Abstract
Bacterial cell growth is a complex process consisting of two distinct phases: cell elongation and septum formation prior to cell division. Although bacteria have evolved several different mechanisms for cell growth, it is clear that tight spatial and temporal regulation of peptidoglycan synthesis is a common theme. In this review, we discuss bacterial cell growth with a particular emphasis on bacteria that utilize tip extension as a mechanism for cell elongation. We describe polar growth among diverse bacteria and consider the advantages and consequences of this mode of cell elongation.
Keywords: cell elongation, polar growth, DivIVA, peptidoglycan
1 Introduction
1.1 The diversity of bacterial morphology
The relative simplicity of certain bacterial cell morphologies and life cycles stands in stark contrast to the complex morphologies shaped by defined developmental processes in many eukaryotes. Consider, for example, the coordinated spatiotemporal pattern of cell proliferation and differentiation involved in the maturation of Drosophila melanogaster from egg to larva to reproductive adult; as opposed to the process of elongation and septation of a single, rod shaped cell for reproduction of Escherichia coli. Organismal growth and reproduction seem comparatively straightforward in the case of the bacterium.
The simplicity of some morphologies notwithstanding, the bacterial domain actually encompasses an extremely broad range of cell shapes [1] (Figure 1). Spirals, spikes, and stalks abound (the list goes on), and these shapes presumably confer selective advantages in particular environments, e.g. uptake of limiting nutrients via elongated stalks in Caulobacter [2, 3]. How, then, is growth coordinated to generate cellular appendages and modified cell body geometries? Stepping back to “simple” bacteria like E. coli, how is cell growth coordinated to drive both elongation and division over the course of the life cycle? Patterns of bacterial growth clearly can vary immensely between species, and even through space and time within the same cell.
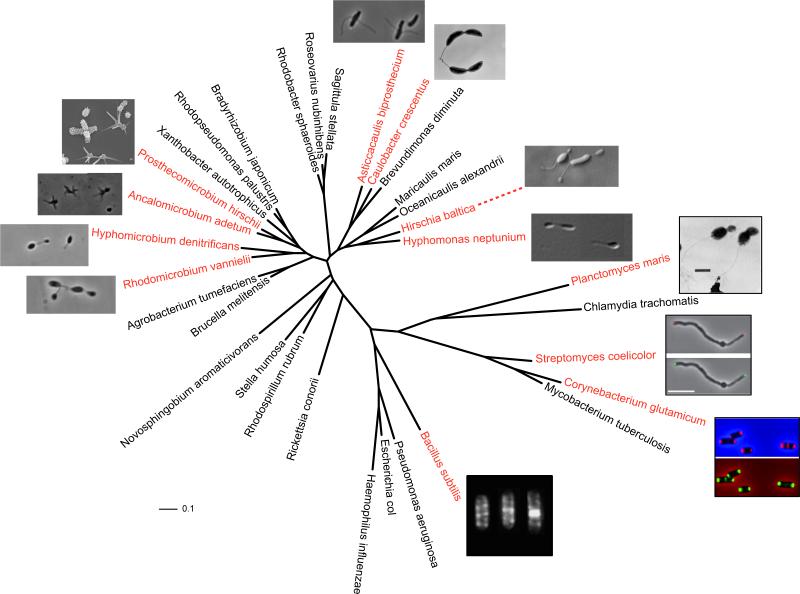
Figure 1.
Phylogeny and morphology of diverse bacteria exhibiting different modes of cell growth. gyrA sequences from representative species were aligned using MUSCLE [11]. RAxML [102] reconstructed the maximum likelihood phylogeny using the JTT amino acid substitution matrix and a four-category discrete gamma distribution of sequence rate variation among sites. Red text indicates species associated with adjacent micrographs. Bacillus subtilis with fluorescent vancomycin staining at progressive stages of the cell cycle; image adapted with permission from [8]. Corynebacterium glutamicum with fluorescent vancomycin staining (red; top panel) and DivIVA localization (green; bottom panel); images reproduced with permission from [25]. Streptomyces coelicolor with fluorescent vancomycin staining (red; top) and DivIVA localization (green; bottom); images reproduced with permission from [41]. Plantomyces maris image reproduced with permission from [4].
1.2 The bacterial cell wall
Before addressing specific modes of bacterial growth, we must identify some aspect of the bacterial cell that provides a useful biological and experimental basis for defining growth. The cell wall, which establishes the boundary of the bacterial cell and maintains its integrity, serves best in this regard.
The composition of the bacterial cell envelope varies between taxa. However, common to most bacteria is a layer of peptidoglycan (often referred to as the sacculus), comprising glycan strands of alternating N-acetylglucosamine and N-acetylmuramic acid residues with peptide cross-bridges, which provides the cell wall with its strength and maintains its shape. A phospholipid bilayer inner membrane lies interior to the peptidoglycan, and a second such outer membrane on the exterior of Gram-negative, but not Gram-positive, bacteria. As the sacculus serves as the structural underpinning of the cell wall in most of the best-studied bacterial model systems, we focus largely on peptidoglycan when considering detailed mechanics of cell growth in this review. The planctomyetes and the closely related chlamydiae represent a notable exception to this paradigm, with defined cell walls containing no peptidoglycan but possibly instead highly crosslinked protein sacculi [4–6]; see Section 2.3, below.
1.3 Zonal, non-polar growth in rod-shaped bacteria
Polar growth is a well-known and widely studied feature of eukaryotic cells. In contrast, many bacteria exhibit relatively simple morphologies that might, at first glance, appear to require little spatial coordination of cell wall growth. For example, the well-studied model species Escherichia coli and Bacillus subtilis form symmetric rods with a single polar axis, whereas Staphylococcus aureus forms non-polar spheres, and division at a central septum in each case gives rise to morphologically symmetric daughter cells.
Several well-studied organisms display highly spatially and temporally structured growth patterns. Between divisions, elongation of rod-shaped bacteria among the Gammaproteobacteria (e.g. Escherichia and Haemophilus), Firmicutes (e.g. Bacillus), and various other diverse taxonomic groups involves incorporation of peptidoglycan along the sidewalls [7–10]. Material at the poles, on the other hand, remains inert, with no indication of new incorporation or turnover of existing material [7, 9, 10]. In Bacillus subtilis, fluorescent vancomycin derivatives label peptidoglycan precursors, thereby identifying regions of active growth via incorporation into newly synthesized cell wall material [8]. Fluorescent vancomycin staining reveals that the sites of growth are structured at a finer spatial resolution into defined, helical bands [8, 11] (Figure 1). In diverse rod-shaped bacteria, the actin-like proteins MreB and, in the case of B. subtilis, Mbl, appear to function in spatially orienting peptidoglycan synthesis machinery to generate and maintain cell shape [8, 12, 13]. These cytoskeletal proteins form helical filaments along which penicillin binding proteins specialized in lateral cell wall growth are active [8, 14, 15, 16].
Upon cell division, growth shifts to the midcell division site, or septum, to generate the nascent poles. The composition and location of the peptidoglycan synthesis and remodeling machinery shift to organize around the well-studied septal FtsZ ring [14, 16–19] (Figure 1). This highly conserved machinery governs division even in spherical bacteria, with local peptidoglycan incorporation at the FtsZ ring despite the fully spherical morphological symmetry of the resulting daughters [20–23].
Taken together, these observations demonstrate that both lateral and septal cell growth involve spatially structured, zonal modes of growth in diverse rod-shaped bacteria. The process of cell wall synthesis is tightly spatiotemporally regulated by specific enzyme complexes targeted to particular modes of growth. These findings belie any notion of diffuse, unstructured growth along the cell wall even in bacteria with relatively uniform and symmetric morphologies.
1.4 Diverse growth patterns for diverse bacterial phenotypes
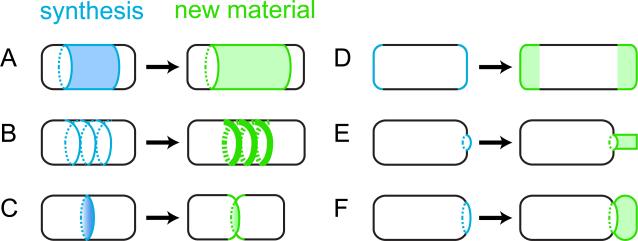
As evidenced by the examples presented above, bacterial growth and reproduction involve temporally regulated cell wall synthesis at specific regions. At a primary level, we can therefore classify growth as zonal rather than diffuse (cf. Figure 2a, 2b).
Figure 2.
Zonal cell wall synthesis establishes various patterns of bacterial growth and division. Blue regions indicate the location of cell wall synthesis activity (left), and green regions the resulting location of new peptidoglycan after synthesis (right). A. Elongation by diffuse synthesis along sidewalls. B. Elongation by synthesis in a spiral pattern along sidewalls (e.g. Bacillus). C. Division by synthesis at the septum (e.g. division at the FtsZ ring in diverse bacteria). D. Polar elongation (e.g. Corynebacterium). E. Stalk formation (e.g. Caulobacter). F. Budding (e.g. planctomycetes).
The zonal pattern of growth inherent even to spherical and rod-shaped bacteria provides a basis for tight, spatiotemporal control over cell wall growth (Figure 2). At the most basic and universal level, repositioning this zonal cell wall synthesis machinery at specific times permits alternation between cell elongation and division (Figure 2b, 2c). Furthermore, polar localization of cell wall synthesis serves to generate appendages (Figure 2e) and even budding modes of reproduction (Figure 2f). Thus, zonal growth potentiates a variety of growth patterns and resulting morphologies.
This review considers different forms of bacterial growth with respect to spatial and temporal patterns. We focus particularly on polar growth, which has received relatively little attention compared with non-polar growth patterns common to most well-studied model systems. A survey of cell wall synthesis in various taxa highlights the diversity of growth modes and underlying mechanisms encountered among the bacteria. We next consider how zonal cell wall synthesis may have facilitated transitions between the various described modes of growth. We conclude by considering selective pressures that may drive the evolution of zonal and polar cell wall synthesis patterns.
2 Polar growth patterns in specific bacterial taxa
2.1 Polar growth occurs among the rod-shaped and filamentous Actinobacteria
The Actinobacteria are comprised of Gram-positive bacteria that display morphological diversity including cocci, rods, and highly branched cells. In contrast to most rod-shaped bacteria whose growth is dependent on MreB rod-shaped and filamentous Actinobacteria grow strictly at the poles by a DivIVA-dependent mechanism. This section highlights what is known about the polar growth of three genera of the Actinobacteria: Corynebacterium, Mycobacterium, and Streptomyces.
2.1.1 Polar cell wall growth in Corynebacterium
Growth of Corynebacterium diphtheriae mitis [24] and Corynebacterium glutamicum [8] is restricted to the cell poles in young cells and to the cells poles and midcell in predivisional cells. In both C. diphtheriae and C. glutamicum, there is little or no recycling of preexisting cell wall material. These observations have led to a proposed model for cell growth in C. glutamicum [8]. In a newborn cell, active peptidoglycan growth is constrained to both cell poles while the remainder of the peptidoglycan along the sidewalls is inert. Once the cell doubles in length, the division machinery localizes to the mid-cell and new peptidoglycan synthesis occurs to allow septum formation. Following cell division, cell growth occurs at both new and old poles to enable cell elongation.
In Gram-positive bacteria, DivIVA is a highly conserved protein that localizes at the cell poles and division site and performs a vast array of functions. In C. glutamicum, DivIVA localizes to the cell poles and to a lesser extent to the mid-cell (Figure 1, bottom panel in C. glutanicum image) [25, 26]. Depletion of the essential DivIVA protein causes a loss of polar cell wall elongation and results in the formation of coccoid cells [25]. In contrast, overexpression of DivIVA causes the protein to accumulate at one cell pole, stimulating polar peptidoglycan synthesis and causing an increase in the pole diameter [25, 26]. Mid-cell DivIVA arrives at that position after chromosome segregation and the initiation of septal peptidoglycan synthesis [25].[27]. These observations indicate that DivIVA is essential for cell elongation, and it has been suggested that DivIVA may oligomerize and act as a scaffold for peptidoglycan synthesis machinery at the cell poles. Functional analyses of the coiled-coiled domains within DivIVA suggest their involvement in self-interaction, localization to the cell poles, and stimulation of peptidoglycan synthesis at the cell poles [28].
DivIVA depletion leads to upregulation of RsmP, which has similarity to eukaryotic intermediate filaments [29]. RsmP is an essential protein and its homologs are unique to the Actinobacteria. RsmP forms filamentous structures in vitro that span from pole to pole, and partial depletion of RsmP in vivo results in the formation of coccoid cells. These data indicate that RsmP is a cytoskeletal element involved in the maintenance of cell shape and polar growth. RsmP is phosphorylated by the essential serine/threonine protein kinase PknA in vitro, and a phosphomimetic form of the RsmP protein accumulates at the cell poles rather than along the length of the cells. Overexpression of PknA causes coccoid morphology while partial depletion causes cell elongation [30]. MurC, an essential ligase required for peptidoglycan biosynthesis, is also phosphorylated by PknA [31]. These observations suggest that phosporylation plays a key role in the regulation of both cell division and polar cell growth.
2.1.2 Polar cell wall growth in Mycobacterium
Nascent peptidoglycan synthesis in M. tuberculosis and M. smegmatis is restricted to the cell poles and mid-cell [32] and depends on an essential DivIVA homolog, Wag31 [33, 34]. Overexpression of Wag31 causes this protein to accumulate at one cell pole where an increase in peptidoglycan synthesis leads to the production of asymmetric bowling pin-shaped cells [34]. Wag31 is primarily localized to the poles where cell elongation occurs and only accumulates at the mid-cell following cytokinesis [33]. Following the completion of cell division, peptidoglycan synthesis occurs at both the new and old poles to allow cell elongation.
The phosphorylation state of Wag31 is modulated by the serine/theronine kinases PknA and PknB [33, 35–37]. Overexpression of wild-type or a phosphomimetic allele of Wag31, but not a phosphoablative allele, leads to the production of short, rounded cells [37]. Expression of a phosphomimetic allele of Wag31 or overexpression of PknB, which increases the pool of phosphorylated Wag31, leads to an increase in polar localization of Wag31 and a subsequent increase in nascent peptidoglycan synthesis [36]. Furthermore, expression of the phosphomimetic allele significantly increases the combined activity of MraY and MurG, transferases involved in the final step of peptidoglycan synthesis [36] and leads to an elevated amount of D-glutamic acid, D-alanine, and N-acetylglucosamine components of peptidoglycan [35]. These results suggest the Wag31 phosphorylation regulates the activity of the peptidoglycan biosynthesis machinery, although it remains unknown if the regulation is direct or indirect.
In addition to modulating the activity of Wag31, the PknA and PknB protein kinases are involved in the regulation of peptidoglycan synthesis. Overexpression of PknA or PknB causes the production of broad cells with nonuniform cell diameters [37]. PknA has been implicated in the phosphorylation of mycobacterial MurD, an essential ligase involved in peptidoglycan synthesis [38], and both PknA and PknB are involved in modulating the phosporylation state of GlmU, which synthesizes a precursor for peptidoglycan [39].
2.1.3 Streptomycetes grow by tip extension
Streptomyces exhibit a complex developmental cycle (for recent reviews see [40, 41]) in which spores germinate into germ tubes that grow by tip extension and branching to form the vegetative mycelium. When nutrient limiting conditions are encountered, aerial hyphae are formed, divided into prespore compartments, and converted into mature spores.
Polar growth of Streptomyces coelicolor during vegetative growth has been recently reviewed [41, 42] and is dependent on the presence of DivIVA. DivIVA is an essential protein, localizes to the sites of nascent peptidoglycan synthesis in the hyphal tips and lateral branches, and overexpression of DivIVA causes bulging at the growing tips of the hyphae (Figure 1) [43]. Furthermore, small foci of DivIVA along the lateral walls mark the future sites for hyphal branching indicating that the presence of DivIVA triggers peptidoglycan synthesis [44]. Notably, this observation indicates that DivIVA promotes the creation of new cell poles from the hyphal sidewall, rather than the sites of cell division.
In addition to DivIVA, a eukaryotic intermediate filament-like protein is involved in tip extension during vegetative growth. FilP forms filaments in vitro, and in vivo the FilPEGFP filaments localize in patches throughout the hyphae [45]. Deletion of FilP causes branching and swelling at the tips of the hyphae suggesting that this protein has a role in regulating peptidoglyan synthesis at the hyphal tips [45].
2.1.4 Common themes in Actinobacterial growth
Cell growth in the Actinobacteria requires precise spatial and temporal targeting of peptidoglycan synthesis. Cell growth requires two distinct phases, polar cell elongation and mid-cell septum formation for cell division. DivIVA, is a key player in the localization and regulation of peptiodoglycan synthesis machinery for cell elongation. The essential protein kinases proteins PknA and PknB modulate the activity of some enzymes involved in peptidoglycan synthesis, suggesting that phosphorylation is likely to be an important means of regulating the timing and location of new peptidoglycan synthesis. Finally, intermediate filament-like proteins likely serve as cytoskeletons and are involved in maintenance of cell shape and polar growth. While many questions remain regarding the mechanisms underlying polar growth within the Actinobacteria, it is clear that common themes are emerging.
2.2 Polar growth in the Alphaproteobacteria
Bacteria exhibiting polar growth are broadly distributed among the Alphaproteobacteria. The Rhizobiales and Caulobacterales clades of Alphaproteobacteria contain genera of classically defined budding bacteria in which polar growth mediates cell elongation In addition, some Alphaproteobacteria in which cell elongation occurs laterally along the sidewall of the cell also produce stalks, requiring a specialized form of focal growth. is required to allow stalk synthesis to occur at specific cellular locations. This section will discuss what is known (and largely unknown) about polar growth for cell elongation and stalk production in the Alphaproteobacteria.
2.2.1 Polar growth among bacteria belonging to the Rhizobiales
Polar growth within the Rhizobiales has been observed primarily within the Hyphomicrobiaceae and Bradyrhizobiaceae families. The genera Hyphomicrobium, Rhodomicrobium, Filomicrobium, Pedomicrobium, and Rhodopseudomonas are comprised of stalked bacteria that grow by budding [46]. Budding bacteria produce daughter cells that are initially smaller than the mother cell and typically increase in diameter during cell growth. Daughter cells are comprised entirely of newly synthesized cell wall material and daughter cell synthesis is usually complete before migration of DNA occurs. Simple observations of these bacteria reveal that the emergence of new cell growth occurs within or at the tips of the stalks [46–48]. In contrast, Prosthecomicrobium and Ancalomicrobium are stalked bacteria that grow by budding; however, new cell growth emerges from the cell body of the mother cell [49]. Since genetic systems are lacking or limited for these bacteria, the mechanisms underlying budding in the Rhizobiales remain unknown.
2.2.1.1 Polar growth in bacteria that bud through stalks
Many of the budding bacteria initially produce a stalk (also called hyphae or prosthecae) at one cell pole and the daughter cell emerges from the tip of the stalk. The striking cell morphologies of the stalked bacteria belonging to the family Hyphomicrobiacae (Figure 1) suggest that these bacteria exhibit polar growth. The most well-studied examples of budding stalked bacteria within the Rhizobiales are Hyphomicrobium [47] and Rhodomicrobium [48], which will be the focus of this section.
Growth of the stalk occurs prior to daughter cell synthesis in Hyphomicrobium and Rhodomicrobium. Hyphomicrobium typically produces a single stalk at one cell pole and a single daughter cell whereas Rhodomicrobium often produces stalks at both poles and these stalks often branch allowing the production of multiple daughter cells (Figure 1) [46–48]. During stalk elongation there is no additional growth of the main cell body [48, 50] and stalk elongation ceases before daughter cell synthesis is initiated. Stalk synthesis resumes from the mother cell-to-stalk junction after division of the daughter cell, indicating that the site of growth alternates between the base and the tip of the stalk at every cell cycle. The presence of a stalk appears to be essential for daughter cell synthesis as budding has not been observed in mother cells without stalks [50, 51].
Following the completion of stalk elongation, the initiation of daughter cell synthesis is observed as a swelling at the tip of the stalk. It remains unknown if growth occurs at the junction of the stalk and cell body, is dispersed throughout the daughter cell, or occurs at the cell pole opposite the stalk. Growth of the daughter cell continues until the cell is similar in size to the mother cell, and then a septum is produced in the stalk, near the daughter cell body [47, 48]. After daughter cell release, a brief period of stalk elongation occurs before the next bud is formed [46, 50]. Both Hyphomicrobium [47] and Rhodomicrobium [48] cells can produce a finite number of daughter cells suggesting that directional growth in which the daughter cell is the recipient of the newly synthesized cell material may adversely impact the reproductive capacity of mother cells (see Section 3.2).
The observations described in this section suggest that growth of bacteria that bud through the stalk requires cell growth at three distinct foci and times: at the junction of the cell body and stalk to allow stalk elongation; at the tip of the stalk to allow daughter cell production; and within the stalk to allow septum formation. The three forms of cell growth appear to be mutually exclusive and are likely to be tightly regulated. While the mechanism and regulation of cell growth in these bacteria remain unknown, it is clear that the bacteria are capable of directional cell growth restricted to specific foci.
2.2.1.2 Polar growth in bacteria that bud through the cell body
The genera Ancalomicrobium and Prosthecomicrobium are comprised of stalked bacteria that grow by budding; however, daughter cells emerge from the cell body of the mother cell rather than the tips of the stalks [49, 52–54]. The complex morphologies of these bacteria (Figure 1) are remarkable, and it is of interest to consider how cell growth can generate these shapes. Ancalomicrobium adetum cells have several long stalks distributed around the cell body, whereas Prosthecomicrobium cells have numerous short stalks surrounding the cell body (Figure 1). The stalks of these bacteria are similar in structure to the stalks of Hyphomicrobium and Rhodomicrobium and are true extensions of the cell body. Despite the morphological differences between A. adetum and Prosthecomicrobium cells, growth appears to be similar [49, 52–54]. Daughter cells emerge repeatedly from a single non-stalked location on the mother cell, typically a cell pole. New stalks are formed de novo on the daughter cells prior to cell division, which occurs when the daughter cell is approximately equal in size to the mother cell.
Unlike most described species of Prosthecomicrobium, P. hirschii produces two distinct morphotypes; a long-stalked cell morphologically similar to Ancalomicrobium and a short-stalked cell typical of the Prosthecomicrobium genus [52]. Both morphotypes grow by budding and daughter cells emerge from the cell pole. Remarkably, daughter cells can form either morphotype, irrespective of the mother cell morphology. The morphology of the mature cells is stable, suggesting that the only way to change morphology is by the production of a daughter cell with a different morphotype. Mother cells typically produce the daughter cells of one morphotype, irrespective of their own morphotype, for several generations.
The inclusion of bacteria within the genus Prosthecomicrobium was initially based largely on morphology. Recent phylogenetic analyses indicate that the genus Prosthecomicrobium is polyphyletic [54, 55], suggesting that the presence of the short-stalked morphology of these bacteria is a poor indicator of species relatedness. Several Prosthecomicrobium species have been reclassified as belonging to either of two novel genera, Vasilyevaea or Bauldia [55]. It is worth noting that these genera are broadly distributed within the order Rhizobiales, suggesting that the ability to grow by budding may be widespread.
2.2.1.3 Does polar growth occur in the Rhizobiaceae and Brucellaceae families?
While polar growth has not been demonstrated for other families within the Rhizobiales, there are indications suggesting that members of the Rhizobiaceae and Brucellaceae families exhibit polar growth. Known modulators of cell elongation, including MreB and DivIVA, are absent from the genomes of sequenced bacteria belonging to these families, suggesting that cell elongation in these bacteria occurs by a mechanism different from either of the two described for rod-shaped bacteria [8, 56]. In addition, mutants have been identified in Brucella abortus [57, 58], Sinorhizobium meliloti [59, 60], and Agrobacterium tumefaciens [61–64] that have branching morphologies. The presence of branching morphologies in mutants of these bacteria may indicate polar or focal growth, as most bacteria with a dispersed mode of cell elongation tend to generate filamentous rather than branched cells. Further support for polar growth of these bacteria is provided by the observation that Sinorhizobium meliloti and Agrobacterium tumefaciens cells, when blocked for cell division, have a strong tendency to form branches [65]. However, branch placement does not appear to be random; branches tend to emerge from the cell pole or the mid-cell, resulting in the production of Y- or T-shaped cells, respectively. It seems quite plausible that the new growth following the cell division block occurs at previous sites of cell growth, suggesting that polar growth may be responsible for cell elongation in these bacteria.
2.2.2 Polar growth among bacteria belonging to the Caulobacterales
Many bacteria belonging to the Caulobacteraceae and Hyphomonadaceae families exhibit polar growth. Bacteria within these families are dimorphic; asymmetric cell division gives rise to two morphologically distinct daughter cells. Within the Hyphomonadaceae, bacteria belonging to the genera Hyphomonas and Hirchia have striking morphological similarity to Hyphomicrobium (See Figure 1 and Section 2.2.1.1). Hyphomonas and Hirchia produce single polar stalks, and daughter cells emerge from the tip of the stalks, suggesting that cell elongation is mediated by polar growth. Within the Caulobacteraceae, many bacteria, including those belonging to the genera Caulobacter and Asticcacaulis, have stalks. The stalks are not reproductive structures and polar growth is not involved in cell elongation, but rather is responsible for stalk synthesis. This section highlights the role of polar growth in cell elongation of Hyphomonas and Hirchia and stalk synthesis of Caulobacter.
2.2.2.1 Polar growth in the budding bacteria Hyphomonas and Hirschia
Much like stalked, budding rhizobiales described above, bacteria of the genera Hyphomonas and Hirschia reproduce by the production of a stalkless daughter bud at the tip of the parent cell stalk [66] (Figure 1). Observations of Hyphomonas neptunium indicate that growth similarly occurs in three spatiotemporally distinct phases: stalk elongation in immature cells, bud formation from the tip of the reproductive cell stalk, and septation to release the immature cell following bud maturation [67]. The transition from stalk elongation to bud formation coincides with the initiation of DNA replication [67], indicating a correspondence between morphological growth transitions and key events in the life cycle.
Detailed electron microscopic analysis of Hyphomonas sp. strains VP-6 and MHS-3 provides insight into the mechanics behind the polar growth process driving bud formation [68]. The cytoplasmic membrane of the mature stalked cell forms a “membrane cap” that partitions the cell body cytoplasm from that of the stalk and the forming bud. A chain of connected, cytoplasmic membrane-bound “pseudovesicles” extends from the membrane cap and into the nascent daughter bud. Though macromolecule movement has not been tracked in live cells, it appears that pseudovesicles mediate translocation of ribosomes and DNA from the mother cell to the bud, thereby providing some of the components required to assemble a viable daughter cell at the distal end of the stalk. These observations offer some insight into how bacteria handle the complexities involved in elaborating a bud spatially removed from the mother cell body. The hyphomonads thus offer a promising model for additional study into highly asymmetric, polarized patterns of growth and reproduction.
2.2.2.2 Polar growth allows stalk formation in Caulobacter
The well-studied bacterium Caulobacter crescentus has a single polar stalk that is a true extension of the cell body and is not involved in reproduction [69]. Interestingly, the localization of nascent peptidoglycan synthesis follows a distinct pattern throughout cell growth [70, 71]. In immature cells, peptidoglycan synthesis occurs laterally along the sidewall. Lateral cell growth likely continues throughout the cell cycle; however, bursts of peptidoglycan synthesis occur at the cell pole to accommodate stalk synthesis and later at the mid-cell of predivisional cells to allow constriction and cell division.
Lateral cell elongation is mediated by MreB, which positions peptidoglycan synthesis proteins in a helical pattern along the sidewall of the cells [71, 72]. Zonal insertion of the newly synthesized peptidoglycan occurs at the mid-cell prior to constriction and is dependent on FtsZ, which recruits at least one peptidoglycan synthesis protein to the mid-cell [70]. A shift from lateral to mid-cell peptidoglycan insertion as the predominant mode of cell elongation occurs when the FtsZ ring is assembled at the mid-cell [70, 71]. In C. crescentus, FtsZ ring assembly occurs before constriction, and zonal growth at the mid-cell is likely to contribute to both cell elongation and division.
In addition to mediating cell elongation and division, targeting of peptidoglycan synthesis machinery is required to enable stalk synthesis. Stalks contain newly synthesized peptidoglycan, which is produced at the junction of the cell pole and the stalk [2, 70, 71]. Interestingly, MreB is required for stalk synthesis [73] and specifically for peptidoglycan synthesis at the cell pole [71]. FtsZ had also been shown to be involved in peptidoglycan synthesis at the cell pole during stalk synthesis [71]. Stalks normally contain crossbands, transverse discs that cross the inner membranes and peptidoglycan and are likely comprised of peptidoglycan [69, 74]. The stalks of cells depleted of FtsZ do not contain crossbands, suggesting that FtsZ is required for transverse synthesis of peptidoglycan at the base of the cell.
These data indicate that the mode of cell growth (i.e. lateral, septal, or polar) may be a consequence of the localization of the protein(s) that target the peptidoglycan synthesis machinery. Thus, it is quite possible that the insertion of peptidoglycan for cell elongation in bacteria occurs by a conserved mechanism but is targeted to different locations (see Section 3.1).
2.3 Planctomycetes
Members of the Planctomycetes and the related Chlamydiae [75] represent a group of related bacteria anciently diverged from the other bacteria considered above, and several distinctive features attest to their separate evolutionary history. Most notably, both planctomycetes [76] and chlamydiae [77–79] lack peptidoglycan. Instead, a proteinaceous cell wall gives the cell envelope its structural integrity [76, 80], with extensive amino acid cross-linking providing the required strength [80–83]. Thus, planctomycetes and chlamydiae utilize cross-linked protein instead of peptidoglycan in a cell wall that nonetheless provides the required strength for maintaining cell shape and integrity.
Planctomycetes exhibit striking morphological features pertinent to polar and asymmetric growth patterns. Though relatively difficult to culture, planctomycetes occur quite frequently in various environments [84, 85]. They are particularly common in surface-associated bacterial communities [86–89], and many planctomycete species grow attached to surfaces via a holdfast anchored at the tip of a long stalk [4] (Figure 1). Much as in other stalked Alphaproteobacteria (caulobacters, hyphomonads, and prosthecate rhizobia), daughter cells are born stalkless with a polar or subpolar flagellum, then later develop a stalk at or near the flagellar pole. The stalk defines a specific zone of growth that is necessarily polar; though many planctomycetes exhibit rounded cell bodies without obvious morphological polarity, stalk biogenesis is polar with respect to the axis established by daughter cell production at the opposite cell body pole. Furthermore, planctomycetes reproduce by budding of the daughter cell (Figure 1) via polar, de novo synthesis of the cell envelope at the site of the daughter bud [4, 90]. This pattern stands in contrast to the zonal but non-polar cell growth observed at the central septum in other round bacteria like Staphylococcus aureus and Streptococcus pneumoniae [20, 22].
We are unaware of any studies into the mechanistic basis of cell wall growth in planctomycetes. However, genomic analysis of planctomycetes and the related chlamydiae indicates at least some conservation of genes responsible for synthesizing and shaping peptidoglycan in other bacteria, including mreB and several muramidase genes [8, 15, 91]. Interestingly, chlamydiae also express several penicillin binding proteins, and sensitivity to antibiotics targeting these proteins suggests they retain a critical functional role, possibly in cross-linking the proteinaceous sacculus [6, 77, 78].
3 Implications of polar growth
3.1 Zonal cell wall synthesis facilitates polar and asymmetric growth
How did polar and asymmetric growth patterns evolve among bacteria? The progenitor of the bacterial lineage was likely rod-shaped [92], though uncertainties regarding the exact ordering of deep-branching lineages in the bacterial phylogeny, as well as limited knowledge of growth pattern details outside of well-studied model systems, preclude speculation about whether cell wall synthesis was originally polar. Regardless, zonal growth permits a ready switch from non-polar to polar cell wall synthesis (and vice versa) even over the life cycle of single bacteria, e.g. in corynebacteria where polar cell elongation alternates with mid-cell septal growth as coordinated by DivIVA (see Section 2.1.1). Zonal growth therefore potentiates a ready evolutionary transition between non-polar and polar modes of growth via the particular localization pattern of cell wall synthesis (Figure 2B, 2D). Contrasting peptidoglycan synthesis along sidewalls during elongation of Bacillus and Escherichia (Section 1.3) with polar elongation in Corynebacterium and Streptomyces (Section 2.1) suggests just such an evolutionary event (Figure 1). Similarly, zonal growth readily generates morphological and reproductive asymmetries: unipolar stalk formation and budding modes of reproduction both result from targeting zonal cell wall synthesis to specific polar regions (Figure 2E 2F). Interestingly, the bacterial phylogeny suggests the independent emergence of stalked cell morphologies at least three times: in planctomycetes, in the stalked Rhizobiaceae (e.g. Hyphomicrobium), and in the Caulobacter/Hyphomonas clade (Figure 1). Thus, zonal cell wall synthesis facilitates ready transitions between various patterns of growth, leading to potentially diverse cell shapes and modes of reproduction simply by modulating the location and timing of growth zones in the cell.
3.2 Why zonal and polar growth?
The apparent ubiquity of zonal growth among bacteria suggests an adaptive advantage to this mode of cell wall synthesis. Considering the numerous distinct proteins required to catalyze the various polymerization and hydrolysis steps involved in peptidoglycan synthesis and remodeling (summarized in [93]), a simple energetic argument proves compelling. Coordinating synthesis on a spatially restricted scaffold greatly enhances the degree of interaction between the components of the machinery and minimizes the number of energetically expensive proteins that must be produced by the cell.
At a fundamental level, cell wall synthesis in defined zonal patterns facilitates the generation of diverse shapes and features. From rod-shaped cell bodies to elaborate stalks and spikes, these shapes figure prominently into bacterial adaptation to the environment [1]. For example, the polar stalk of C. crescentus functions in nutrient uptake from the environment. Enhanced stalk growth under conditions of low phosphate permit the bacterium to scavenge limiting phosphate more efficiently [2, 3], providing a link between directed, polar cell wall growth and the cell's fitness in the current environment.
Zonal growth may also enable independent regulation of specific modes of growth. For example, in H. influenzae and E. coli, distinct peptidoglycan synthetic complexes function in H. influenzae elongation and division at different times in the life cycle [14, 16, 94, 95]. Zonal growth through interactions between different enzyme complexes and their unique scaffolds could help to ensure the proper type, location, and timing of new peptidoglycan incorporation. Observations of specific interactions between cytoskeletal scaffolding proteins and peptidoglycan synthesis enzymes [12, 71, 72, 96] support this notion.
Finally, zonal growth tends to generate reproductive asymmetries that may prove advantageous in purging accumulated cellular damage via cell “aging.” By bundling damaged macromolecules into an “old” cell upon division, bacterial lineages may persist through the attendant rejuvenation of “young” cells under conditions where damage accumulation would drive to extinction populations where damage is partitioned equally [97]. Even in rod-shaped bacteria like E. coli and B. subtilis with apparent morphologically symmetric division by binary fission, zonal cell wall synthesis at the septum defines a young pole distinct from the extant, old cell pole with its inert peptidoglycan [7, 9, 98], and the poles themselves are indeed morphologically asymmetric [99]. Tracking these cells through time, an aging cell that repeatedly inherits the old pole at each division shows a concomitant increase in damage accumulation and decrease in growth rate [100, 101]. Notably, while zonal growth serves to establish polar asymmetry in morphologically symmetric bacteria, even stronger reproductive asymmetry should occur with asymmetric polar growth as observed in the various budding bacteria described earlier in this review. Despite reports of Hyphomicrobium and Rhodomicrobium cells ceasing to divide after producing a limited number of buds [47, 48], aging in these organisms awaits detailed study.
4. Perspectives
Bacterial cell growth is, in essence, mediated by spatiotemporal regulation of peptidoglycan synthesis. In many rod-shaped bacteria for which growth has been studied in detail, MreB-like proteins comprise a helical scaffold that drives the insertion of new peptidoglycan along the lateral cell wall. Notably, the insertion of peptidoglycan occurs not at random locations, but rather at discrete positions. While often viewed as an exception to the rule, it is becoming increasingly clear that many rod-shaped bacteria employ a mode of cell elongation in which the insertion of peptidoglycan occurs at constrained locations, such as the cell pole. Both helical and polar growth require the targeting of peptidoglycan synthesis machinery to specific cellular locations, such that both modes of growth can be accurately described as zonal.
In this review, we have highlighted the prevalence of obligate polar growth within three large and anciently diverged clades of bacteria: Actinobacteria, Alphaproteobacteria, and Planctomycetes. Within these groups, the targeting of cell wall synthesis machinery to the cell pole occurs by diverse mechanisms. While DivIVA is involved in recruiting the peptidoglycan synthesis machinery to the cell pole in the Actinobacteria, homologs of this protein are abent in the Alphaproteobacteria and Planctomycetes. This observation supports the hypothesis that polar growth evolved independently and repeatedly within the bacteria. Given that the details of cell wall synthesis have only been explored in a limited subset of bacterial taxa, the prevalence of polar growth among bacteria remains to be determined. Indeed, its occurrence among groups spanning such a broad phylogenetic spectrum suggests that further study may identify ancient and widely inherited mechanisms of polar growth.
Why has polar growth evolved among the bacteria? It seems likely that this mode of growth confers an advantage in particular environments. Polar growth has allowed diverse bacterial cell shapes to emerge, including the stalked bacteria, where the presence of the stalks may help bacteria to survive in oligotrophic environments by increasing nutrient uptake. In addition, the asymmetric allocation of newly synthesized peptidoglycan to the daughter cell may serve to rejuvenate nascent cells and prevent the accumulation of molecular damage in a cell lineage. Further research into the details of cell wall growth in a more phylogenetically diverse set of organisms will better inform our understanding of the prevalence, evolutionary history, and ecological importance of polar and other zonal growth patterns in bacteria.
Acknowledgements
We thank Clay Fuqua, Chao Jiang, H. Velocity Hughes and Cecile Berne for critical reading of the manuscript. Y.V.B. was funded by the National Institutes of Health (GM051986 and GM077648) and by the National Science Foundation (MCB0731950). P.B. and D.K. were supported by National Institutes of Health National Research Service Awards AI072992 and GM083581, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schmidt JM, Stanier RY. The development of cellular stalks in bacteria. J Cell Biol. 1966;28:423–36. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad Sci U S A. 2006;103:11772–7. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ward N, Staley J, Fuerst J, Giovannoni S, Schlesner H, Stackebrandt E. In: The Order Planctomycetales, Including the Genera Planctomyces, Pirellula, Gemmata and Isosphaera and the Candidatus Genera Brocadia, Kuenenia and Scalindua. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes: Springer; New York: 2006. pp. 757–93. [Google Scholar]
- [5].Kalayoglu M, Byrne G. In: The Genus Chlamydia—Medical. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes: Springer; New York: 2006. pp. 741–54. [Google Scholar]
- [6].Ghuysen JM, Goffin C. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob Agents Chemother. 1999;43:2339–44. doi: 10.1128/aac.43.10.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mobley HL, Koch AL, Doyle RJ, Streips UN. Insertion and fate of the cell wall in Bacillus subtilis. J Bacteriol. 1984;158:169–79. doi: 10.1128/jb.158.1.169-179.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–76. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- [9].de Pedro MA, Quintela JC, H√∂ltje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–34. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schlaeppi JM, Schaefer O, Karamata D. Cell wall and DNA cosegregation in Bacillus subtilis studied by electron microscope autoradiography. J Bacteriol. 1985;164:130–5. doi: 10.1128/jb.164.1.130-135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–32. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- [13].Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987;169:4935–40. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alaedini A, Day RA. Identification of two penicillin-binding multienzyme complexes in Haemophilus influenzae. Biochem Biophys Res Commun. 1999;264:191–5. doi: 10.1006/bbrc.1999.1509. [DOI] [PubMed] [Google Scholar]
- [15].Jones LJ, Carballido-L√≥pez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–22. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- [16].Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen JC, Weiss DS, Ghigo JM, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol. 1999;181:521–30. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weiss DS, Pogliano K, Carson M, Guzman LM, Fraipont C, Nguyen-Disteche M, et al. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–81. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- [19].Errington J, Daniel RA, Scheffers D-J. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pinho MG, Errington J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol. 2003;50:871–81. doi: 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- [21].Lara B, Rico AI, Petruzzelli S, Santona A, Dumas J, Biton J, et al. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol. 2005;55:699–711. doi: 10.1111/j.1365-2958.2004.04432.x. [DOI] [PubMed] [Google Scholar]
- [22].Morlot C, Zapun A, Dideberg O, Vernet T. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol Microbiol. 2003;50:845–55. doi: 10.1046/j.1365-2958.2003.03767.x. [DOI] [PubMed] [Google Scholar]
- [23].Briles EB, Tomasz A. Radioautographic evidence for equatorial wall growth in a gram-positive bacterium. Segregation of choline-3H-labeled teichoic acid. J Cell Biol. 1970;47:786–0. doi: 10.1083/jcb.47.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Umeda A, Amako K. Growth of the surface of Corynebacterium diphtheriae. Microbiol Immunol. 1983;27:663–71. doi: 10.1111/j.1348-0421.1983.tb00629.x. [DOI] [PubMed] [Google Scholar]
- [25].Letek M, Ordonez E, Vaquera J, Margolin W, Flardh K, Mateos LM, et al. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol. 2008;190:3283–92. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ramos A, Honrubia MP, Valbuena N, Vaquera J, Mateos LM, Gil JA. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum. Microbiology. 2003;149:3531–42. doi: 10.1099/mic.0.26653-0. [DOI] [PubMed] [Google Scholar]
- [27].Valbuena N, Letek M, Ordonez E, Ayala J, Daniel RA, Gil JA, et al. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol Microbiol. 2007;66:643–57. doi: 10.1111/j.1365-2958.2007.05943.x. [DOI] [PubMed] [Google Scholar]
- [28].Letek M, Fiuza M, Ordonez E, Villadangos AF, Flardh K, Mateos LM, et al. DivIVA uses an N-terminal conserved region and two coiled-coil domains to localize and sustain the polar growth in Corynebacterium glutamicum. FEMS Microbiol Lett. 2009;297:110–6. doi: 10.1111/j.1574-6968.2009.01679.x. [DOI] [PubMed] [Google Scholar]
- [29].Fiuza M, Letek M, Leiba J, Villadangos AF, Vaquera J, Zanella-Cleon I, et al. Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shaped morphology in Corynebacterium glutamicum. J Biol Chem. 2010;285:29387–97. doi: 10.1074/jbc.M110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fiuza M, Canova MJ, Zanella-Cleon I, Becchi M, Cozzone AJ, Mateos LM, et al. From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J Biol Chem. 2008;283:18099–112. doi: 10.1074/jbc.M802615200. [DOI] [PubMed] [Google Scholar]
- [31].Fiuza M, Canova MJ, Patin D, Letek M, Zanella-Cleon I, Becchi M, et al. The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum. J Biol Chem. 2008;283:36553–63. doi: 10.1074/jbc.M807175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chauhan A, Lofton H, Maloney E, Moore J, Fol M, Madiraju MV, et al. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol Microbiol. 2006;62:132–47. doi: 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- [33].Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–35. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- [34].Nguyen L, Scherr N, Gatfield J, Walburger A, Pieters J, Thompson CJ. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J Bacteriol. 2007;189:7896–910. doi: 10.1128/JB.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hamasha K, Sahana MB, Jani C, Nyayapathy S, Kang CM, Rehse SJ. The effect of Wag31 phosphorylation on the cells and the cell envelope fraction of wild-type and conditional mutants of Mycobacterium smegmatis studied by visible-wavelength Raman spectroscopy. Biochem Biophys Res Commun. 2010;391:664–8. doi: 10.1016/j.bbrc.2009.11.117. [DOI] [PubMed] [Google Scholar]
- [36].Jani C, Eoh H, Lee JJ, Hamasha K, Sahana MB, Han JS, et al. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 2010;10:327. doi: 10.1186/1471-2180-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Thakur M, Chakraborti PK. Ability of PknA, a mycobacterial eukaryotic-type serine/threonine kinase, to transphosphorylate MurD, a ligase involved in the process of peptidoglycan biosynthesis. Biochem J. 2008;415:27–33. doi: 10.1042/BJ20080234. [DOI] [PubMed] [Google Scholar]
- [39].Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J Mol Biol. 2009;386:451–64. doi: 10.1016/j.jmb.2008.12.031. [DOI] [PubMed] [Google Scholar]
- [40].Elliot MA, Buttner MJ, Nodwell JR. Multicellular Development in Streptomyces. In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. ASM Press; Washington, D.C.: 2008. pp. 419–38. [Google Scholar]
- [41].Flardh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- [42].Flardh K. Cell polarity and the control of apical growth in Streptomyces. Curr Opin Microbiol. 2010;13:758–65. doi: 10.1016/j.mib.2010.10.002. [DOI] [PubMed] [Google Scholar]
- [43].Flardh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol Microbiol. 2003;49:1523–36. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- [44].Hempel AM, Wang SB, Letek M, Gil JA, Flardh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol. 2008;190:7579–83. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bagchi S, Tomenius H, Belova LM, Ausmees N. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol Microbiol. 2008;70:1037–50. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hirsch P. Budding bacteria. Annu Rev Microbiol. 1974;28:391–444. doi: 10.1146/annurev.mi.28.100174.002135. [DOI] [PubMed] [Google Scholar]
- [47].Moore RL. The biology of Hyphomicrobium and other prosthecate, budding bacteria. Annu Rev Microbiol. 1981;35:567–94. doi: 10.1146/annurev.mi.35.100181.003031. [DOI] [PubMed] [Google Scholar]
- [48].Whittenbury R, Dow CS. Morphogenesis and differentiation in Rhodomicrobium vannielii and other budding and prosthecate bacteria. Bacteriol Rev. 1977;41:754–808. doi: 10.1128/br.41.3.754-808.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Staley JT. Prosthecomicrobium and Ancalomicrobium: new prosthecate freshwater bacteria. J Bacteriol. 1968;95:1921–42. doi: 10.1128/jb.95.5.1921-1942.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Moore RL, Hirsch P. First generation synchrony of isolated Hyphomicrobium swarmer populations. J Bacteriol. 1973;116:418–23. doi: 10.1128/jb.116.1.418-423.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Murray RG, Douglas HC. The reproductive mechanism of Rhodomicrobium vannielii and the accompanying nuclear changes. J Bacteriol. 1950;59:157–67. doi: 10.1128/jb.59.2.157-167.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Staley JT. Prosthecomicrobium hirschii, a New Species in a Redefined Genus. Int J Syst Bacteriol. 1984;34:304–8. [Google Scholar]
- [53].Jenkins C, Stanley PM, Stanley JT. Genus Ancalomicrobium. In: Brenner DJ, Krieg NR, Stanley JT, Garrity GM, editors. Bergey's Manual of Systematic Bacteriology. Williams & Wilkins; Baltimore: 2005. pp. 494–7. [Google Scholar]
- [54].Jenkins C, Rainey FA, Ward NL, Stanley JT. Genus Prosthecomicrobium. In: Brenner DJ, Krieg NR, Stanley JT, Garrity GM, editors. Bergey's Manual of Systematic Bacteriology. Williams & Wilkins; Baltimore: 2005. pp. 538–43. [Google Scholar]
- [55].Yee B, Oertli GE, Fuerst JA, Staley JT. Reclassification of the polyphyletic genus Prosthecomicrobium to form two novel genera, Vasilyevaea gen. nov. and Bauldia gen. nov. with four new combinations: Vasilyevaea enhydra comb. nov., Vasilyevaea mishustinii comb. nov., Bauldia consociata comb. nov. and Bauldia litoralis comb. nov. Int J Syst Evol Microbiol. 2010;60:2960–6. doi: 10.1099/ijs.0.018234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–22. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Robertson GT, Reisenauer A, Wright R, Jensen RB, Jensen A, Shapiro L, et al. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J Bacteriol. 2000;182:3482–9. doi: 10.1128/jb.182.12.3482-3489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol. 2002;43:945–60. doi: 10.1046/j.1365-2958.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- [59].Kobayashi H, De Nisco NJ, Chien P, Simmons LA, Walker GC. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol Microbiol. 2009;73:586–600. doi: 10.1111/j.1365-2958.2009.06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cheng J, Sibley CD, Zaheer R, Finan TM. A Sinorhizobium meliloti minE mutant has an altered morphology and exhibits defects in legume symbiosis. Microbiology. 2007;153:375–87. doi: 10.1099/mic.0.2006/001362-0. [DOI] [PubMed] [Google Scholar]
- [61].Fujiwara T, Fukui S. Unidirectional growth and branch formation of a morphological mutant, Agrobacterium tumefaciens. J Bacteriol. 1974;120:583–9. doi: 10.1128/jb.120.2.583-589.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Su S, Stephens BB, Alexandre G, Farrand SK. Lon protease of the alpha-proteobacterium Agrobacterium tumefaciens is required for normal growth, cellular morphology and full virulence. Microbiology. 2006;152:1197–207. doi: 10.1099/mic.0.28657-0. [DOI] [PubMed] [Google Scholar]
- [63].Fujiwara T, Fukui S. Isolation of morphological mutants of Agrobacterium tumefaciens. J Bacteriol. 1972;110:743–6. doi: 10.1128/jb.110.2.743-746.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kahng LS, Shapiro L. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J Bacteriol. 2001;183:3065–75. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Latch JN, Margolin W. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J Bacteriol. 1997;179:2373–81. doi: 10.1128/jb.179.7.2373-2381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Poindexter JS. Dimorphic Prosthecate Bacteria: The Genera Caulobacter, Asticcacaulis, Hyphomicrobium, Pedomicrobium, Hyphomonas and Thiodendron. In: Dworkin M, Falkow S, editors. The prokaryotes : a handbook on the biology of bacteria. 3rd ed. Springer; New York: 2006. pp. 72–90. [Google Scholar]
- [67].Wali TM, Hudson GR, Danald DA, Weiner RM. Timing of swarmer cell cycle morphogenesis and macromolecular synthesis by Hyphomicrobium neptunium in synchronous culture. J Bacteriol. 1980;144:406–12. doi: 10.1128/jb.144.1.406-412.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zerfas PM, Kessel M, Quintero EJ, Weiner RM. Fine-structure evidence for cell membrane partitioning of the nucleoid and cytoplasm during bud formation in Hyphomonas species. J Bacteriol. 1997;179:148–56. doi: 10.1128/jb.179.1.148-156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–95. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–52. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- [71].Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–88. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- [72].White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–33. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- [73].Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J Bacteriol. 2005;187:544–53. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jones HC, Schmidt JM. Ultrastructural study of crossbands occurring in the stalks of Caulobacter crescentus. J Bacteriol. 1973;116:466–70. doi: 10.1128/jb.116.1.466-470.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Teeling H, Lombardot T, Bauer M, Ludwig W, Glockner FO. Evaluation of the phylogenetic position of the planctomycete Rhodopirellula baltica SH 1 by means of concatenated ribosomal protein sequences, DNA-directed RNA polymerase subunit sequences and whole genome trees. Int J Syst Evol Microbiol. 2004;54:791–801. doi: 10.1099/ijs.0.02913-0. [DOI] [PubMed] [Google Scholar]
- [76].König E, Schlesner H, Hirsch P. Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol. 1984:200–5. [Google Scholar]
- [77].Barbour AG, Amano K, Hackstadt T, Perry L, Caldwell HD. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J Bacteriol. 1982;151:420–8. doi: 10.1128/jb.151.1.420-428.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].How SJ, Hobson D, Hart CA. Studies in vitro of the nature and synthesis of the cell wall of Chlamydia trachomatis. Curr Microbiol. 1984;10:269–74. [Google Scholar]
- [79].Fox A, Rogers JC, Gilbart J, Morgan S, Davis CH, Knight S, et al. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect Immun. 1990;58:835–7. doi: 10.1128/iai.58.3.835-837.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hatch TP. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J Bacteriol. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Liesack W, König H, Schlesner H, Hirsch P. Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch Microbiol. 1986:361–6. [Google Scholar]
- [82].Newhall VWJ, Jones RB. Disulfide-Linked Oligomers of the Major Outer Membrane Protein of Chlamydiae. J Bacteriol. 1983;154:998–1001. doi: 10.1128/jb.154.2.998-1001.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Melgosa MP, Kuo CC, Campbell LA. Outer membrane complex proteins of Chlamydia pneumoniae. FEMS Microbiol Lett. 1993;112:199–204. doi: 10.1111/j.1574-6968.1993.tb06448.x. [DOI] [PubMed] [Google Scholar]
- [84].Morris RM, Longnecker K, Giovannoni SJ. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol. 2006;8:1361–70. doi: 10.1111/j.1462-2920.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- [85].Buckley DH, Huangyutitham V, Nelson TA, Rumberger A, Thies JE. Diversity of Planctomycetes in Soil in Relation to Soil History and Environmental Heterogeneity. Appl Environ Microbiol. 2006;72:4522–31. doi: 10.1128/AEM.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hempel M, Blume M, Blindow I, Gross EM. Epiphytic bacterial community composition on two common submerged macrophytes in brackish water and freshwater. BMC Microbiol. 2008;8:58. doi: 10.1186/1471-2180-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].DeLong EF, Franks DG, Alldredge AL. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–34. [Google Scholar]
- [88].Longford SR, Tujula NA, Crocetti GR, Holmes AJ, Holmstr√∂m C, Kjelleberg S, et al. Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat Microb Ecol. 2007;48:217–29. [Google Scholar]
- [89].Bengtsson MM, Ovreas L. Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol. 2010;10:261. doi: 10.1186/1471-2180-10-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tekniepe BL, Schmidt JM, Starr MP. Immunoferritin labeling shows de novo synthesis of surface components in buds of a prokaryote belonging to mirphotype IV of the Blastocaulis-Planctomyces group. Curr Microbiol. 1982;7:1–6. [Google Scholar]
- [91].Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer K-H, et al. Characterization and Evolution of Cell Division and Cell Wall Synthesis Genes in the Bacterial Phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and Phylogenetic Comparison with rRNA Genes. J Bacteriol. 2008;190:3192–202. doi: 10.1128/JB.01797-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Siefert JL, Fox GE. Phylogenetic mapping of bacterial morphology. Microbiology. 1998;144(Pt 10):2803–8. doi: 10.1099/00221287-144-10-2803. [DOI] [PubMed] [Google Scholar]
- [93].Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3:601–10. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- [94].Spratt BG, Pardee AB. Penicillin-binding proteins and cell shape in E. coli. Nature. 1975;254:516–7. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- [95].Spratt BG. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Vollmer W, von Rechenberg M, Holtje JV. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem. 1999;274:6726–34. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- [97].Ackermann M, Chao L, Bergstrom CT, Doebeli M. On the evolutionary origin of aging. Aging Cell. 2007;6:235–44. doi: 10.1111/j.1474-9726.2007.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Dworkin J. Cellular polarity in prokaryotic organisms. Cold Spring Harb Perspect Biol. 2009;1:a003368. doi: 10.1101/cshperspect.a003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Itan E, Carmon G, Rabinovitch A, Fishov I, Feingold M. Shape of nonseptated Escherichia coli is asymmetric. Phys Rev E. 2008;77:061902. doi: 10.1103/PhysRevE.77.061902. [DOI] [PubMed] [Google Scholar]
- [100].Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci U S A. 2008;105:3076–81. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]