its DNA-binding domain (DBD, residues 102 to 292) encoded by the sequence from exon 4 to exon 8 (1–2). In normal cells, p53 is tightly controlled and kept at low level. A wide variety of signals involved in DNA damage, oncogenic stress, hypoxia and cellular distress activates p53 primarily through post-translational modifications that result in an augmented level of p53 protein and its transactivation activity. Ubiquitin ligase MDM2 that interacts with and recruits p53 to the ubiquitin-proteasome regulates p53 degradation (1, 3).
p53 function is always compromised in most tumors, as a result of somatic TP53 mutations followed by loss of heterozygosity (LOH) during the course of carcinogenesis (1, 4). The frequencies of p53 mutations vary considerably between cancer types, ranging from ~10% in hematopoietic malignancies to 50–70% in ovarian, colorectal and head and neck cancers (1, 4). The majorities of p53 mutants in human cancers abrogate sequence-specific DNA-binding to the promoter element of the p53-responsive genes. Moreover, p53 mutants confer a dominant-negative activity over the remaining wild-type allele by functionally inactive hetero-oligomers of the mutants with the wild-type protein (1, 3). Increasing evidence indicates that many p53 mutants also gain new oncogenic properties that are independent from wild-type p53 (1, 3). p53 mutants that promote tumor progression and resistance to therapies become the most common prognostic indicator for both tumor recurrence and cancer death (1, 4–5). Restoration of p53 function that has been succeeded in regression of tumors represents a critical approach to treat cancers (1, 6). This review highlights resuscitation of wild-type p53 expression by targeting ceramide glycosylation, a novel approach eradicating mutant p53 cancers.
Reactivation of p53 pathway in tumor suppression
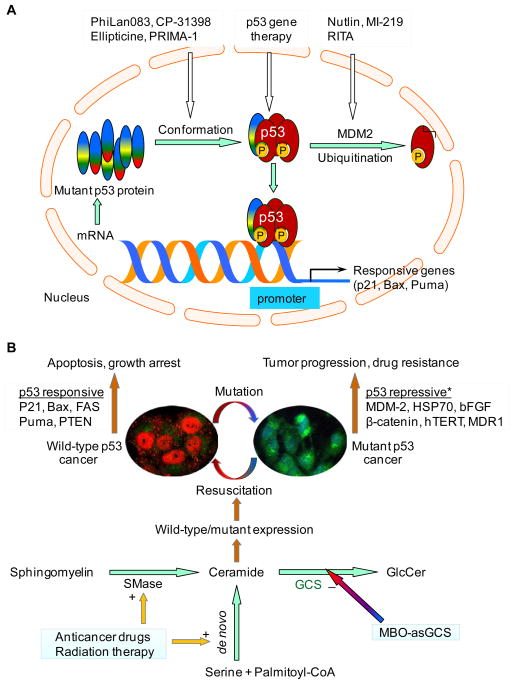
Most malignant tumors that disrupt p53 signaling pathways remain addicted to p53 mutants. Various strategies have been successfully developed to reconstitute p53 functions in order to abrogate tumor progression (1, 3, 6–8). Based on the action sites, these strategies can be briefly listed into three groups: replacement of wild-type p53 by gene therapy, augmenting of wild-type p53 by inhibition of MDM2-mediated degradation and reactivation of mutant p53 by alteration of protein conformation, as sketched in Fig. 1A.
Figure 1.
Reactivation of p53 tumor suppressor. A. Current strategies in reactivation of p53. B. Targeting ceramide glycosylation to resuscitate p53 expression. Ceramide modulates the expression of p53 to resuscitate wild-type p53 (phosphorylated, red fluorescence in the cell nucleus) and p53-dependent apoptosis, thus sensitizes mutant p53 tumors to therapies. Silencing of glucosylceramide synthase (GCS, green fluorescence in Golgi apparatus) with MBO-asGCS disrupts ceramide glycosylation to enhance endogenous ceramide. (+), increasing enzyme activity or synthesis; (−), inhibiting enzyme activity or synthesis. *, these genes are upregulated by mutant p53 in cancer cells.
Restoration of p53 function by introduction of wild-type p53 gene alone has been demonstrated sufficient to cause regression of several different types of tumors in mice (1, 6). The gene therapy for p53 replacement delivered by adenoviral vectors to human tumors has presented very promising in a number of clinical trials (9–10). The p53 gene therapy (Gendicine, Advexin) administered locally has shown at least partial clinical responses as monotherapy, and has increased the effectiveness of radiation therapy and chemotherapy. Expression of p53 transgene has occurred at high levels and is associated with the activation of other genes in the p53 pathway after treatments (9). Although these studies indicate proof-of-principle for p53 replacement, it is also noted that adenoviral p53 gene therapy by intraperitoneal injection could not significantly improve standard chemotherapy in ovarian cancers harboring p53 mutants (10). Several factors including inefficient systemic delivery, non-specific immune responses and p53 mutants in cancers may limit the efficacy and application of p53 gene therapy (1, 10) (Fig. 1A).
A group of small molecules have been applied to target the p53-MDM2 protein interaction in order to enhancing wild-type p53 protein (7, 11–13). Wild-type p53 usually presents at low level, due to its short half-life (15–30 min) that is primarily maintained by ubiquitin-mediated degradation. As a crucial negative regulator, MDM2 works together with MDMX to ubiquitinate p53 for proteasome-mediated degradation. Nutlin and MI-219 that accumulate wild-type p53 protein by binding to MDM2 and blocking p53-MDM2 interaction have shown activities against human xenografts in preclinical models (11–12). RITA (reactivation of p53 and induction of tumor apoptosis) accumulates wild-type p53 by binding to p53 protein and preventing p53 interaction with MDM2 (7, 13). Consequently, these compounds induce the expression of p53-responsive genes and trigger apoptosis in various tumor cells expressing wild-type p53 (11–13). The dominate-negative effects and gain-of-functions of p53 mutants that present in almost half of cancer cases may severely compromise the effectiveness of these compounds that target p53-MDM2 interaction (1, 5, 10) (Fig. 1A).
Restoration of wide-type function to mutant p53 tumors has been attempted extensively in modulation of the protein conformation (3, 8). The binding of wild-type p53 to DNA element primarily depends on its protein structure that is divided into a β-sandwich scaffold, and a DNA-binding surface including a loop-sheet-helix (LSH) motif and two loops (L2 and L3) (3). Compound PhiLan083 binds with high affinity to a crevice created by the Cys220 of p53 mutant and stabilizes the β-sandwich that serves as a basic scaffold for the DNA-binding surface (7–8, 14). On the other hand, another compound P53R3 restores the sequence-specific DNA binding of both His273 and His175 of mutant p53 in DNA-binding surface (15). CP-31398 prevents unfolding of wild-type or mutant p53 and stabilizes p53 through reduced ubiquitination. In these ways, CP-31398 induces the expression of p53-responsive genes, such as p21, but also induces p53-independent cells death (3, 7, 16). Ellipticine enhances the sequence-specific DNA binding and transcription activity of mutant p53, consequently induces mutant p53-dependent cell death (1, 3, 7). The PRIMA-1 (p53 reactivation and induction of massive apoptosis) restores wild-type conformation to mutant p53 protein by covalent binding to and modifying the thiol groups of His175 and His273 in the core domain (8). The more potential PRIMA-1 analogue APR-246 that inhibits human tumor growth and is able to synergize with chemotherapeutic drugs is currently tested in a clinical trial (8, 17). Additionally, synthetic peptides (C369–383, C361–382) corresponding to the C-terminal residues of p53 can alter p53 protein conformation by allosteric mechanism and restore p53 transactivation in cancer cells, since the C-terminal negative-regulatory domain of p53 locks the unphosphorylated p53 tetramer in a inactive state (7). The compounds in this category directly target particular types of p53 mutants and can be developed specific therapeutic agents to cancers harboring these mutants. p53 binds to promoters of the responsive genes in tetramers, the hetero-tetramer of wild-type with mutants or homo-tetramer of mutants may interfere the effects of these compounds on p53 transactivation. Whether these compounds are able to mediate the formation of p53 tetramer requires to be studied in p53 mutant cells (Fig. 1A).
Resuscitation of wild-type p53 expression by disrupting ceramide glycosylation
TP53 mutation usually is heterozygous either in germ line or somatic cells (1, 4). This may provide opportunities modulating the transcription and posttranscriptional process to restore wild-type p53 expression in mutant p53 cancer cells. Little is known whether p53 expression can be restored in p53-mutant tumors (1, 7–8), until lately reporting that suppression of glucosylceramide synthase restores wild-type p53 expression and p53-dependent apoptosis in the mutant cancer cells (18). These intriguing findings shed light on the insights of p53 mutation in transcription and post-transcriptional process and the role of sphingolipid in expression regulation, and provide an novel approach to target cancers harboring p53 mutants.
It is speculated that p53 mutants can be corrected in gene expression level as well as posttranslational modification; and the latter has been broadly demonstrated (1, 3, 8). Heterozygous TP53 that is able to express either wild-type or mutant heterogeneous nuclear RNA (hnRNA) has been detected in most of mutant p53 tumors, even not of all (1, 4–5). More likely, mutant p53 cancer cells generate heterozygous hnRNA from the transcription using both wild-type and mutant DNA, however, these cells produce mutant protein after translation. NCI/ADR-RES and OVCAR-8 cancer cells are mutant p53 cell lines that dominantly express the p53 mutants with a deletion of 7- and 6-amino acids (encoded by exon 5) within the DBD (18–19), respectively. These deleted mutants lack p53 transactivation activity and lead these cells resistance to apoptosis induced by DNA-damage (18–19). Analyses of hnRNA and mRNA, it was found that both NCI/ADR-RES and OVCAR-8 cell lines expressed the same p53 hnRNA as in wild-type cells, even though these p53 mutant cells could not generate wild-type p53 mRNA when they were exposed to doxorubicin (18). Interestingly, wild-type p53 mRNA and the phosphorylated p53 protein (at Ser15) presented in the p53 mutant cells after the treatment of mixed backbone oligonucleotide against human GCS (MBO-asGCS) and doxorubicin exposure. Consequently, the functional p53 activated the expression of p53-responsive genes and induced apoptosis in the p53 mutant cells (18). This study as proof of concept indicates that dysfunctional regulation in transcription and post-transcriptional process is an important cause for p53 mutants in cancer cells (Fig. 1B). Its finding brings many questions, and further investigations should answer which types and locations of p53 mutations are resulted from dysfunctional regulation of transcription/posttranscriptional process, and what are the molecular mechanisms underlying these. Based on these, we would be able to develop therapeutic approaches to correct or resuscitate the wild-type p53 expression in cancers harboring p53 mutants.
Suppression of glucosylceramide synthase (GCS) has resuscitated p53-dependent apoptosis in p53 mutant cells, indicating active sphingolipids play a role in turning the expression of mutant to wild-type p53 (18) (Fig. 1B). MBO-asGCS that silenced GCS expression significantly increased the levels of phosphorylated p53 (pp53, at Ser15 in DBD) and p53-responsive genes including p21Waf1/Cip1, Bax and Puma with dose-dependent manner in p53 mutant cells (18). Restored p53-dependent apoptosis by MBO-asGCS dramatically sensitized mutant p53 cancer to doxorubicin-induced apoptosis in cells and animal studies (18). Resuscitation of p53 expression with GCS suppression was confirmed in NCI/ADR-RES cells treated with D-PDMP (d-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol), a GCS inhibitor. Cellular immunofluorescent staining revealed that MBO-asGCS treatment dramatically decreased GCS protein in Golgi apparatus where ceramide glycosylation occurred, consequently that increased nuclear phosphorylated p53 in NCI/ADR-RES cells exposed to doxorubicin (18) (Fig. 1B). As GCS catalyzes ceramide glycosylation, converting ceramide to glucosylceramide, disrupting ceramide glycosylation by silencing of GCS can increase ceramide and decrease glucosylceramide and other glycosphingolipids (20), as sketched in Fig. 1B. Ceramide is an active sphingolipid and plays critical roles in processing of apoptosis and other cellular functions (21–24). It was found that in NCI/ADR-RES cells, MBO-asGCS treatments significantly increased endogenous ceramide in dose-dependent fashion and that were directly associated with p53 resuscitation. Phosphorylated wild-type p53 were accumulated in nucleus while endogenous ceramide appeared in cytoplasm of NCI/ADR-RES cells after disruption of ceramide glycosylation. Fumonisin B1 (FB1) treatments that inhibited ceramide synthase in the de novo pathway eliminated the effects of MBO-asGCS on restoration of p53; exogenous C6-ceramide, but not C6-dihydroceramide presented the resuscitation of p53 pathways as MBO-asGCS in p53 mutant cells (18). It is not clear how ceramide modulates p53 resuscitation, but several studies suggest that ceramide play a role in mediating posttranscriptional processing, as it alters the isoform expression of caspase-9 and bcl-x in cancer cells (25–26).
Targeting ceramide glycosylation is an alternative approach to restore p53 function and improve cancer treatment. Overexpression of GCS that confers cell resistance to apoptosis becomes a potential marker predicting tumor response to chemotherapy and clinical progression (27–29). Inhibition of GCS by gene silencing or tamoxifen leads p53-mutant cancer cells to apoptosis (20, 28, 30). It has been reported that p53 mutants upregulate the expression of MDR1, hERT, bFGF, bcl-xl, HSP70, and other genes, and down-regulate the expression of FAS, PTEN and others, gaining the oncogenic functions to promote tumor progression (1, 3, 7). Coincidently, glycosphingolipids (globo-series, ganglio-series) produced after ceramide glycosylation upregulate the expression of MDR1, hERT, bFGF and HSP70 in cancer cells, enhancing cells growth and resistance to therapies (21, 27). On the other hand, inhibition of glycosphingolipid synthesis or increased endogenous ceramide represses the expression of MDR1, hERT, bFGF and HSP70, and increases the expression of FAS, PTEN, p21, Bax and Puma, thus promotes cell cycle arrest, apoptosis and sensitization to cancer therapies (18, 21, 27). Although it is unclear how frequently GCS is overexpressed with p53 mutation in cancers, disrupting ceramide glycosylation resuscitates p53 expression that directly enhances p53-responsive genes and simultaneously inhibits p53-repressive genes might be a very effective therapeutic approach for these cancer patients (Fig. 1B).
Implications and Future Direction
TP53 mutation presents in LOH, and 90% of p53 mutants in tumors occur within DBD encoded from exons 4–8. Suppression of GCS can restore p53 expression in cells with a deleted mutation of exon-5 (18), and this approach may be able to resuscitate p53 in cells with point mutant (for instance R172H exon 5, unpublished data). It has been disclosed that transcription and posttranscriptional process are crucial for p53 mutation, even little is known about regulation mechanisms by which cells specifically express gene like p53 using identical allele and by particular posttranscriptional process to determine wild-type protein or mutant one in tumors. Further studies in these will characterize therapeutic targets restoring p53 signal pathway to mutant p53 addicted tumors. High levels of GCS and mutant p53 are coincidently detected in several drug-resistant ovarian cancer cells lines (18). Investigating the association of GCS and mutant p53 in drug resistance and the molecular mechanism by which ceramide mediates p53 restoration in mutant p53-barrier cancers may lead to discovering effective approaches to improve cancer treatment.
Acknowledgments
This work was supported by United States Public Health Service/NIH grant P20 RR16456 from the NCRR, and Department of Defense Breast Cancer Research Program DAMD17-01-1-0536.
References
- 1.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 2.Stein T, Crighton D, Warnock LJ, Milner J, White RJ. Several regions of p53 are involved in repression of RNA polymerase III transcription. Oncogene. 2002;21:5540–7. doi: 10.1038/sj.onc.1205739. [DOI] [PubMed] [Google Scholar]
- 3.Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat Rev Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 4.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 5.Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–67. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 6.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 7.Chen F, Wang W, El-Deiry WS. Current strategies to target p53 in cancer. Biochem Pharmacol. 2010;80:724–30. doi: 10.1016/j.bcp.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 8.Wiman KG. Pharmacological reactivation of mutant p53: from protein structure to the cancer patient. Oncogene. 2010;29:4245–52. doi: 10.1038/onc.2010.188. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Zheng D. An update on gene therapy in China. Curr Opin Mol Ther. 2009;11:547–53. [PubMed] [Google Scholar]
- 10.Zeimet AG, Marth C. Why did p53 gene therapy fail in ovarian cancer? Lancet Oncol. 2003;4:415–22. doi: 10.1016/s1470-2045(03)01139-2. [DOI] [PubMed] [Google Scholar]
- 11.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–8. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 13.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–8. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 14.Joerger AC, Ang HC, Fersht AR. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci U S A. 2006;103:15056–61. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinmann L, Wischhusen J, Demma MJ, Naumann U, Roth P, Dasmahapatra B, et al. A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2008;15:718–29. doi: 10.1038/sj.cdd.4402301. [DOI] [PubMed] [Google Scholar]
- 16.Tang X, Zhu Y, Han L, Kim AL, Kopelovich L, Bickers DR, et al. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest. 2007;117:3753–64. doi: 10.1172/JCI32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao W, Chen M, Zhao X, Kumar R, Spinnler C, Thullberg M, et al. PRIMA-1Met/APR-246 induces wild-type p53-dependent suppression of malignant melanoma tumor growth in 3D culture and in vivo. Cell Cycle. 2011;10:301–7. doi: 10.4161/cc.10.2.14538. [DOI] [PubMed] [Google Scholar]
- 18.Liu YY, Patwardhan GA, Bhinge K, Gupta V, Gu X, Jazwinski SM. Suppression of Glucosylceramide Synthase Restores p53-Dependent Apoptosis in Mutant p53 Cancer Cells. Cancer Res. 2011;71:2276–85. doi: 10.1158/0008-5472.CAN-10-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogretmen B, Safa AR. Expression of the mutated p53 tumor suppressor protein and its molecular and biochemical characterization in multidrug resistant MCF-7/Adr human breast cancer cells. Oncogene. 1997;14:499–506. doi: 10.1038/sj.onc.1200855. [DOI] [PubMed] [Google Scholar]
- 20.Patwardhan GA, Zhang QJ, Yin D, Gupta V, Bao J, Senkal CE, et al. A new mixed-backbone oligonucleotide against glucosylceramide synthase Sensitizes multidrug-resistant tumors to apoptosis. PLoS One. 2009;4:e6938. doi: 10.1371/journal.pone.0006938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patwardhan GA, Liu YY. Sphingolipids and expression regulation of genes in cancer. Prog Lipid Res. 2011;50:104–14. doi: 10.1016/j.plipres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santana P, Pena LA, Haimovitz-Friedman A, Martin S, Green D, McLoughlin M, et al. Acid sphingomyelinase- deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell. 1996;86:189–99. doi: 10.1016/s0092-8674(00)80091-4. [DOI] [PubMed] [Google Scholar]
- 23.Dbaibo GS, Pushkareva MY, Rachid RA, Alter N, Smyth MJ, Obeid LM, et al. p53- dependent ceramide response to genotoxic stress. J Clin Invest. 1998;102:329–39. doi: 10.1172/JCI1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 25.Massiello A, Roesser JR, Chalfant CE. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–2. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- 26.Shultz JC, Goehe RW, Wijesinghe DS, Murudkar C, Hawkins AJ, Shay JW, et al. Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res. 2010;70:9185–96. doi: 10.1158/0008-5472.CAN-10-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YY, Gupta V, Patwardhan GA, Bhinge K, Zhao Y, Bao J, et al. Glucosylceramide synthase upregulates MDR1 expression in the regulation of cancer drug resistance through cSrc and beta-catenin signaling. Mol Cancer. 2010;9:145. doi: 10.1186/1476-4598-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YY, Han TY, Giuliano AE, Cabot MC. Ceramide glycosylation potentiates cellular multidrug resistance. FASEBJ. 2001;15:719–30. doi: 10.1096/fj.00-0223com. [DOI] [PubMed] [Google Scholar]
- 29.Itoh M, Kitano T, Watanabe M, Kondo T, Yabu T, Taguchi Y, et al. Possible role of ceramide as an indicator of chemoresistance: decrease of the ceramide content via activation of glucosylceramide synthase and sphingomyelin synthase in chemoresistant leukemia. Clin Cancer Res. 2003;9:415–23. [PubMed] [Google Scholar]
- 30.Furlong SJ, Mader JS, Hoskin DW. Lactoferricin-induced apoptosis in estrogen-nonresponsive MDA-MB-435 breast cancer cells is enhanced by C6 ceramide or tamoxifen. Oncol Rep. 2006;15:1385–90. [PubMed] [Google Scholar]