Conspectus
Riboswitches, which were discovered in the first years of the XXI century, are gene-regulatory mRNA domains that respond to the intracellular concentration of a variety of metabolites and second messengers. They control essential genes in many pathogenic bacteria, and represent a new class of biomolecular target for the development of antibiotics and chemical-biological tools. Five mechanisms of gene regulation are known for riboswitches. Most bacterial riboswitches modulate transcription termination or translation initiation in response to ligand binding. All known examples of eukaryotic riboswitches and some bacterial riboswitches control gene expression by alternative splicing. The glmS riboswitch, widespread in Gram-positive bacteria, is a catalytic RNA activated by ligand binding. Its self-cleavage destabilizes the mRNA of which it is part. Finally, one example of trans-acting riboswitch is known. Three-dimensional (3D) structures have been determined of representatives of thirteen structurally distinct riboswitch classes, providing atomic-level insight into their mechanisms of ligand recognition. While cellular and viral RNAs in general have attracted interest as potential drug targets, riboswitches show special promise due to the diversity and sophistication of small molecule recognition strategies on display in their ligand binding pockets. Moreover, uniquely among known structured RNA domains, riboswitches evolved to recognize small molecule ligands. Structural and biochemical advances in the study of riboswitches provide an impetus for the development of methods for the discovery of novel riboswitch activators and inhibitors. Recent rational drug design efforts focused on select riboswitch classes have yielded a small number of candidate antibiotic compounds, including one active in a mouse model of Staphylococcus aureus infection. The development of high-throughput methods suitable for riboswitch-specific drug discovery is ongoing. A fragment-based screening approach employing equilibrium dialysis that may be generically useful has had early success. Riboswitch-mediated gene regulation is widely employed by bacteria; however, only the thiamine pyrophosphate-responsive riboswitch has thus far been found in eukaryotes. Thus, riboswitches are particularly attractive as targets for antibacterials. Indeed, antimicrobials with previously unknown mechanisms have been found to function by binding riboswitches and leading to aberrant gene expression.
1. RNA as a drug target
Although RNA might appear to lack chemical diversity, and thus not be a promising drug target, closer inspection reveals that it has many attractive characteristics. Like proteins, RNA can fold into intricate 3D structures with pockets and cavities that have the potential to bind ligands specifically1. Many RNAs undergo extensive structural rearrangement upon associating with small-molecules, RNAs or proteins (e.g. 2–5), and therefore exist in multiple distinct conformations that can be targeted. In addition, many cellular RNAs are subject to extensive post-transcriptional modification, further increasing their chemical versatility (reviewed in 6).
The majority of antibiotics in clinical use target the ribosome, and structures of ribosome–antibiotic complexes reveal that nearly all bind the rRNAs, rather than ribosomal proteins7, thereby validating RNA as a drug target. The aminoglycoside neomycin has been shown to bind several RNAs in addition to rRNA. It prevents charging of tRNAPhe by its cognate aminoacyl-tRNA synthetase at concentrations well below those of other aminoglycoside inhibitors8,9. It also binds the trans-activating response (TAR) and Rev response element (RRE) RNAs of HIV-1. Neomycin inhibits the interaction between TAR and the protein Tat that enhances transcription of full-length HIV-1 RNA10, and binds to RRE with KD ~100 nM to disrupt the Rev–RRE interaction that facilitates nuclear-cytoplasmic export of the viral RNA11. Neomycin has also been shown to inhibit catalytic RNAs, such as group I introns, RNase P, and the hammerhead and hepatitis delta virus ribozymes12–15, by displacing specific Mg2+ ions.
2. Discovery and characterization of riboswitches
Isolation of aptamers preceded discovery of riboswitches. Aptamers are nucleic acids selected in vitro to bind specific ligands (small molecules, nucleic acids, proteins) by subjecting pools of as many as 1015 different RNAs or DNAs to iterative rounds of selection and amplification. A variety of aptamers and catalytic nucleic acids have been discovered thus. The ligand selectivity of aptamers has led to their application as biosensors and therapeutics (reviewed in 16). In addition, several aptamers have been shown to function as small molecule-responsive in vivo genetic switches, that is, as artificial riboswitches17,18.
To date, natural riboswitches responsive to 17 different small molecule metabolites, one divalent cation, and one second messenger have been described (Table 1, reviewed in 19). Biochemical and phylogenetic analyses show that many of these have a substructure that is necessary and sufficient to bind specifically to their cognate ligand and is also typically the element with the highest sequence conservation. This is often called the ‘aptamer domain’ of the riboswitch.
Table 1.
Riboswitches with available structural information
| Class | Ligand(s) | Bound structure(s)* [X,N,S] | Free structures(s) [X,N,S]* | Fold† | Expression platform structural info | References |
|---|---|---|---|---|---|---|
| cyclic diguanylate (c-di-GMP) | c-di-GMP | X, S | S | 3HJ | - | 64,65 |
| flavin mononucleotide (FMN) | FMN | X, S | S | 6HJ | - | 3,32 |
| glmS | glucosamine-6-phosphate (GlcN6P) | X | X | 3Ψ, 3HJ | - | 66–69 |
| glycine | glycine | X, S | X, S | 3HJ | - | 70,71 |
| lysine | lysine | X, S | X, S | 5HJ | - | 3,50,72 |
| m-box (magnesium) | Mg2+ | X, S | S | 2 × 3HJ | - | 73 |
| preQ1 | 7-aminomethyl-7-deazaguanine (preQ1), 7-cyano-7-deazaguanine (preQ0) | X, N | N | Ψ | N | 74–77 |
| purine | adenine, guanine, 2′-deoxyguanosine (dG)‡, hypoxanthine | X, N | N | 3HJ | - | 51,52,78,79 |
| SAH | S-adenosylhomocysteine | X | - | Ψ | - | 80 |
| SAM-I | S-adenosylmethionine (SAM) | X, S | S | 4HJ, Ψ | - | 3,22,81 |
| SAM-II | SAM | X | - | Ψ | X | 23 |
| SAM-III (SMK box) | SAM | X, N | N, S | 3HJ | N, S | 24,82 |
| thi-box (TPP) | thiamine pyrophosphate (TPP) | X, N, S | N, S | 3HJ | - | 3,38,83–86 |
X, X-ray crystallography; N, NMR; S, SAXS (small-angle X-ray scattering).
HJ, helix-junction; Ψ, pseudoknot.
dG recognition analyzed with an engineered guanine riboswitch with reduced affinity.
In order to regulate gene expression, binding of ligand to the aptamer domain must be communicated to the machinery of gene expression. The majority of known bacterial riboswitches function by transcriptional or translational attenuation, in which the aptamer domain and either a terminator stem-loop or a Shine-Dalgarno element compete for folding and accessibility. Mechanisms unique to specific riboswitches have also been described. Eukaryotic and some bacterial riboswitches can modulate alternative splicing through ligand binding. A single bacterial Class I SAM riboswitch is trans-acting20. Finally, the glmS riboswitch undergoes ligand binding-induced self-cleavage that destabilizes the mRNA in which it resides, leading to mRNA degradation by RNase J1 (reviewed in 19,21). In most cases, transduction of the ligand binding signal to a change in gene expression relies on an RNA segment that has been termed the ‘expression platform’. Depending on the riboswitch, the expression platform can be an RNA segment downstream of the aptamer domain or can overlap partially or wholly with it.
3D structural information is available for thirteen different riboswitch classes (Table 1). The majority of studies have elucidated the structure of ligand-bound aptamer domains, although some have characterized ligand-free states or the conformation of expression platforms. Overall, the structures available at present do not suggest phylogenetic relationships between riboswitch classes, except in the instances where structurally near-identical riboswitches bind closely related ligands (e.g. the purine riboswitches, Table 1). From a purely structural standpoint, riboswitch aptamer domains have been classified based on whether they fold around a multi-helical junction, a pseudoknot, or combinations of these. Consistent with the diversity of RNA folds employed by different riboswitch classes, the molecular details of their ligand recognition also vary between classes. Even riboswitches of different classes that bind the same ligand (e.g. the SAM-binding riboswitches, Table 1) recognize their ligand differently. For instance, the SAM-I riboswitch flanks SAM with two helical stacks, the SAM-II riboswitch encapsulates SAM inside an RNA triplex, and the SAM-III riboswitch intercalates SAM in a three-way junction22–24. The riboswitches differ in their recognition of the methionine moiety of SAM: SAM-I forms extensive contacts with it, SAM-II forms fewer, and SAM-III makes no contacts.
3. Riboswitches as drug targets
Riboswitches are attractive targets for new drugs for three principal reasons. First, riboswitches evolved to recognize small molecules. Although numerous RNA domains have been experimentally targeted for drug discovery, small molecule binding by these RNAs is often fortuitous, not their physiologic function25. Ligands targeting such RNAs can exhibit poor selectivity26. Second, with the exception of the TPP riboswitch, known riboswitches occur predominantly in bacteria, not eukaryotes. If eukaryotes employ riboswitches, it is likely that these will be distinct from those of bacteria, minimizing cross-reactivity of bacterial riboswitch-targeted ligands. Third, known riboswitches respond to ubiquitous and essential metabolites and second messengers, and are often associated with mRNAs encoding proteins essential for survival or pathogenesis. Often, a riboswitch and its association with specific genes are highly conserved across phylogeny27 underscoring their physiologic importance.
4. Serendipitously discovered riboswitch-targeting compounds
Recent studies on four anti-bacterials and anti-fungals for which the mechanism of action was unknown have revealed that they target riboswitches25. These serendipitously discovered riboswitch-targeting compounds are roseoflavin, pyrithiamine, L-aminoethylcysteine and DL-4-oxalysine.
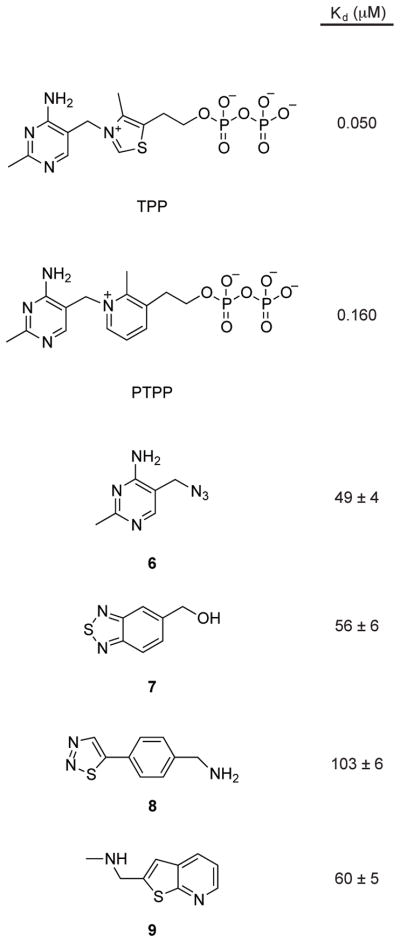
Roseoflavin, an analog of riboflavin and FMN, is a pigment from Streptomyces davawensis with antimicrobial activity28. Roseoflavin inhibits the growth of several Gram-positive bacteria, and roseoflavin-resistant mutants overproduce riboflavin29. In Gram-positive bacteria, all genes involved in riboflavin synthesis are under the control of a single FMN riboswitch30,31.
Roseoflavin has been shown directly to bind the aptamer domain of the FMN riboswitch32 (Figure 1) and to reduce the expression of a reporter gene placed under control of an FMN riboswitch in Bacillus subtilis33,34. Mutations within the aptamer domain of the FMN riboswitch are present in roseoflavin-resistant bacteria, disrupt roseoflavin binding, and result in an impaired regulatory response in the presence of either riboflavin or roseoflavin, reducing bacterial survival33.
Figure 1.
A. The Fusobacterium nucleatum FMN riboswitch32 (PDB 3F4E) and, B. its FMN binding site. C. Mg2+ (purple sphere) mediates FMN-RNA interactions. D. Superposition of FMN (RNA, green; FMN, magenta) and roseoflavin (RNA, cyan; roseoflavin, pink, PDB 3F4H) bound to the riboswitch.
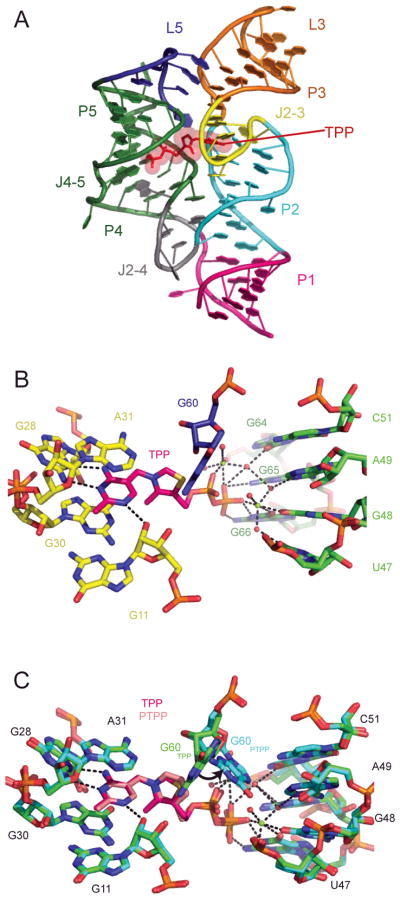
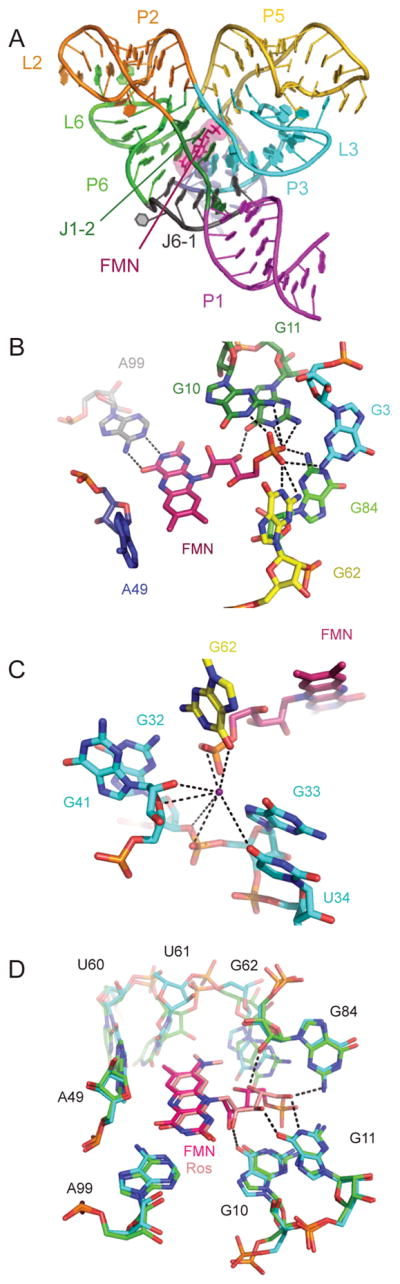
Pyrithiamine (PT) is an analog of thiamine toxic to fungi and bacteria in which the thiazole ring is replaced by a pyridinium ring35,36. Like thiamine, PT is pyrophosphorylated in vivo to pyrithiamine pyrophosphate (PTPP)37. PTPP binds the TPP riboswitch (Figure 2) with an affinity similar to that of TPP. In vivo, PT (presumably acting as PTPP) inhibits the expression of reporter genes placed under the control of a TPP riboswitch35. PT-resistant bacteria have mutations within the TPP riboswitch that disrupt ligand binding to the aptamer domain, and result in the aberrant regulation of genes involved in TPP metabolism35. Crystallographic studies show that while PT binds the TPP riboswitch, it stabilizes the bound conformation of the aptamer domain poorly38. In contrast, the PTPP-bound aptamer domain is highly stabilized39. Interestingly, PTPP binding leads to a local rearrangement in the ligand-binding pocket of the TPP riboswitch. In the TPP complex, the riboswitch does not recognize the central thiazole ring of TPP. In contrast, in the PTPP complex, the RNA rearranges to make specific contacts with the central pyridinium ring of PTPP. Thus, PTPP binding drives the RNA into a conformation that it does not adopt in the cognate complex, reminiscent of the recognition of a unique inactive conformation of the Abl kinase by the anti-cancer drug Gleevec40. This demonstrates that drug specificity may arise from RNA conformations absent in the cognate complex of a riboswitch.
Figure 2.
A. The Arabidopsis thaliana TPP riboswitch39 with TPP (red) bound (PDB 3D2G). B. Mg2+ and waters (green and red spheres, respectively) in the ligand binding site. C. Superposition of TPP (RNA, green; TPP, magenta) and PTPP (RNA, cyan; PTPP, pink, PDB 3D2V) bound to the RNA. Black arrow denotes movement of G60 between the two structures.
L-aminoethylcysteine (AEC) and DL-4-oxalysine are lysine analogs that inhibit the growth of some Gram-positive bacteria. They bind to the lysC riboswitch of B. subtilis (Figure 3) and repress expression of a lysine riboswitch-regulated reporter gene in B. subtilis41. Mutations within the lysC riboswitch are present in AEC-resistant mutants of B. subtilis42 and Escherichia coli43. While these studies suggest that AEC and DL-4-oxalysine have an antibacterial effect by targeting the lysine riboswitch25, it is possible that the primary antimicrobial action of these analogs is a riboswitch-independent mechanism in which the lysyl-tRNA synthetase (LysRS) is inhibited by AEC, inhibiting protein synthesis. In this scenario, AEC resistance conferred by mutations to the riboswitch leads to more effective competition of lysine with AEC for binding to LysRS, reflecting an indirect effect of the lysine riboswitch-mediated increase in aspartokinase levels and cellular concentration of lysine44.
Figure 3.
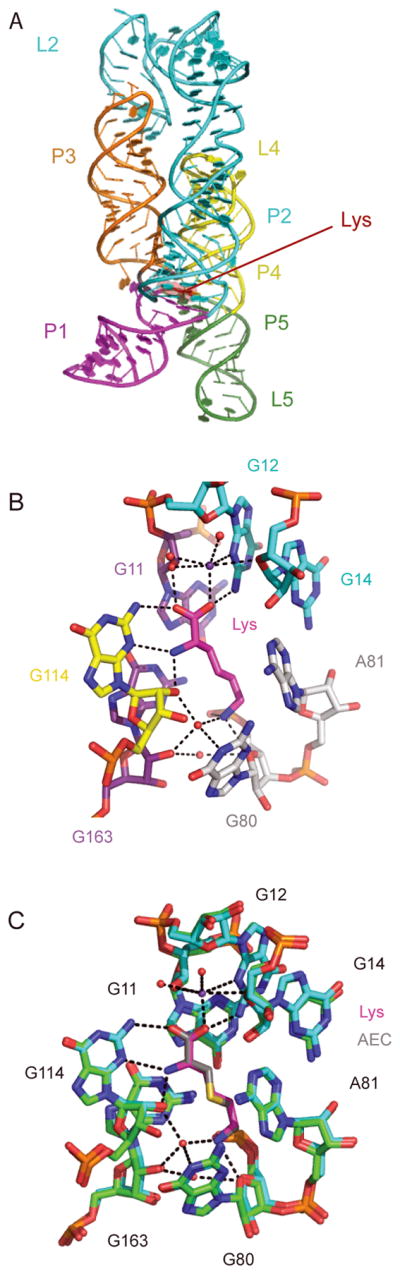
A. The Thermotoga maritima lysine riboswitch50 (PDB 3DIL) bound to lysine (red). B. K+ and waters (purple and red spheres, respectively) mediate lysine-RNA interactions. C. Superposition of the lysine (RNA; green; lysine, magenta) and AEC (RNA, cyan; AEC, gray, PDB 3DIG) bound riboswitch structures.
5. Challenges for riboswitch-targeted drug discovery
A challenge in anti-riboswitch drug discovery is to establish whether compounds that bind riboswitches in vitro are deleterious to pathogens in vivo, and if so, that they exert their effect through riboswitch binding. This is non-trivial because riboswitches respond to essential, often abundant, metabolites and second messengers. Thus, the importance of riboswitches (and the effect of their inhibitors) cannot be evaluated by depleting cells of their cognate ligands. For instance, the glmS ribozyme-riboswitch responds to intracellular glucosamine-6-phosphate (GlcN6P) levels. Since GlcN6P is essential for bacterial cell wall synthesis, depleting bacterial cell strains incapable of its biosynthesis of exogenous GlcN6P causes death, without providing evidence for the effect of glmS riboswitch inactivation. Conversely, replacing the single genomic copy of the riboswitch with a version that is incapable of GlcN6P-induced activation impairs sporulation in B. subtilis. While important, this experiment does not establish whether glmS riboswitch hyper-activation (and consequent down-regulation of the mRNA whose stability it controls) results in a decrease in phenotypic fitness for the bacterium (reviewed in 21). Other riboswitches are present in multiple genomic loci in a given organism, or occur in tandem, making experimental verification of their physiologic importance and effectiveness as drug targets even more difficult. Orthogonal riboswitch mutants that are not activated by natural compounds but respond specifically to a synthetic ligand45 represent a potential means to tackle this challenge.
The ubiquity of the cognate ligands for riboswitches raises the issues of cross-reactivity and toxicity for potential anti-riboswitch drugs. Compounds that target riboswitches have been shown to interact with mammalian proteins. The TPP riboswitch ligand PT inhibits rat thiamine pyrophosphokinase46. The lysine riboswitch ligand AEC is incorporated into proteins following transport into mammalian cells47. Two features of known riboswitches mitigate toxicity concerns. First, riboswitches employ recognition mechanisms different from those of proteins that recognize the same small molecules. Second, while there are eukaryotic gene regulatory RNAs that switch conformation in a ligand-dependent manner (e.g. 48), none of the known riboswitches are present in human, lowering the likelihood of cross-reactivity of new riboswitch-targeted compounds.
The emergence of resistance is a concern with any antibiotic. Bacteria express proteins that act on metabolites recognized by riboswitches. Mutations to these proteins or their regulators can confer resistance to anti-riboswitch compounds that resemble the natural ligand25. For instance, B. subtilis can evolve resistance to PT by overexpressing a thiaminase49. Resistance to compounds known to interact with riboswitches has been witnessed to evolve through mutations to the riboswitch35. However, this resistance mechanism may be less prevalent than for protein targets. A point mutation would only confer resistance in cases in which a riboswitch of one class does not regulate multiple genes or operons essential for survival25. In addition, if the drug candidates were chemical analogs of the natural ligand, such mutations would likely disrupt binding of the natural ligand. This would impact riboswitch function, potentially decreasing bacterial fitness.
A consideration in targeting of riboswitches is that many of them appear to be under kinetic, rather than thermodynamic control. Many assays employed for drug discovery, especially those that are high-throughput, screen for candidates with high equilibrium association constants. Optimal assays for the discovery of novel riboswitch ligands may have to monitor directly the effect of candidates on the expression of genes under riboswitch control, rather than measuring the free energy of the small molecule–RNA interaction (reviewed in 19).
6. Progress in riboswitch-targeted drug discovery
Structure-guided rational design, high-throughput screening methods, and riboswitch-specific assays have been applied to the discovery of novel riboswitch-targeted drugs. These efforts have produced compounds with antibacterial activity in vivo that appear to function by targeting riboswitches.
Blount and colleagues identified L-lysine analogs that bind to the lysine riboswitch and inhibit B. subtilis growth41. They evaluated the affinity of twelve L-lysine analogs to the riboswitch, and found that the RNA is intolerant to changes in most of the amino acid, except at the Cγ position. In addition, the Nε could also be modified, as long as the amine still carried a positive charge and could function as a hydrogen bond donor. Five of the twelve analogs tested bound to the lysC riboswitch from B. subtilis. Of these five, three (L-4-oxalysine and compounds 1 and 2 in Figure 4) inhibited B. subtilis growth in minimal medium, one (compound 3) only inhibited a lysine auxotroph strain, and one did not inhibit growth. Two additional analogs did not bind the riboswitch and inhibited bacterial growth through riboswitch-independent mechanisms. The three compounds that bound the riboswitch and inhibited B. subtilis growth were determined to function at least in part by riboswitch-mediated repression of the B. subtilis enzyme aspartokinase II41. Structure determination of AEC, L-4-oxalysine, and L-homoarginine (compound 3) bound to the riboswitch shows how the environment of Cγ and Nε can accommodate substituents (Figure 3). In the case of the latter, the substituents displace water molecules, and induce subtle rearrangements of the binding pocket50.
Figure 4.
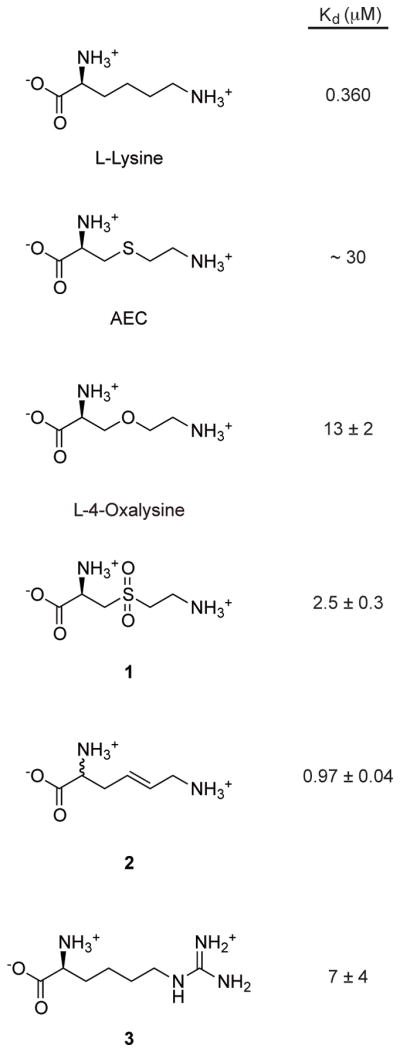
L-lysine analogs that bind the lysine riboswitch: L-aminoethylcysteine (AEC), L-4-oxalysine, L-3-[(2-Aminoethyl)-sulfonyl]-alanine (1), DL-trans-2, 6-Diamino-4-hexenoic acid (2) and L-homoarginine (3). KD values from refs 41,63.
Structures of the ligand-bound guanine riboswitch51,52 suggested that substitutions at guanine positions C2 and C6 would be tolerated53 (Figure 5). Sixteen analogs with modifications at these positions were tested for binding, inhibition of bacterial growth, and repression of a reporter gene. Assessment of affinity yielded KD values between 500 pM and 3.3 μM; one analog was found to have a KD lower than that of guanine. These results confirm that the guanine C2 and C6 positions are available for modification. Five analogs were found to reduce bacterial growth by greater than 50%, and three were found to completely inhibit growth. Only one of these (compound 4, Figure 5), however, was found to repress expression of a guanine riboswitch-regulated reporter gene. The poor correlation between in vitro binding and bacterial growth inhibition may reflect bioavailability and compound stability53.
Figure 5.
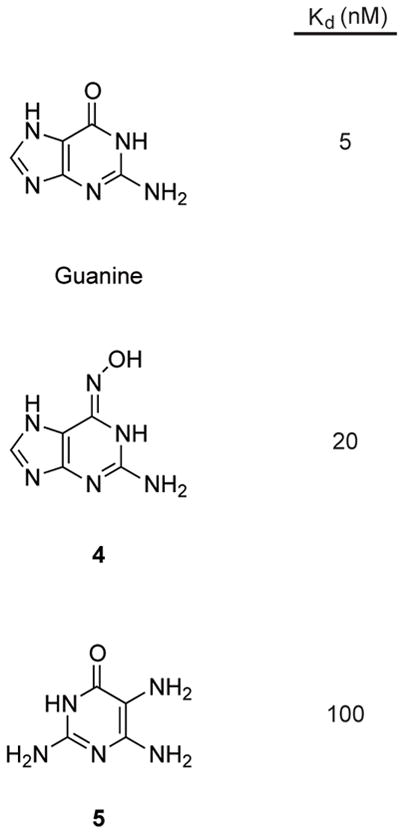
Guanine analogs that bind the guanine riboswitch: (Z)-2-amino-1H-purin-6(9H)-one oxime (4) and 2,5,6-triaminopyrimidin-4-one (5). KD values from refs 53,54.
A second study targeting the guanine riboswitch indicates that this RNA is a viable target for new antibiotics. Mulhbacher et al.54 devised two pyrimidine analogs that would be predicted, on the basis of the crystal structures, to bind to the RNA. In vitro binding was demonstrated, both pyrimidines were found to repress B. subtilis growth in minimal media, and to modulate reporter gene expression. However, only one of these pyrimidines (compound 5, Figure 5) inhibited growth of S. aureus, a bacterium that possesses a four-ORF operon under the control of the guanine riboswitch. When tested against broad range of Gram-positive bacteria, compound 5 was found to inhibit growth of organisms that have the gene guaA (encoding GMP synthetase) under riboswitch control. Specificity was supported by lack of growth inhibition of an E. coli strain lacking guanine riboswitches. Encouragingly, it was found that multi-drug resistant bacterial strains were sensitive to the compound, suggesting that its antimicrobial action employs a mechanism not shared with previously known antibiotics. Finally, the compound was tested in vivo using a mouse model with S. aureus-infected mammary glands, where it was found to inhibit bacterial growth54.
The T-box discriminates between charged and uncharged tRNAs and modulates transcriptional antitermination (reviewed in 55). Oxazolidinones have been known to bind to rRNA, and Means et al. screened a small library of derivatives for their ability to bind and modulate the conformation of an RNA that mimics the antiterminator stem loop of a T-box56. In a subsequent study they optimized some candidates to find compounds with increased solubility57. In vitro transcription antitermination assays identified one compound that led to reduced tRNA-dependent antitermination and another that led to increased tRNA-independent antitermination. An interesting feature of these compounds is that they are polar but not ionic, suggesting that specificity does not arise from interactions with the RNA backbone.
Two assays suitable for high-throughput discovery of activators of the glmS ribozyme-riboswitch have been described. One monitors glmS ribozyme cleavage through fluorescence polarization. Fluorescein is attached covalently 5′ to the self-cleavage site of a cis-acting glmS riboswitch. Ligand-induced self-cleavage results in a short, fluorescent oligonucleotide. The smaller size of the latter results in increased decorrelation of polarization58. The second utilizes FRET (fluorescence resonance energy transfer). Ligand-induced ribozyme cleavage of a oligonucleotide containing FRET donor and acceptor groups on either end releases the FRET acceptor at the 5′ end of the oligonucleotide, while the FRET donor-containing fragment remains bound to the ribozyme. This method was employed to screen ~1000 compounds. However, only glucosamine, an analog of the natural ligand GlcN6P, was found to induce efficient cleavage59.
Development of a generic, high-throughput method for screening compounds for identification of small molecule riboswitch ligands is highly desirable. A promising approach is fragment-based screening using equilibrium dialysis60,61. Screening with chemically and structurally diverse fragments instead of larger molecules allows for chemical space to be explored more efficiently using relatively small libraries (on the order of 100–1000 fragments), and increases the likelihood of finding a hit62. When employed against the E. coli thiM TPP riboswitch, this method identified 20 hits out of a screen of ~1300 fragments, all of which were confirmed with water-LOGSY NMR and ITC (select fragment hits, compounds 6–9 in Figure 6). However, in an in vitro translation screen, these fragments failed to affect gene expression significantly. The authors propose that this may be due to the small size of the fragments, which likely only occupy a portion of the ligand binding site and are insufficient to induce the switching necessary to activate the riboswitch, or decreased affinity of ligands to riboswitches in their mRNA context. Further development of the fragments into larger and more potent molecules may lead to viable new ligands for the TPP riboswitch, making fragment-based screening a generic method for discovering new riboswitch-targeting ligands.
Figure 6.
The TPP analog PTPP and select fragments that bind the TPP riboswitch: 5-(azidomethyl)-2-methylpyrimidin-4-amine (6), benzo[c][1,2,5]thiadiazol-5-ylmethanol (7), (4-(1,2,3-thiadiazol-5-yl)phenyl)methanamine (8) and N-methyl-1-(thieno[2,3-b]pyridin-2-yl)methanamine (9). KD values from refs 35,61.
RNA has been considered a drug target since the discovery that aminoglycosides target rRNA. Riboswitches are exciting targets for novel antibiotics and chemical tools due to their structural sophistication, specificity, and their function as genetic regulators of essential bacterial genes. Previously reported antibiotics and newly identified compounds that function through riboswitches emphasize progress made in the field and provide a foundation for future discovery of new riboswitch-targeting compounds.
Acknowledgments
The authors thank Professor C. Abell, Professor A. Smith and M. Warner (Departments of Chemistry, Plant Sciences and Clinical Biochemistry, respectively) of the University of Cambridge for discussions. K.E.D. was supported by a scholarship from the Winston Churchill Foundation and an NIH-Oxford-Cambridge Ph.D. studentship. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute.
Biographies
Katherine Deigan received her B.A at the University of North Carolina - Chapel Hill under the direction of Kevin Weeks, and her M.Phil. at the University of Cambridge in the lab of Ian Brierley as a Churchill Scholar. She is currently pursuing a Ph.D. at the University of Cambridge and the National Institutes of Health as an NIH-Oxford-Cambridge Scholar under the supervision of Adrian Ferré-D’Amaré and Chris Abell.
Adrian Ferré-D’Amaré received his Ph.D. from The Rockefeller University. After a postdoctoral stay at Yale University, he joined the faculty of the Fred Hutchinson Cancer Research Center (FHCRC) in 1999. In 2008 he was appointed Investigator of the Howard Hughes Medical Institute (HHMI). He resigned from the FHCRC and HHMI in 2011 to pursue opportunities in molecular biophysics and riboregulation at the National Heart, Lung and Blood Institute.
References
- 1.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 2.Xiao H, Murakami H, Suga H, Ferré-D’Amaré AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- 3.Baird NJ, Ferré-D’Amaré AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferré-D’Amaré AR. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- 5.Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferré-D’Amaré AR. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol Cell. 2006;24:535–545. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Ferré-D’Amaré AR. RNA-modifying enzymes. Curr Op Struct Biol. 2003;13:49–55. doi: 10.1016/s0959-440x(02)00002-7. [DOI] [PubMed] [Google Scholar]
- 7.Sutcliffe JA. Improving on nature: antibiotics that target the ribosme. Curr Op Microbiol. 2005;8:534–542. doi: 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Mikkelsen NE, Johansson K, Virtanen A, Kirsebom LA. Aminoglycoside binding displaces a divalent metal ion in a tRNA-neomycin B complex. Nat Struct Biol. 2001;8:510–514. doi: 10.1038/88569. [DOI] [PubMed] [Google Scholar]
- 9.Walter F, Pütz J, Giegé R, Westhof E. Binding of tobramycin leads to conformational changes in yeast tRNAAsp and inhibition of aminoacylation. EMBO J. 2002;21:760–768. doi: 10.1093/emboj/21.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faber C, Sticht H, Schweimer K, Rösch P. Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin B. J Biol Chem. 2000;275:20660–20666. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 11.Kirk S, Luedtke N, Tor Y. Neomycin-acridine conjugate: A potent inhibitor of Rev-RRE binding. J Am Chem Soc. 2000;122:980–981. [Google Scholar]
- 12.von Ahsen U, Davies J, Schroeder R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 13.Stage TK, Hertel KJ, Uhlenbeck OC. Inhibition of the hammerhead ribozyme by neomycin. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen NE, Brännvall M, Virtanen A, Kirsebom LA. Inhibition of RNase P RNA cleavage by aminoglycosides. Proc Natl Acad Sci USA. 1999;96:6155–6160. doi: 10.1073/pnas.96.11.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers J, Chang AH, von Ahsen U, Schroeder R, Davies J. Inhibition of the self-cleavage reaction of the human hepatitis delta virus ribozyme by antibiotics. J Mol Biol. 1996;259:916–925. doi: 10.1006/jmbi.1996.0369. [DOI] [PubMed] [Google Scholar]
- 16.Joyce GF. Forty years of in vitro evolution. Angew Chem. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 17.Suess B, Weigand JE. Engineered riboswitches - overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 18.Xiao H, Edwards TE, Ferré-D’Amaré AR. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol. 2008;15:1125–1137. doi: 10.1016/j.chembiol.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Lau MW, Ferré-D’Amaré AR. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 21.Ferré-D’Amaré AR. The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA. Q Rev Biophys. 2010;43:423–447. doi: 10.1017/S0033583510000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montange RK, Batey RT. Structure of the S-denosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A. Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 26.Hermann T, Tor Y. RNA as a target for small-molecule therapeutics. Expert Opin Ther Pat. 2005;15:49–62. [Google Scholar]
- 27.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otani S, Takatsu M, Nakano M, Kasai S, Miura R. Roseoflavin, a new antimicrobial pigment from Streptomyces. J Antibiot. 1974;27:86–87. [PubMed] [Google Scholar]
- 29.Burgess C, O’connell-Motherway M, Sybesma W, Hugenholtz J, van Sinderen D. Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods. Appl Environ Microbiol. 2004;70:5769–5777. doi: 10.1128/AEM.70.10.5769-5777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 31.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–13. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–94. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott E, Stolz J, Lehmann M, Mack M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 2009;6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- 35.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Robbins WJ. The pyridine analog of thiamin and the growth of fungi. Proc Natl Acad Sci USA. 1941;27:419–422. doi: 10.1073/pnas.27.9.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwashima A, Wakabayashi Y, Nose Y. Formation of pyrithiamine pyrophosphate in brain tissue. J Biochem. 1976;79:845–847. doi: 10.1093/oxfordjournals.jbchem.a131138. [DOI] [PubMed] [Google Scholar]
- 38.Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J Am Chem Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- 40.Schindller T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 41.Blount K, Wang J, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Chen NY, Paulus H. Identification of aecA mutations in Bacillus subtilis as nucleotide substitutions in the untranslated leader region of the aspartokinase II operon. J Gen Microbiol. 1991;137:1135–1143. doi: 10.1099/00221287-137-5-1135. [DOI] [PubMed] [Google Scholar]
- 43.Patte JC, Akrim M, Méjean V. The leader sequence of the Escherichia coli lysC gene is involved in the regulation of LysC synthesis. FEMS Microbiol Lett. 1998;169:165–170. doi: 10.1111/j.1574-6968.1998.tb13313.x. [DOI] [PubMed] [Google Scholar]
- 44.Ataide SF, Wilson SN, Dang S, Rogers TE, Roy B, Banerjee R, Henkin TM, Ibba M. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem Biol. 2007;2:819–827. doi: 10.1021/cb7002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon N, Duncan JN, Geerlings T, Dunstan MS, Mccarthy JEG, Leys D, Micklefield J. Reengineering orthogonally selective riboswitches. Proc Natl Acad Sci USA. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koedam JC. The mode of action of pyrithiamine as an inductor of thiamine deficiency. Biochim Biophys Acta. 1958;29:333–344. doi: 10.1016/0006-3002(58)90192-6. [DOI] [PubMed] [Google Scholar]
- 47.Di Girolamo M, Di Girolamo A, Cini C, Coccia R, De Marco C. Thialysine utilization for protein synthesis by CHO cells. Physiol Chem Phys Med NMR. 1986;18:159–164. [PubMed] [Google Scholar]
- 48.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas AL, Laun NP, Begley TP. Thi20, a remarkable enzyme from Saccharomyces cerevisiae with dual thiamin biosynthetic and degradation activities. Bioorg Chem. 2005;33:338–344. doi: 10.1016/j.bioorg.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–7. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 52.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JN, Blount K, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-Box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Means J, Katz S, Nayek A, Anupam R, Hines J, Bergmeier S. Structure-activity studies of oxazolidinone analogs as RNA-binding agents. Bioorg Med Chem Lett. 2006;16:3600–3604. doi: 10.1016/j.bmcl.2006.03.068. [DOI] [PubMed] [Google Scholar]
- 57.Anupam R, Nayek A, Green NJ, Grundy FJ, Henkin TM, Means JA, Bergmeier SC, Hines JV. 4,5-disubstituted oxazolidinones: high affinity molecular effectors of RNA function. Bioorg Med Chem Lett. 2008;18:3541–3544. doi: 10.1016/j.bmcl.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayer G, Famulok M. High-throughput-compatible assay for glmS riboswitch metabolite dependence. ChemBioChem. 2006;7:602–604. doi: 10.1002/cbic.200500490. [DOI] [PubMed] [Google Scholar]
- 59.Blount K, Puskarz I, Penchovsky R, Breaker RR. Development and application of a high-throughput assay for glmS riboswitch activators. RNA Biol. 2006;3:77–81. doi: 10.4161/rna.3.2.3102. [DOI] [PubMed] [Google Scholar]
- 60.Chen L, Cressina E, Leeper FJ, Smith AG, Abell C. A fragment-based approach to identifying ligands for riboswitches. ACS Chem Biol. 2010;5:355–8. doi: 10.1021/cb9003139. [DOI] [PubMed] [Google Scholar]
- 61.Cressina E, Chen L, Abell C, Leeper FJ, Smith AG. Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chem Sci. 2011;2:157–165. [Google Scholar]
- 62.Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 63.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–97. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulshina N, Baird NJ, Ferré-D’Amaré AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein DJ, Ferré-D’Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 67.Klein DJ, Wilkinson SR, Been MD, Ferré-D’Amaré AR. Requirement of helix P2.2 and nucleotide G1 for positioning of the cleavage site and cofactor of the glmS ribozyme. J Mol Biol. 2007;373:178–189. doi: 10.1016/j.jmb.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klein DJ, Been MD, Ferré-D’Amaré AR. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc. 2007;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- 69.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garst AD, Héroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–51. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dann CE, Wakeman C, Sieling C, Baker S, Irnov I, Winkler W. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 74.Klein D, Edwards T, Ferré-D’Amaré A. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang M, Peterson R, Feigon J. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2010;39:653–655. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 76.Rieder U, Kreutz C, Micura R. Folding of a transcriptionally acting preQ1 riboswitch. Proc Natl Acad Sci USA. 2010;107:10804–10809. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spitale R, Torelli A, Krucinska J, Bandarian V, Wedekind J. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noeske J, Schwalbe H, Wöhnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edwards AL, Reyes FE, Héroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16:2144–2155. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free-state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson RC, Smith AM, Fuchs RT, Kleckner IR, Henkin TM, Foster MP. Tuning riboswitch regulation through conformational selection. J Mol Biol. 2011;405:926–938. doi: 10.1016/j.jmb.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–71. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 85.Noeske J, Richter C, Stirnal E, Schwalbe H, Wöhnert J. Phosphate-group recognition by the aptamer domain of the thiamine pyrophosphate sensing riboswitch. ChemBioChem. 2006;7:1451–1456. doi: 10.1002/cbic.200600151. [DOI] [PubMed] [Google Scholar]
- 86.Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. The ligand-free state of the TPP riboswitch: a partially folded RNA structure. J Mol Biol. 2010;396:153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]