Abstract
Metabolite identification in the complex NMR spectra of biological samples is a challenging task due to significant spectral overlap and limited signal to noise. In this study we present a new approach, RANSY (Ratio Analysis NMR Spectroscopy), which identifies all the peaks of a specific metabolite based on the ratios of peak heights or integrals. We show that the spectrum for an individual metabolite can be generated by exploiting the fact that the peak ratios for any metabolite in the NMR spectrum are fixed and proportional to the relative numbers of magnetically distinct protons. When the peak ratios are divided by their coefficient of variations derived from a set of NMR spectra, the generation of an individual metabolite spectrum is enabled. We first tested the performance of this approach using one-dimensional (1D) and two-dimensional (2D) NMR data of mixtures of synthetic analogues of common body fluid metabolites. Subsequently, the method was applied to 1H NMR spectra of blood serum samples to demonstrate the selective identification of a number of metabolites. The RANSY approach, which does not need any additional NMR experiments for spectral simplification, is easy to perform and has the potential to aid in the identification of unknown metabolites using 1D or 2D NMR spectra in virtually any complex biological mixture.
Keywords: NMR, metabolomics, ratio analysis, peak assignment
Introduction
Nuclear magnetic resonance (NMR) based metabolomics is increasingly used in numerous studies and applications that include drug response, early disease diagnosis, toxicity and nutritional studies, and basic systems biology using a variety of complex biological samples.1–10 A number of reviews have been published that describe advancements in the field.11–16 NMR spectroscopy is an important analytical technique in metabolomics primarily because of its quantitative nature and high reproducibility. Owing to the complexity of the NMR spectra of biological samples, however, obtaining information on low concentration metabolites is challenging due to the high degree of spectral overlap. Nevertheless, these species are important to analyze because their changing concentrations may distinguish biological status, such as health and disease.
Numerous advances in NMR methods and databases have so far been made to aid the analysis of complex NMR spectra, as well as to identify specific metabolites and pathways associated with the onset of various diseases.17–20 Some important approaches for peak assignments in the complex NMR spectra using NMR databases include targeted profiling of complex NMR spectra using mathematical modeling of the pure compound spectra to fit the experimental data,21 the COLMAR approach that screens chemical shift lists or cross sections of multidimensional NMR spectra,22 and annotation of metabolites using a statistical index called Spin-Assign p-value.23 Methods such as statistical total correlation spectroscopy (STOCSY),24 selective TOCSY,25,26 and isotope enhanced methods27–32 offer approaches to simplify NMR spectra of complex samples such as biofluids and cells. In addition, spectra simplification in terms of increased resolution along the indirect dimension can be achieved through covariance NMR spectroscopy, introduced by Bruschweiler and coworkers.33, 34,35
Among the methods applicable to metabolomics, STOCSY24 has proven to be a very useful and general data analysis method to identify related metabolite peaks from complex spectra. The auto-correlation matrix of a set of NMR spectra identifies peaks from the same metabolite because these peaks show high correlation, which can simplify assignments. In addition, from the STOCSY analysis peaks from other metabolites in the same or different pathways that are also correlated appear in the spectrum, which provides benefits but also complicates the analysis. STOCSY has successfully been extended to analyze spectra obtained from different nuclei36 and even different analytical techniques, such as NMR, cryoflow LC-NMR and UPLC-MS.37–39 Correlation was also used to group variables in complex spectra and prove interpretability of latent variables for metabolic biomarker recovery.40 However, due to the often substantial number of high correlation values, it can be difficult to find the meaningful correlations without choosing a threshold empirically. Clean spectra for individual metabolites are sometimes difficult to achieve.
Here we propose an alternative method to detect metabolite signals selectively from the complex 1D and 2D spectra. The new approach involves an analysis of the ratio between a selected peak and all the other peaks in the spectrum. The mean of each ratio is calculated across the multiple spectra in the data set, and is then divided by the standard deviation of this ratio to generate a spectrum that clearly illustrates the relationship among the peaks from the same molecular species. The newly generated spectrum contains clean peaks with good signal to noise; identification can then be easily performed by comparing the ratio spectrum with a standard spectrum from a database. The RANSY method therefore allows easier identification of individual molecules in overlapped spectral regions without the need for additional experiments. The same methodology can also be applied to simplify 2D spectra. For example, in the spectra generated by isotope tagging methods, metabolites with multiple functional groups can be identified easily.30 Because of intrinsic characteristics of the calculated ratio, no data normalization is needed.
Theory
RANSY is based on the fact that the ratio between two NMR peak intensities or areas from the same metabolite will be equal to the relative number of magnetically nonequivalent spins and will be constant across all spectra; and therefore the standard deviation of this ratio will in theory be zero, or very close to zero. Although spectral noise contributes to some variation in this ratio, the noise contribution is still relatively small. Of course, in highly overlapped spectra, it is the contributions to the peaks in the spectra from a particular metabolite that will have fixed ratios. On the other hand, if the two peaks originate from different metabolites, their ratio should vary across different spectra. Thus, the standard deviation of the ratio of peaks from separate molecules across all the spectra will be typically large, except in those rare cases where these metabolites are very highly correlated.
In order to obtain the ratio spectrum of an unknown metabolite with only one known peak, the ratios between all the other points in each spectrum and the known peak is calculated. Each of these ratios is then divided by its coefficient of variation (1/CV), i.e., the quotient of the ratio’s standard deviation and its mean. Since the ratio’s standard deviation is used as the denominator, small standard deviations will produce a very large reciprocal value, generating a peak, but large standard deviations will give small numbers, similar to noise values. The reciprocal of the CV is used instead of the standard deviation because CV is dimensionless and standard deviations can only be compared in the context of the mean value.
The RANSY algorithm is as follows. We denote the ith spectrum of a set of n spectra as a vector Xi. The jth data point of m total points in that spectrum is denoted as Xi,j. We designate one peak of interest from a given metabolite (the kth point in spectrum i, Xi,k) as the “driving” peak. The first step is to calculate all the ratios between all the other points and the driving peak; we can denote the ratio matrix as D, which is an n*m matrix. Each point in that matrix is defined as:
| (1) |
Next, in order to generate a ratio spectrum, which is an m-element vector R, the quotient of means and standard deviations across columns of D is calculated. Thus, the jth element of the 1D RANSY spectrum (vector R), is calculated as follows:
| (2) |
However, since the standard deviation is zero for the driving peak itself, the corrected ratio for the driving peak Xk is infinite. In order not to sacrifice the driving peak, we assign the corrected ratio of that peak equal to the maximum of R. So the final RANSY spectrum is given by equation (2) except the driving peak Rk, which is given by:
All ratio analyses were performed on a personal computer using R version 2.10.0 software.
Experimental Methods
Chemicals
Thirty-three synthetic analogs of human serum metabolites (see Supplemental Information Table S-1) and the NMR standard, TSP (trimethylsilylpropionic acid-d4, sodium salt), were purchased from Sigma-Aldrich (St. Louis, MO). Deuterium oxide (D2O, 99.9% D) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). Chemicals for 15N-ethanolamine tagging, 4-(4,6-dimethoxy[1,3,5]triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM) and 15N-ethanolamine were purchased from Cambridge Isotope Laboratories (Andover, MA), and Isotech (Miamisburg, OH), respectively. All chemicals were used without further purification.
Serum samples
100 serum samples for 1D NMR were obtained from Innovative Research, Inc. (Novi, MI), and 550 μL of each sample was used for the CPMG experiment.
For 1D RANSY experiments, 15 standard samples were prepared by mixing 15 μL TSP (5 mM in D2O) with stock solutions of the 33 metabolites (20 mM each). Volumes for each of the standards were chosen randomly within a range, and their final concentrations can be found in Supplemental Information Table S-1. Concentrations of the standards were varied by nearly two orders of magnitude (0.03-2 mM), and were chosen to match roughly with their physiological concentrations in blood or urine.41 A 285 μL solution of 0.5 M phosphate buffer in D2O (pH=7.4) was added to each standard mixture to minimize peak shifts. After mixing, 550 μL of each sample was used to acquire the NMR spectra. A second set of 10 sample mixtures containing 15 different metabolites was also prepared in a similar manner, except that the concentration ranges for each of these metabolites was varied from 0.03 to 2 mM (see Supplemental Information Table S-2).
For 2D RANSY experiments, 15 standard mixtures were prepared by mixing 100 μL water with stock solutions of the 33 metabolites (20 mM each) to obtain the final concentrations. These 15 samples were derivatized with 15N ethanolamine, according to the procedure described previously.30 Briefly, 3 μL 15N labeled ethanolamine (50 μM) was added to each sample. After adjusting the pH to 7.0 with 1M HCl, 21 mg DMT-MM was added to the solution. The reaction mixture was kept at room temperature for 4 hrs with stirring to complete the reactions. In order to maintain 15N amide protonation, the pH was adjusted to 5.0 and the solution volume was adjusted to 600 μL by adding water prior to 2D NMR experiments, of which 550 μL was used for NMR experiments. A blank sample was also prepared by the same procedure as above but without adding any metabolites.
NMR Spectroscopy
1D NMR experiments for the 15 standard mixture samples were performed on a Bruker Avance-III-800 spectrometer equipped with a room temperature 1H inverse detection Z-gradient probe. NMR data were acquired using the 1D NOESY pulse sequence with water presaturation. 128 scans with 16 k time domain data points were collected using a spectral width of 12,800 Hz. An exponential weighting function corresponding to 1.0 Hz line broadening was applied to the free induction decay (FID) before Fourier transformation. The spectra were then phased, baseline corrected and referenced to TSP (δ=0.000 ppm) using Bruker Topspin 3.0 software.
1H NMR experiments were obtained for 100 commercial serum samples using a standard 1D CPMG (Carr-Purcell-Meiboom-Gill) pulse sequence coupled with water presaturation on a Bruker Avance-500 spectrometer equipped with a TXI gradient cryoprobe. 128 scans with 16 k time domain data points were collected using a spectral width of 6,000 Hz. An exponential weighting function corresponding to 1.0 Hz line broadening was applied to the FIDs before Fourier transformation. The acquired spectra were then phased, baseline corrected and referenced to alanine (δ=1.479 ppm) using Bruker’s Topspin 3.0 software.
Two dimensional experiments for standard samples were performed at 298 K on a Bruker Avance-III-800 spectrometer as well. 1H-15N 2D heteronuclear single quantum coherence (HSQC) experiments employed an INEPT transfer delay of 5.5 ms corresponding to a 1JNH coupling of 90 Hz. Spectral widths of approximately 10 kHz for the 1H dimension and 5 kHz for 15 N were used at 800 MHz. A total of 128 free induction decays of 2048 data points each were collected in the indirect (t1) dimension with 32 transients per increment. 15N decoupling during the direct detection dimension (t2) was achieved using globally optimized alternating-phase rectangular pulses (GARP). The resulting 2D data were zero-filled to 1024 points in the t1 dimension after forward linear prediction to 512 points. The spectra were then phased, baseline corrected and integrated using Bruker Topspin 3.0 software as described earlier.30, 42
Results and Discussion
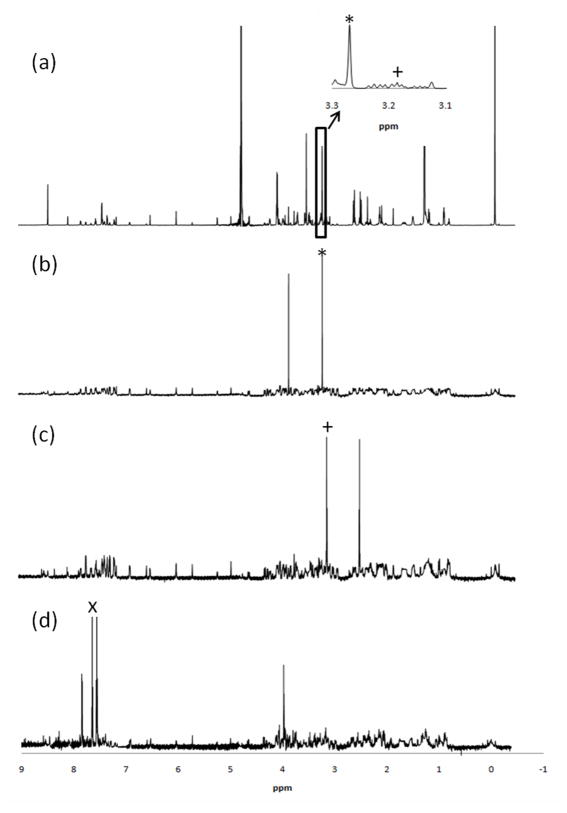
The RANSY approach was first applied to analyze a set of 15 standard metabolite mixtures, and some of these results are shown in Figure 1. The 1D NMR spectrum of a mixture of 33 standard compounds can be seen in Figure 1(a). Some regions in this spectrum are quite crowded even in this relatively simple mixture of standard compounds. RANSY was applied to the collection of 15 samples by selecting one of the isolated metabolite peaks as the “driving peak.” For example, in Figure 1(b) the driving peak is at 3.26 ppm and corresponds to the upfield peak of the methyl singlet of betaine. The other singlet at 3.91 ppm also appears in the RANSY spectrum at the correct position. In Figure 1(c), the driving peak is at 3.18 ppm, and corresponds to the downfield triplet of β-alanine. The other triplet appears in the RANSY spectrum at the expected position (2.56 ppm) and intensity. Both betaine and β-alanine are quite low in concentration and their resonances sit in relatively crowded regions, as shown in the expanded region of Figure 1(a). Nevertheless, one can easily generate their spectra by RANSY. The example shown in Figure 1(d) is hippuric acid, which has several peaks in the aromatic region, and a doublet at 3.96 ppm. Three multiplets could be found in the aromatic region, centered at 7.81, 7.64 and 7.54 ppm, corresponding to hydrogens located on the phenyl ring. Using the center peak at 7.64 ppm as the driving peak, the RANSY spectrum reveals the other two peaks plus a doublet centered at 3.96 ppm, which is due to hippuric acid’s CH2 group, and also had its correct chemical shift. In the spectra of Figure 1, the noise levels are significantly lower when compared with the peaks generated by RANSY, and the RANSY spectra are very similar to standard spectra of these metabolites. The relative peak heights are also close to those for the standards, indicating that the standard deviations for the ratios from the same compounds are small and very similar to one another. As will be shown below, using a larger number samples will further improve the signal to noise ratio (vide infra). In order to investigate RANSY on mixtures in which the signals vary over a larger dynamic range, we also record spectra of 10 mixtures of 15 metabolites where the concentrations all varied over a (non-biological) range of 0.03-2 mM (Supplemental Table S-2). The RANSY spectra of the same three metabolites (betaine, β-alanine and hippuric acid) were calculated by the driving peaks mentioned above and shown in Supplemental Figure S-1. Similar RANSY results were obtained with this larger concentration range, with the one difference being that the noise appeared to be more evenly distributed in this case.
Figure 1.
(a) 1H NMR spectrum of a mixture of 33 standard compounds; (b)-(d) Selective detection of (b) betaine, (c) β-alanine and (d) hippuric acid by the RANSY method; (b), (c) and (d) were generated from 15 1H NMR spectra of mixtures, and the driving peaks are indicated by an asterisk, plus sign and letter X respectively. The inset in (a) shows the crowded region for driving peaks of betaine and β-alanine.
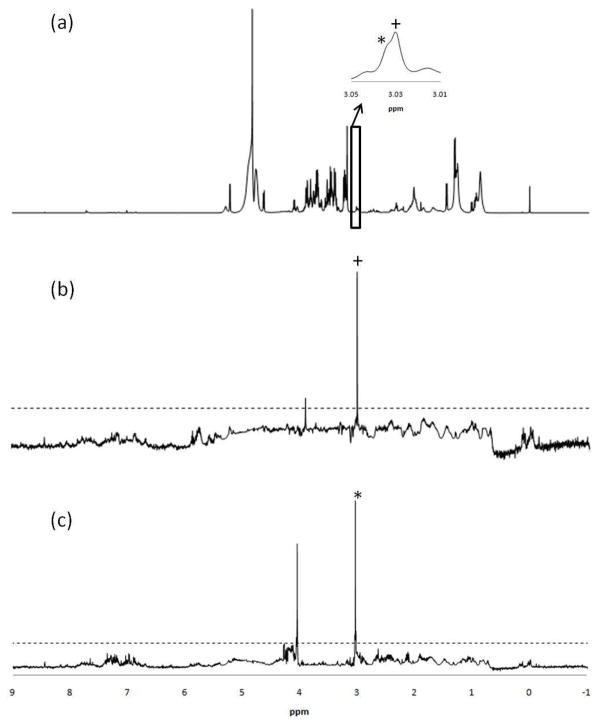
RANSY was then applied to a set of 100 serum samples. In Figure 2(a) the CPMG spectrum of serum is shown prior to the application of RANSY. Subsequently, RANSY was applied to the upfield peaks of creatinine and creatine centered at 3.03 and 3.02 ppm, respectively, as shown in Figure 2(a). These two metabolites were chosen because their upfield peaks are very close to one another. They are somewhat difficult to identify in serum since they both often have overlapping singlets, and these signals are somewhat weak due to their low concentrations in serum (~30 μM for adults). The RANSY spectra for both the metabolites are shown in Figure 2(b) and (c); it is very clear that the lowfield singlet centered at 4.05 ppm is associated with creatinine, and the singlet at 3.92 ppm is associated with creatine. While the RANSY spectra for both the metabolites are clear, there is some baseline noise generated by the large number of small peaks in the baseline of the serum spectra. One can, however, distinguish the RANSY peaks from baseline noise by applying a minimum threshold value across the spectrum. A threshold of R=6 for the calculated ratio of Eq. 2 can be applied to the spectrum of Figure 2(b) and (c), and indicated by the dash line. It can be seen that the all the noise peaks in both figures are below that threshold.
Figure 2.
(a) 1H NMR spectrum of serum, obtained using the CPMG pulse sequence; (b) Selective detection of creatine by the RANSY method. The driving peak is at 3.02 ppm (indicated by the plus sign); (c) Selective detection of creatinine driven by the singlet at 3.03 ppm (indicated by the asterisk). The noise threshold value of R = 6 is indicated by the dashed horizontal line in both (b) and (c). The inset in (a) shows the two very closely spaced peaks for creatine and creatinine used as driving peaks.
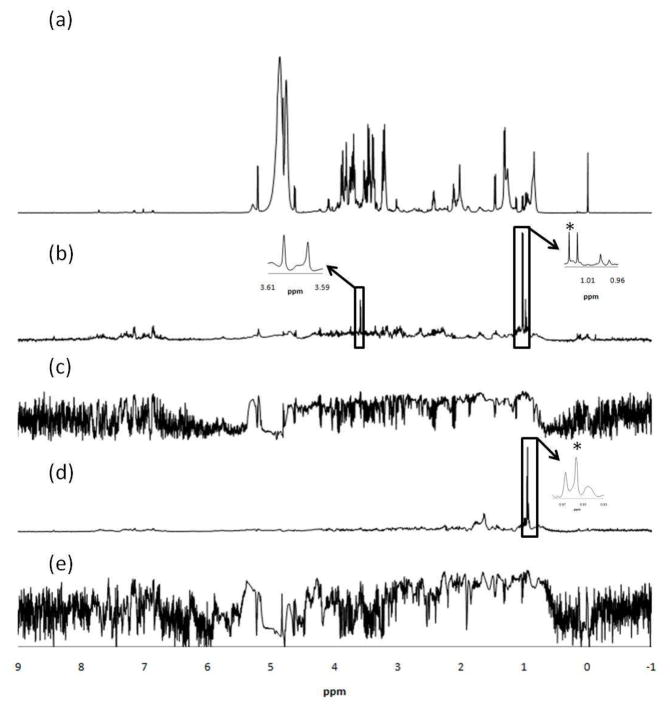
To test RANSY further in serum samples, the method was also applied to identify resonances from valine and leucine. Both of the upfield peaks for valine and leucine come from methyl groups that overlap with isoleucine. In addition, leucine’s upfield triplet centered at 0.95 ppm overlaps with the tail of a strong, broad lipid peak. The chosen driving peaks were at 1.02 ppm for valine and 0.94 ppm for leucine. Correlation spectra were also calculated using the same driving peak for comparison. The valine RANSY spectrum shown in Figure 3(b) shares two upfield doublets from the two methyl groups, and another doublet at around 3.6 ppm, which was from the α-CH proton. The RANSY method is not perfect, as the multiplet at 2.25 ppm (R=3.1) was missing due to its low intensity and some peak shifting observed in the CPMG spectra. For the RANSY spectrum calculated for leucine, we can see in Figure 3(d) that another multiplet centered at 1.70 ppm appears for leucine, which comes from the β-CH2 and γ-CH protons. However, the multiplet centered at 3.71 ppm was missing (R=0.69) compared with its standard spectrum, because of the very strong signal intensity and overlap from glucose. To compare RANSY with the conventional statistical correlation method, correlation spectra based on Pearson correlation coefficients were calculated using the same driving peaks for valine and leucine. Peaks in the spectra as shown in Figure 3(c) and (e) thus represent correlation coefficients between the vector of the driving peak and those of all other peaks. While the RANSY results are not perfect in these cases, they are still much better than the correlation method calculated and shown in Figure 3(c) and (e), in which most of the peaks can hardly be seen.
Figure 3.
1H NMR spectrum of serum (a), obtained using the CPMG pulse sequence; (b) Selective detection of valine from the serum 1H NMR spectrum by RANSY. The identified peaks around 3.6 ppm are from the α-CH proton; (c) Selective detection of valine from the serum 1H NMR spectrum by statistical correlation; (d) Selective detection of leucine from the complex serum 1H NMR spectrum by RANSY. The identified multiplet around 1.70 ppm is from the β-CH2 and γ-CH protons ; (e) Selective detection of leucine from the complex serum 1H NMR spectrum by statistical correlation. The driving peaks are indicated by asterisks.
In general, in order to calculate the RANSY spectra it is necessary to use multiple spectra to calculate the relative CV values. To evaluate the effect of limited sample numbers, an analysis of the RANSY spectra of serum was performed using different numbers of spectra. The RANSY spectrum for valine was calculated by averaging the results from using each of the four peaks from the two valine doublets located at 1.04 and 0.98 ppm as the driving peaks. First, 5 samples were randomly picked from the 100 commercial serum samples. Next, additional and randomly chosen samples were added to the original 5 samples and the RANSY spectrum was recalculated. The results of applying RANSY to different numbers of spectra, from 5 to 80 are shown in Supplemental Figure S-2. When only 5 samples were used, the two doublets were not of the same height and at least one peak was distorted. The addition of 2 spectra in the RANSY calculation (a total of 7 spectra) resulted in much better resolution for the two doublet peaks and the peak shape also improved. When 20 samples or more were used, the peak became well shaped and showed no additional changes when additional samples were added to the RANSY calculation. Overall, the differences in the RANSY spectra between 20 and 80 samples were minor and not easy to identify. From this example, we expect that 20 samples would often be sufficient to well deconvolute metabolites in complex samples such as serum.
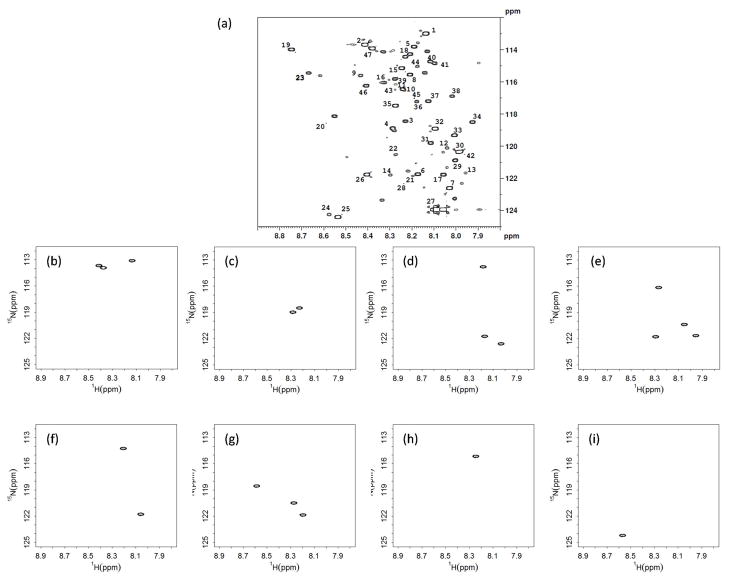
RANSY was also applied to 2D HSQC spectra of isotope tagged metabolite mixtures. 15 mixtures each containing different concentrations of the same 33 metabolites were tagged with 15N ethanolamine as described in the experimental section. Of these 33 metabolites, 3 metabolites (isocitric acid, citric acid and cis-aconitic acid) have 3 carboxyl groups, and 3 metabolites (L-glutamic acid, oxalic acid and malic acid) have 2 carboxyl groups, while the remaining metabolites have only one carboxyl group each. Compared with the library,30 all peaks were observed as expected except for one peak each for L-glutamic acid and oxalic acid. A typical isotope enhanced 2D HSQC spectrum is shown in Figure 4(a). A few minor peaks can also be seen, which we determined to be due to the impurities present in the starting materials, or in a few cases, incomplete derivatization of standard compounds with multiple functional groups (especially OH groups). Similar issues are sometimes present in GC-MS analysis employing derivatization as well. All 47 metabolite peaks detected in every spectrum were integrated, and the RANSY analysis was performed for all pairs of peaks by an in-house developed R code (See Supplemental Information). Whether the two peaks are associated with the same metabolite can be determined by RANSY method. The algorithm used to calculate individual metabolite spectra is as follows: if two peaks showed a RANSY R value above the threshold mutually, we regard them as associated with a single metabolite. And a third peak will be added to that metabolite if and only if it showed high R values when using both peaks as the driving peak. Following this approach, 2D spectra of individual metabolites could then be generated. In a few examples as shown in Figure 4(b) through (i): the peaks labeled as 1, 2 and 47 in Figure 4(a) were identified as coming from lactic acid; peaks 3 and 4 from glucuronic acid; peaks 5, 6 and 7 from citric acid; peaks 11, 12, 13 and 14 from isocitric acid (including its OH group that was also derivatized); peaks 17 and 18 from malic acid and peaks 20, 21and 22 from cis-aconitic acid. All of these peaks were detected with a threshold value, R≥6.5. Two examples of single peak metabolites, leucine (peak 15) and betaine (peak 24) are also shown here. Interestingly, the ratio threshold of 6.5 is close to what we have used for 1D RANSY, which indicates at least some generality of this threshold value. Overall, 30 out of 33 metabolites can be assigned correctly from this approach (Supplementary Table S-3).The metabolites not detected correctly were oxalic acid, L-glutamic acid and 4-hydroxyl-L-proline. The first two metabolites only showed one peak in some of the spectra due to low concentration and incomplete reaction. One of the two peaks of 4-hydroxyl-L-proline (Peak 10 in Figure 4(a)) overlapped with a background peak and became distorted. The other peaks thus identified from RANSY match very well with the peak assignments for the metabolites using the 15N-tagging.30 By this method, the spectra can be substantially simplified leading to better assignments. Considering that 20 samples or more improve the RANSY analysis, as discussed before, a larger sample set will improve the discrimination between peaks from the same metabolite and background peaks, and thus metabolites with multiple functional groups will be easier to identify.
Figure 4.
(a) A 1H-15N HSQC spectrum of a standard mixture containing 33 metabolites with identifications and concentrations given in Table S-1. Examples of metabolites detected by RANSY: (b) lactic acid (peaks 1,2,47); (c) glucuronic acid (3,4); (d) citric acid (5,6,7); (e) isocitric acid (11,12,13,14); (f) malic acid (17,18); (g) cis-aconitic acid (20,21,22); (h) leucine (15); and (i) betaine (24). Individual HSQC spectra (b)-(i) were generated by thresholding the RANSY ratio of peaks above a value, R ≥ 6.5.
Conclusions
NMR spectra of complex biosamples can pose difficulties in assignment and analysis. Based on the relationship of spin correlated peaks, RANSY can provide a useful approach to help solve this problem. Using peak ratios to analyze the relationship between intrinsically correlated signals is a new approach that can be applied to complex spectral analysis problems. While a variety of correlation methods have been proved to be quite useful in this effort, the RANSY approach is quite complementary and has some advantages. For example, no additional experiments are needed to apply the RANSY method, it is computationally efficient, and does not require enormous numbers of samples. The clearly visible spectra generated by RANSY compare well to correlation calculations. No normalization is needed before analysis. And in principle, RANSY should be applicable to the identification of unknown species, even those which are not present in spectral libraries. As shown here, RANSY analysis results in an efficient recovery of metabolite peak information from complex 1D and 2D spectra of standard mixtures as well as blood samples. Peak identification can be facilitated with the development of RANSY.
Supplementary Material
Acknowledgments
This work is supported by the NIH/NIGMS (Grant 1R01 GM085291). The authors also thank Dr. Shanaiah Murthy at Matrix-Bio, Inc, and the Purdue Interdepartmental NMR Facility (PINMRF) for their assistance with NMR experiments.
References
- 1.Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Le Net JL, Baker D, Walley RJ, Everett JR, Nicholson JK. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 2.Viant MR, Rosenblum ES, Tjeerdema RS. Environ Sci Technol. 2003;37:4982–4989. doi: 10.1021/es034281x. [DOI] [PubMed] [Google Scholar]
- 3.Pears MR, Cooper JD, Mitchison HM, Mortishire-Smith RJ, Pearce DA, Griffin JL. J Biol Chem. 2005;280:42508–42514. doi: 10.1074/jbc.M507380200. [DOI] [PubMed] [Google Scholar]
- 4.Choi YH, Kim HK, Linthorst HJM, Hollander JG, Lefeber AWM, Erkelens C, Nuzillard JM, Verpoorte R. J Nat Prod. 2006;69:742–748. doi: 10.1021/np050535b. [DOI] [PubMed] [Google Scholar]
- 5.Soininen P, Kangas AJ, Wurtz P, Tukiainen T, Tynkkynen T, Laatikainen R, Jarvelin MR, Kahonen M, Lehtimaki T, Viikari J, Raitakari OT, Savolainen MJ, Ala-Korpela M. Analyst. 2009;134:1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Gowda GAN, Asiago V, Shanaiah N, Barbas C, Raftery D. Anal Biochem. 2009;385:392–392. doi: 10.1016/j.ab.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giskeodegard GF, Grinde MT, Sitter B, Axelson DE, Lundgren S, Fjosne HE, Dahl S, Gribbestad IS, Bathen TF. J Proteome Res. 2010;9:972–979. doi: 10.1021/pr9008783. [DOI] [PubMed] [Google Scholar]
- 8.Asiago VM, Alvarado LZ, Shanaiah N, Gowda GAN, Owusu-Sarfo K, Ballas RA, Raftery D. Cancer Res. 2010;70:8309–8318. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, Capstick V, Faught W, Sawyer MB. Clin Cancer Res. 2010;16:5835–5841. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- 10.Weljie AM, Bondareva A, Zang P, Jirik FR. J Biomol NMR. 2011;49:185–193. doi: 10.1007/s10858-011-9486-4. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson JK, Connelly J, Lindon JC, Holmes E. Nat Rev Drug Discovery. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 13.Griffin JL. Curr Opin Chem Biol. 2003;7:648–654. doi: 10.1016/j.cbpa.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Serkova NJ, Niemann CU. Expert Rev Mol Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 15.Gowda GAN, Zhang SC, Gu HW, Asiago V, Shanaiah N, Raftery D. Expert Rev Mol Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang SC, Gowda GAN, Ye T, Raftery D. Analyst. 2010;135:1490–1498. doi: 10.1039/c000091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viant MR. Biochem Biophys Res Commun. 2003;310:943–948. doi: 10.1016/j.bbrc.2003.09.092. [DOI] [PubMed] [Google Scholar]
- 18.Fan TWM, Lane AN. J Biomol NMR. 2011;49:267–280. doi: 10.1007/s10858-011-9484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, MacInnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao HY, Markley JL. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 22.Robinette SL, Zhang FL, Bruschweiler-Li L, Bruschweiler R. Anal Chem. 2008;80:3606–3611. doi: 10.1021/ac702530t. [DOI] [PubMed] [Google Scholar]
- 23.Chikayama E, Sekiyama Y, Okamoto M, Nakanishi Y, Tsuboi Y, Akiyama K, Saito K, Shinozaki K, Kikuchi J. Anal Chem. 2010;82:1653–1658. doi: 10.1021/ac9022023. [DOI] [PubMed] [Google Scholar]
- 24.Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J, Blancher C, Gauguier D, Lindon JC, Holmes E, Nicholson J. Anal Chem. 2005;77:1282–1289. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- 25.Sandusky P, Raftery D. Anal Chem. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- 26.Sandusky P, Raftery D. Anal Chem. 2005;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi J, Shinozaki K, Hirayama T. Plant Cell Physiol. 2004;45:1099–1104. doi: 10.1093/pcp/pch117. [DOI] [PubMed] [Google Scholar]
- 28.Shanaiah N, Desilva MA, Gowda GAN, Raftery MA, Hainline BE, Raftery D. Proc Natl Acad Sci U S A. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane AN, Fan TWM. Metabolomics. 2007;3:79–86. [Google Scholar]
- 30.Ye T, Mo HP, Shanaiah N, Gowda GAN, Zhang SC, Raftery D. Anal Chem. 2009;81:4882–4888. doi: 10.1021/ac900539y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSilva MA, Shanaiah N, Gowda GAN, Rosa-Perez K, Hanson BA, Raftery D. Magn Reson Chem. 2009;47:S74–S80. doi: 10.1002/mrc.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye T, Zhang SC, Mo HP, Tayyari F, Gowda GAN, Raftery D. Anal Chem. 2010;82:2303–2309. doi: 10.1021/ac9024818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F, Bruschweiler R. J Am Chem Soc. 2004;126:13180–13181. doi: 10.1021/ja047241h. [DOI] [PubMed] [Google Scholar]
- 34.Bruschweiler R, Zhang F. J Chem Phys. 2004;120:5253–5260. doi: 10.1063/1.1647054. [DOI] [PubMed] [Google Scholar]
- 35.Zhang FL, Bruschweiler-Li L, Bruschweiler R. J Am Chem Soc. 2010;132:16922–16927. doi: 10.1021/ja106781r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coen M, Hong YS, Cloarec O, Rhode CM, Reily MD, Robertson DG, Holmes E, Lindon JC, Nicholson JK. Anal Chem. 2007;79:8956–8966. doi: 10.1021/ac0713961. [DOI] [PubMed] [Google Scholar]
- 37.Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, Rainville P, Stumpf CL, Nicholson JK. Anal Chem. 2006;78:363–371. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- 38.Crockford DJ, Lindon JC, Cloarec O, Plumb RS, Bruce SJ, Zirah S, Rainville P, Stumpf CL, Johnson K, Holmes E, Nicholson JK. Anal Chem. 2006;78:4398–4408. doi: 10.1021/ac060168o. [DOI] [PubMed] [Google Scholar]
- 39.Cloarec O, Campbell A, Tseng LH, Braumann U, Spraul M, Scarfe G, Weaver R, Nicholson JK. Anal Chem. 2007;79:3304–3311. doi: 10.1021/ac061928y. [DOI] [PubMed] [Google Scholar]
- 40.Blaise BJ, Shintu L, Elena B, Emsley L, Dumas ME, Toulhoat P. Anal Chem. 2009;81:6242–6251. doi: 10.1021/ac9007754. [DOI] [PubMed] [Google Scholar]
- 41.www.hmdb.ca
- 42.Gowda GAN, Tayyari F, Ye T, Suryani Y, Wei SW, Shanaiah N, Raftery D. Anal Chem. 2010;82:8983–8990. doi: 10.1021/ac101938w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.