Abstract
Background & objectives
Multiple drug resistance in epilepsy is a common problem and one third of epilepsy patients remain non responsive to antiepileptic drug (AED) therapy. In this study we aimed to investigate the relationship between the genetic polymorphism of cytochrome P450 genes, namely CYP2C9 and CYP2C19 with multiple drug resistance in epilepsy patients.
Methods:
A total of 402 patients with epilepsy were enrolled in this study; 128 were drug resistant and 274 were drug responsive. The peripheral blood samples of the patients with epilepsy were collected. Drug compliance was confirmed in 20 per cent patient population using HPLC. Genotyping of CYP2C9 (*2 and *3), and CYP2C19 (*2 and *3) was carried out by PCR-RFLP.
Results:
The genotype frequencies of CYP2C9 430 C>T (*2 variant) and CYP2C9 1075 A>C (*3 variant) did not differ significantly in drug resistant versus responsive patients. After combining CYP2C9 *2 and CYP2C9 *3, the frequency of CYP2C9*1/*3 was significantly lower in drug resistant as compared to drug responsive epilepsy patients (P=0.03, OR=0.53, 95%CI=0.30-0.95). Similarly, combined frequency of all the slow and poor metabolizer variants (2C9 *1/*2, *1/*3 and *2/*3) was also lower as compared to drug resistant group (P=0.03, OR=0.60, 95% CI 0.38-0.96). There was no significant differences in genotypic or allelic distribution of CYP2C19*2 while CYP2C19*3 was monomorphic in northern Indian population.
Interpretation & conclusions:
Our results demonstrated significant involvement of CYP2C9 genetic variants in the modulation of epilepsy pharmacotherapy confirming the important role of CYP2C9 mutants preventing epilepsy patients from developing drug resistance.
Keywords: Cytochrome, drug refractory, epilepsy, P450, pharmacogenetics, single nucleotide polymorphism
Epilepsy is one of the most common paroxysmal and heterogeneous neurological disorder affecting over 42 million people worldwide with distinct symptoms, aetiology and prognosis. According to WHO estimate, worldwide eight per 1000 people have epilepsy1. Despite the availability of several antiepileptic drugs, critical challenges remain in the treatment of epilepsy. Even after all possible therapeutic interventions, drug resistance has been reported in up to one-third of the patient2.
Genotype-phenotype relationship in epilepsy is fairly complex but studies have shown involvement of genetic factors affecting prognosis and treatments in epilepsy3. Drug metabolism represents the prominent pathway both in qualitative and quantitative elimination of drugs including anti epileptic drugs (AEDs). It is accomplished by the hepatic (metabolism) and/or renal (excretion) route which comprises phase I (e.g., oxidative reactions catalyzed by various cytochrome P-450 enzymes) and phase II (e.g., conjugations like glucuronidation) reactions. Cytochrome P450 (CYP450) family are major phase-I drug metabolizing enzymes (DMEs) which are responsible for the metabolism of both endogenous and exogenous compounds. The polymorphisms of the important CYP450 genes such as CYP2C9, CYP2C19, CYP2D6 and CYP2E1 have been studied extensively in a large number of populations and show significant heterogeneity in the frequency of different alleles/genotypes and the resulting metabolizer phenotypes. Two CYP2C9 alleles that produce a phenotype of poor metabolism occur in 11 and 8 per cent of whites but only 3 and 0.8 per cent of blacks4. Several groups have shown the genetic variants/mutations in the CYP2C9 and CYP2C19 as determinants of significantly impaired metabolism of their substrates including commonly prescribed AEDs5–8. A study from south India reported that CYP2C9*2 and *3 mutant alleles caused decreased hydroxylation of phenytoin in vivo, whereas the mutant alleles of CYP2C19 played only a minor role in the metabolism of phenytoin in subjects9. Others have also reported strong association between CYP2C9 allelic variants and phenytoin dose requirement10. Genetic polymorphisms of CYP2C19 have also been shown to contribute to the pharmacokinetic variability of phenytoin and phenobarbital. Genetic variants which lead to poor metabolizers phenotype of CYP2C19 are relatively common in Asian groups7,11. Though previous studies have correlated drug doses with genetic variants but these reports did not correlate it drug responsiveness. The variations in CYP450 DMEs genes could influence inter-individual variation in AEDs metabolism that could be responsible for drug responsive or drug resistant phenotype; therefore, we investigated the effect of CYP2C9 and CYP2C19 genetic variants on AED and multiple drug resistance in north Indian patients with epilepsy.
Material & Methods
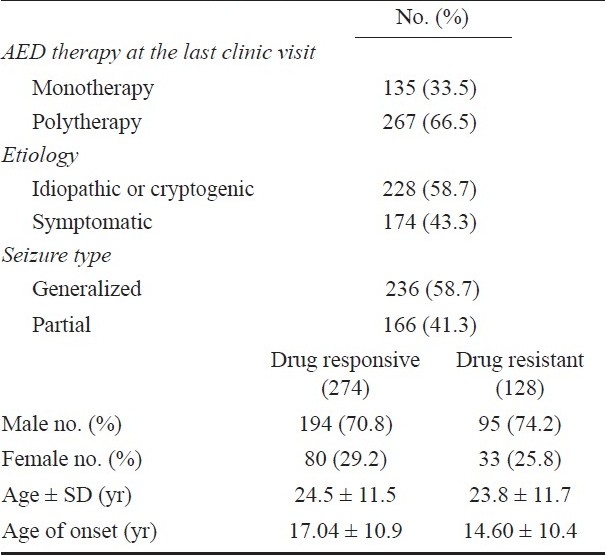
Patients and controls: A total of 402 randomly selected patients with epilepsy were included in this study who were recruited from department of Neurology, Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGIMS), Lucknow, India during August 2005 to September 2009. The study was approved by local ethics committee. Exclusion criteria included severe adverse drug reactions; poor compliance with AEDs, unreliable record of seizure frequency, history of pseudo-seizures, alcohol or drug abuse, or any other malignant diseases such as brain tumour, secondary metastasis, hepatic failure or renal failure. An informed consent was signed by each participant or responsible adult and they were personally interviewed for information on ethnicity, seizure frequency, and duration of seizure, compliance and other habits. Among the patients with epilepsy, 71.9 per cent were male and 28.1 per cent were female. Epilepsy was classified as symptomatic and idiopathic. Seizures were classified as generalized or partial (Table I). Drug responsiveness of epilepsy patients was determined from clinical records and personal interview of patients. All drug refractory and drug responsive patients were of same ethnic origin. AED levels were measured in 20 per cent patients to confirm drug compliance. Mean carbamazepine, phenytoin, and valproate levels were 8.26 ± 5.25, 11.27 ± 8.12, and 68.0 ± 36.22 μg/ml, respectively.
Table I.
Demographic profile of epilepsy patients
Definition of drug resistance and responsiveness: The main criterion for drug resistance was the occurrence of at least four seizures over a period of one year with three appropriate antiepileptic drugs at maximum tolerated doses12,13. Patients who had undergone surgeries for seizure control were considered drug refractory irrespective of their outcome after surgery. The epilepsy patients who had complete freedom from seizures for at least one year from last follow up visit were considered drug responsive.
Laboratory protocols: Sample collection, genotyping and HPLC methods: Blood samples (5 ml) were taken in EDTA vials from patients and DNA was isolated using salting out method with slight modifications14. The plasma was separated and stored at -20°C for drug level assay. A total of 804 chromosomes of 402 epilepsy patients, were genotyped. Genotyping for CYP2C9 and CYP2C19 was performed using PCR-RFLP method as reported previously15–17. Restriction endonuclease digested products were separated using 10-15 per cent poly-acryl amide gel electrophoresis and genotyping patterns were recorded. Primer sequences and product sizes are depicted in Table II. For quality control, 20 per cent of the samples were re-genotyped by different laboratory personnel and results found were 100 per cent concordant to those previously observed.
Table II.
List of primers used in the study

The plasma was separated and stored at -20°C for drug level assay. HPLC Instrument (Perkin Elmer series 410, USA) equipped with rheodyne injector 20 μl loop capacity and LC 290 UV/VIS detector and LCI 100 Integrator with C18 hypersil column; octadecylsilane (ODS) 250×4.6 mm with particle size of 5μ was used. The drugs lamotrigine, phenobarbitone, phenytoin and carbamazepine were extracted using chloroform as organic solvent. The extract was evaporated at 35°C under the continuous stream of nitrogen and reconstituted with 0.1 ml of methanol; 20 μl of the sample at 1.4 ml/min flow rate was injected. The elutes were read at λ=255 nm wavelength in UV detector in single run and corresponding peaks were analyzed according to standards used. For valproate, drug was extracted in exaction reagent of 50μl volume (0.5 g 4-bromophenacyl bromide + 0.125 g DiCyclohexane-18-crown-6 dissolved in 50 ml of acetonitrile) and 20 μl sample was injected into HPLC column at 1.8 ml/min flow rate and elute read at λ=254 nm. All these methods were specific for these respective drugs and no interference was found in blank plasma at the retention time of these drugs. The sensitivity of the methods was, 1 μg/ml for carbamazepine, lamotrigine, phenytoin, phenobarbitone and 10 μg/ml for valproate.
Statistical analysis: The relationship between various genotypes and responsiveness was examined using binary logistic regression. Association was expressed as odds ratios (OR) or risk estimates with 95 per cent confidence intervals (CI). Association was considered significant at P<0.05. All analyses were performed using SPSS statistical analysis software, Version 15.0 (SPSS, Chicago, IL, USA). Sample size was calculated using the CaTS-Power Calculator (www.sph.umich.edu/csg/abecasis/CaTS). The power of study was 80 per cent and relative risk for power calculation was set at 2.
Results
The mean age of the patients was 24.30 ± 11.62 yr and it was not different between the drug-resistant (23.8 ± 11.7 yr) and drug-responsive (24.5 ± 11.5) patients. The onset of the first seizure in the responders (14.60 ± 10.4 yr) was significantly at an early age compared to non-responders (17.04 ± 10.9 yr; P=0.04). Among all, 174 (43.3%) had symptomatic epilepsy while the remaining 228 (56.7%) were idiopathic or cryptogenic. The types of seizure were generalized in 58.7 per cent (236) and partial in 41.3 per cent (166) patients and were not different between the responder and non-responder groups (P=0.23 OR = 1.29, 95% CI = 0.84–1.99). The correlation of AED levels with genotypes was not done as AED levels were assayed in smaller number of patients.
None of the patients were homozygous for the 2C9*2 or 2C9*3. The genotype frequencies of CYP2C9 430 C>T (*2 variant) did not differ significantly in drug resistant versus responsive patients. Significant differences were also not observed at allele level for 430 T variant (OR = 0.67, 95% CI = 0.11-0.96, P=0.36) (Table IIIa).
Table III (a).
CYP2C9 and CYP2C19: Genotype and allele frequencies in drug resistant versus drug responsive patients with epilepsy
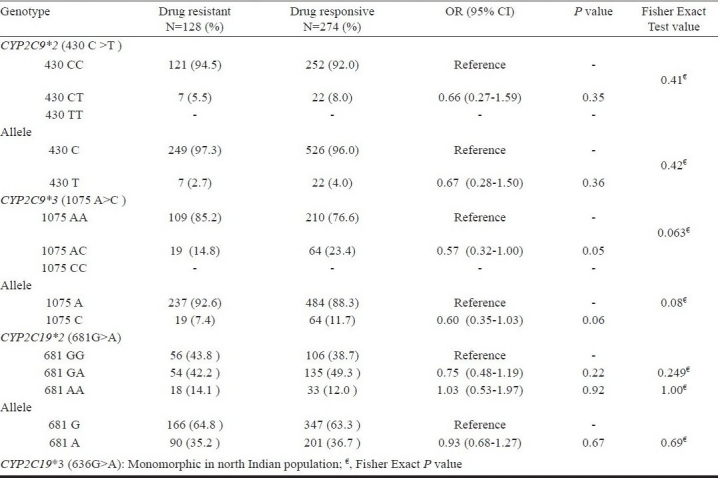
CYP2C9 1075 A>C (*3 variant) polymorphism showed marginal significant difference between patients having multiple drug resistance and drug responsive patients for AT genotype (OR = 0.57, 95%CI=0.32-1.00; P=0.05). The CYP2C9*3 variant allele frequency was also higher in drug responsive patients compared to resistant patients (P=0.06; OR = 0.60, 95%CI=0.35-0.1.03) and appears to be contributing towards lower risk for developing multiple drug resistance in epilepsy (Table IIIa).
On combining CYP2C9*2 and CYP2C9*3, we found the frequency of variant genotypes (CYP2C9*1/*3) which are known to result in slow metabolizer phenotype was significantly lower in drug resistant group as compared to drug responsive group (Table IIII). The genotype CYP2C9*1/*3 was significantly protective for drug resistance in epilepsy (P=0.03, OR=0.53, 95%CI=0.30-0.95). When all the slow metabolizer (*1/*2, *1/*3) and poor metabolizer (*2/*3) phenotypes were compared, it was found that variant genotypes prevented epilepsy patients for developing drug resistance (P=0.03, OR=0.60, 95% CI 0.38-0.96; Table-IIIb). Though, after applying Bonferroni correction for multiple testing, the total significance goes away but according to Fisher exact test, P value remained significant.
Unlike CYP2C9, the genotype frequencies of CYP2C19*2 681G>A did not differ significantly in drug resistant compared to responsive patients for GA (OR = 0.75, 95%CI=0.48-1.19; P=0.22) and AA (OR = 1.03, 95%CI=0.53-1.97; P=0.92) (Table III a) genotypes. At allele level also, it did not show any significant differences between the groups. The CYP2C19*3 636G>A polymorphism was found to be monomorphic in our population.
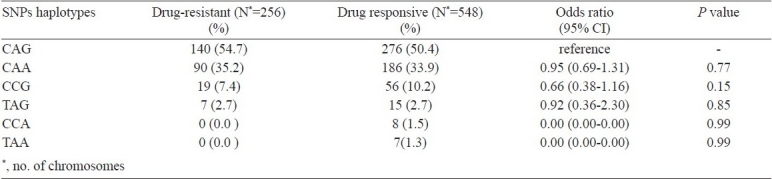
After using Expectation-Maximization algorithm six haplotypes were observed in drug responsive and four in drug resistant epilepty patients. These haplotypes were compared (Table IV) for the difference in the distribution between both the groups. However, no significant differences were observed among responders and non responders for given haplotypes.
Table III (b).
Combined CYP2C9*2 genetic variants: genotype and allele frequencies
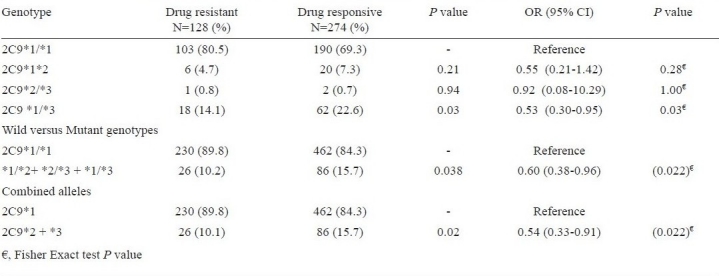
Table IV.
Haplotype frequencies of CYP2C9*2, *3 and CYP2C19*2 gene polymorphism in drug resistant and drug responsive epilepsy patients
Discussion
In this study, out of four genetic polymorphisms of the CYP2C9 and CYP2C19, we observed the protective effects of genetic variants of CYP2C9 (*2 and *3). Slow and poor metabolizer phenotypes in epilepsy patients prevented them for developing multiple drug resistance. No association was found with CYP2C19*2 and CYP2C19*3 turned out to be monomorphic in our population. These findings indicate important role of CYP2C9 variants in conferring multiple-drug resistant phenotype against AEDs. Our findings are concordant with the findings of a report from Southern India which showed major role of CYP2C9 variants in phenytoin hydroxylation while CYP2C19 had a minor role in the metabolism9.
Our results support previous observations which identified CYP2C9 as major drug metabolizer for commonly prescribed AEDs. CYP2C9 and CYP2C19 are known to encode clinically important enzymes that metabolize numerous therapeutic drugs with a narrow therapeutic index. CYP2C9 is responsible for the hydroxylation of up to 90 per cent of serum phenytoin, while CYP2C19 is partially related to phenytoin metabolism18. These enzymes are polymorphically expressed and most of the variants result in decreased metabolism of the respective substrates. Of all the AEDs, mainly phenytoin and partially carbamazepine and phenobarbital undergo significant metabolism by cytochrome P450 isoenzymes with significant genetic polymorphisms in CYP2C9, CYP2C19 and CYP3A4. Earlier studies have also observed strong association between CYP2C9 genotype and phenytoin maintenance dose requirement10,15,19. Van der Weide et al10 found that CYP2C9 allelic variants affected phenytoin dose requirement, for patients carrying at least one mutant CYP2C9 allele, the mean phenytoin dose required to achieve a therapeutic serum concentration was about 37 per cent lower than the mean dose required by wild-type individuals. Another similar study from Taiwan revealed that the CYP2C9 and CYP2C19 polymorphisms have dramatic effects on the population pharmacokinetic parameters of phenytoin20. On the basis of these observations, we propose that slow metabolizer phenotype could have advantage over fast metabolizer phenotype in epilepsy pharmacotherapy. It is likely that patients with slow metabolizer phenotype will require lesser drug doses to control seizures as compared to epilepsy patients with fast metabolism. Therefore, patients with extensive metabolism are more likely to become drug resistant during the course of antiepileptic treatment.
The frequency distribution of CYP2C9 and CYP2C19 gene polymorphism at genotype and allele level in the drug-responsive group in our study was similar to those reported earlier in the healthy Indian populations21–23. We found that CYP219*3 is monomorphic in northern Indians, so it does not contribute to inter-individual variation of drug metabolizing phenotype in our population of drug resistant and responsive epilepsy patients. This variant allele has not been reported from Caucasian population; however, a report from India identified CYP2C19*3 allele, with frequency distribution of 2.7 per cent in Tamilian population24.
The drug resistance is a complex phenotype resulting from contribution of numerous genes. In addition to drug metabolizing enzymes, differences in intestinal and blood brain barrier multi drug transporters; MDR1 (ABCB1) and MRP2 expression also influence carbamazepine and phenytoin disposition and may account for inter-individual pharmacokinetic variability12,25,26. Kerb et al27 performed a combined analysis of variable alleles of CYP2C9, CYP2C19 and ABCB1 gene which revealed that the number of mutant CYP2C9 alleles is a major determinant, but the number of ABCB1*T alleles further contributes to the prediction of phenytoin plasma levels.
There were some limitations to our study. First, environmental factors also have an important role in determining drug response. We did not have enough data for BMI, smoking, alcohol intake etc. therefore, we could not include these factors in our analysis. Second, we could measure drug levels only in 100 randomly selected epilepsy patients. It was not possible to correlate AED levels with individual genotypes because sample size was small for each subgroup.
In addition, there are various other factors which influence responsiveness to newly administered AED treatment and are highly dependent on past treatment history. Factors such as type of epilepsy, duration of seizure, and number of seizures prior to initiation of drug therapy28 may also be responsible for differences in drug responsiveness. Recently, drug targets have emerged as major class of genes responsible for drug responsiveness that include but are not limited to sodium channels, calcium channels, potassium channels, and GABA receptors3,29. So, the overall genetic effect of these genes may have a greater role in determining drug responsiveness rather than the effect of a single gene and its genetic polymorphisms.
In conclusion, our results suggest that CYP2C9*3 but not CYP2C19 genetic polymorphisms appear to be associated with overall drug responsive behaviour in multiple drug resistant epilepsy in northern Indian population.
Acknowledgments
The study was supported by a grant received from the Department of Biotechnology (DBT) to BM and fellowships provided by CSIR to RL and DST to RK, Government of India, New Delhi. We thankfully acknowledge support from Dr N.J. Gogtay, KEM Hospital, Mumbai, for drug level assays.
References
- 1.WHO. World Health Organization: epilepsy: epidemiology, aetiology and prognosis. WHO Factsheet. 2001:165. [Google Scholar]
- 2.Sisodiya SM Genetics of drug resistance. Epilepsia. 2005;46(Suppl 10):33–8. doi: 10.1111/j.1528-1167.2005.00356.x. [DOI] [PubMed] [Google Scholar]
- 3.Depondt C. The potential of pharmacogenetics in the treatment of epilepsy. Eur J Paediatr Neurol. 2006;10:57–65. doi: 10.1016/j.ejpn.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Xie HG, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv Drug Deliv Rev. 2002;54:1257–70. doi: 10.1016/s0169-409x(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 5.Patsalos PN, Froscher W, Pisani F, van Rijn CM. The importance of drug interactions in epilepsy therapy. Epilepsia. 2002;43:365–85. doi: 10.1046/j.1528-1157.2002.13001.x. [DOI] [PubMed] [Google Scholar]
- 6.Klotz U. The role of pharmacogenetics in the metabolism of antiepileptic drugs: pharmacokinetic and therapeutic implications. Clin Pharmacokinet. 2007;46:271–9. doi: 10.2165/00003088-200746040-00001. [DOI] [PubMed] [Google Scholar]
- 7.Yukawa E, Mamiya K. Effect of CYP2C19 genetic polymorphism on pharmacokinetics of phenytoin and phenobarbital in Japanese epileptic patients using Non-linear Mixed Effects Model approach. J Clin Pharm Ther. 2006;31:275–82. doi: 10.1111/j.1365-2710.2006.00712.x. [DOI] [PubMed] [Google Scholar]
- 8.Tate SK, Depondt C, Sisodiya SM, Cavalleri GL, Schorge S, Soranzo N, et al. Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci USA. 2005;102:5507–12. doi: 10.1073/pnas.0407346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosemary J, Surendiran A, Rajan S, Shashindran CH, Adithan C. Influence of the CYP2C9 and CYP2C19 polymorphisms on phenytoin hydroxylation in healthy individuals from south India. Indian J Med Res. 2006;123:665–70. [PubMed] [Google Scholar]
- 10.van der Weide J, Steijns LS, van Weelden MJ, de Haan K. The effect of genetic polymorphism of cytochrome P450 CYP2C9 on phenytoin dose requirement. Pharmacogenetics. 2001;11:287–91. doi: 10.1097/00008571-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Rosemary J, Adithan C. The pharmacogenetics of CYP2C9 and CYP2C19: ethnic variation and clinical significance. Curr Clin Pharmacol. 2007;2:93–109. doi: 10.2174/157488407779422302. [DOI] [PubMed] [Google Scholar]
- 12.Lakhan R, Misra UK, Kalita J, Pradhan S, Gogtay NJ, Singh MK, et al. No association of ABCB1 polymorphisms with drug-refractory epilepsy in a north Indian population. Epilepsy Behav. 2009;14:78–82. doi: 10.1016/j.yebeh.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqui A, Kerb R, Weale ME, Brinkmann U, Smith A, Goldstein DB, et al. Association of multidrug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. N Engl J Med. 2003;48:1442–8. doi: 10.1056/NEJMoa021986. [DOI] [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aynacioglu AS, Brockmoller J, Bauer S, Sachse C, Guzelbey P, Ongen Z, et al. Frequency of cytochrome P450 CYP2C9 variants in a Turkish population and functional relevance for phenytoin. Br J Clin Pharmacol. 1999;48:409–15. doi: 10.1046/j.1365-2125.1999.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh K, Inoue K, Yanagiwara S, Kyoya H, Suzuki T. A rapid and simple detection of genetic defects responsible for the phenotypic polymorphism of cytochrome P450 2C19. Biol Pharm Bull. 1999;22:77–9. doi: 10.1248/bpb.22.77. [DOI] [PubMed] [Google Scholar]
- 17.Sviri S, Shpizen S, Leitersdorf E, Levy M, Caraco Y. Phenotypic-genotypic analysis of CYP2C19 in the Jewish Israeli population. Clin Pharmacol Ther. 1999;65:275–82. doi: 10.1016/S0009-9236(99)70106-2. [DOI] [PubMed] [Google Scholar]
- 18.Fritz S, Lindner W, Roots I, Frey B M, Küpfer A Stereochemistry of aromatic phenytoin hydroxylation in various drug hydroxylation phenotypes in humans. J Pharmacol Exp Ther. 1987;241:615–22. [PubMed] [Google Scholar]
- 19.Anderson GD. Pharmacokinetic, pharmacodynamic, and pharmacogenetic targeted therapy of antiepileptic drugs. Ther Drug Monit. 2008;30:173–80. doi: 10.1097/FTD.0b013e318167d11b. [DOI] [PubMed] [Google Scholar]
- 20.Hung CC, Lin CJ, Chen CC, Chang CJ, Liou HH. Dosage recommendation of phenytoin for patients with epilepsy with different CYP2C9/CYP2C19 polymorphisms. Ther Drug Monit. 2004;26:534–40. doi: 10.1097/00007691-200410000-00012. [DOI] [PubMed] [Google Scholar]
- 21.Lamba JK, Dhiman RK, Kohli KK. CYP2C19 genetic mutations in North Indians. Clin Pharmacol Ther. 2000;68:328–35. doi: 10.1067/mcp.2000.109365. [DOI] [PubMed] [Google Scholar]
- 22.Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics. 2009;19:170–9. doi: 10.1097/FPC.0b013e32831ebb30. [DOI] [PubMed] [Google Scholar]
- 23.Zainuddin Z, Teh LK, Suhaimi AW, Ismail R. Malaysian Indians are genetically similar to Caucasians: CYP2C9 polymorphism. J Clin Pharm Ther. 2006;31:187–91. doi: 10.1111/j.1365-2710.2006.00727.x. [DOI] [PubMed] [Google Scholar]
- 24.Adithan C, Gerard N, Vasu S, Rosemary J, Shashindran CH, Krishnamoorthy R. Allele and genotype frequency of CYP2C19 in a Tamilian population. Br J Clin Pharmacol. 2003;56:331–3. doi: 10.1046/j.1365-2125.2003.01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon C, Stieger B, Kullak-Ublick GA, Fried M, Mueller S, Fritschy JM, et al. Intestinal expression of cytochrome P450 enzymes and ABC transporters and carbamazepine and phenytoin disposition. Acta Neurol Scand. 2007;115:232–42. doi: 10.1111/j.1600-0404.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 26.Lakhan R, Kumari R, Misra UK, Kalita J, Pradhan S, Mittal B. Differential Role of Sodium channels SCN1A and SCN2A gene polymorphisms with epilepsy and multiple drug resistance in north Indian population. Brit J Clin Pharmacol. 2009;68:214–20. doi: 10.1111/j.1365-2125.2009.03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerb R, Aynacioglu AS, Brockmoller J, Schlagenhaufer R, Bauer S, Szekeres T, et al. The predictive value of MDR1, CYP2C9, and CYP2C19 polymorphisms for phenytoin plasma levels. Pharmacogenomics J. 2001;1:204–10. doi: 10.1038/sj.tpj.6500025. [DOI] [PubMed] [Google Scholar]
- 28.Schiller Y, Najjar Y. Quantifying the response to antiepileptic drugs: effect of past treatment history. Neurology. 2008;70:54–65. doi: 10.1212/01.wnl.0000286959.22040.6e. [DOI] [PubMed] [Google Scholar]
- 29.Loscher W, Klotz U, Zimprich F, Schmidt D. The clinical impact of pharmacogenetics on the treatment of epilepsy. Epilepsia. 2009;50:1–23. doi: 10.1111/j.1528-1167.2008.01716.x. [DOI] [PubMed] [Google Scholar]


