Abstract
Background & objectives:
Emergence and spread of drug resistant Mycobacterium tuberculosis is a serious threat to tuberculosis (TB) control programme. Therefore, the objective of this study was to genotype drug-resistant M. tuberculosis strains isolated from patients in Sichuan, China, using Mycobacterial Interspersed Repetitive Units (MIRU) for epidemiological analysis.
Methods:
Drug-resistance testing of M. tuberculosis isolates from pulmonary TB patients was confirmed by proportion method. Twelve MIRU loci were analyzed on 80 drug-resistant and 9 susceptible isolates by polymerase chain reaction and agarose gel electrophoresis. Hunter-Gaston discriminatory index (HGI) values were determined for each 12 MIRU loci for the evaluation of their discrimination power.
Results:
Among 12 MIRU loci examined, polymorphic bands could be generated on 11 loci. Sixty five isolates had distinct MIRU patterns, while other 24 belonged to 8 clusters and resistant to at least one anti-TB drug tested. The association between the MIRU patterns and the mutation patterns of drug-resistance relevant target genes was not significant among the drug-resistant isolates.
Interpretation & conclusions:
The results showed that with a satisfactory discrimination power exhibited, the 12 loci based MIRU typing could be a valuable tool for epidemiological studies in M. tuberculosis isolates from Sichuan.
Keywords: Drug resistance, genotyping, MIRU, Mycobacterium tuberculosis, Sichuan
Tuberculosis (TB) is still a leading cause of adult mortality arising from a single infectious agent globally. It is estimated that nearly a third of the world population is infected with Mycobacterium tuberculosis, among which 8 - 10 million individuals develop active disease and nearly two million people die annually1. According to the report of World Health Organization2 in 2008, China is one of the high burden countries, with TB incidence 101/100,000, ranking second only to India in the total number of new tuberculosis cases. The main challenge for TB control in China is the most pressing task due to the spreading of multidrug-resistant (MDR) strains of M. tuberculosis, defined as resistance to at least isoniazid (INH) and rifampin (RIF).
Molecular epidemiological study is essential to provide the estimation of tuberculosis infection among epidemiologically linked patients, the forecast of next epidemic outbreak, and the analysis of risk factors associated with tuberculosis infection. Based on DNA fingerprinting, such a study is capable of tracing the infection routine and determining the risk factors for transmission3–5. IS6110 RFLP typing has been considered as the gold standard in the molecular epidemiology of TB6–8, but this approach is technically demanding and requires weeks to culture a certain amount of isolates in order to obtain the necessary quantities of DNA9,10. In addition, for isolates with less than 6 copy numbers of IS6110, it provides insufficient discrimination power11. Spoligotyping, based on the direct repeat (DR) region of M. tuberculosis, has the advantage compared with IS6110: firstly, only a small amount of DNA sample is needed for clinical examination and strain testing from liquid culture; secondly, only one digital number is used to express the results; thirdly, it can be used for genotyping of isolates with less than 6 copies of IS6110. However, the discrimination power of spoligotyping may not be high enough thus a second genotyping method is usually required when spoligotyping is applied. Additionally, it usually needs commercial kits for analysis12. Mycobacterial Interspersed Repetitive Unit (MIRU) typing has been increasingly used to solve these problems because it is easy to perform, takes less time and requires low amount of DNA extracts13,14. Furthermore, it allows the high-throughput analysis of clinical isolates15. Additionally, the results are expressed as numerical codes and therefore, easy to be compared and exchanged with those from studies carried on different locations and population. Currently, MIRU typing is often based on 12 MIRU loci13,15–17 and it has been integrated into TB control systems at a national scale, for example, in the USA14.
Molecular epidemiology of tuberculosis has been studied in some areas in China15,16. The previous study showed that the MIRU typing could serve as the first-line typing methods in epidemiological study of M. tuberculosis from the east China. However, a few efforts have been made for the collection of DNA fingerprint data based on the mutation profiles of isolates from western region of China such as Sichuan Province, where the prevalence of TB is much higher than the average level in China due to the existence of MDR-TB, HIV-positive individuals, and poverty18. Total cases of TB in Sichuan, a province located in the southwest of China with nearly 87 millions populations, were about 272,000 in 2006 and the new cases found in 2006 were more than 70,000 (unpublished data, Sichuan Centre for Disease and Prevention). Although since April 30, 2004, DOTS (Directly observed treatment, short-course) has been executed at county level for up to 100 per cent of TB patients under surveillance programme guided by Department of Heath of Sichuan Province, the incidence of TB is still hazardous, and no study on molecular genotyping of M. tuberculosis isolate from Sichuan has been reported so far. Previously, we reported molecular mutation type of drug-relevant target genes on drug-resistant M. tuberculosis isolates from Sichuan and showed the existence of geographic variation on the molecular genetic mechanism of drug-resistance19. This study was undertaken to genotype M. tuberculosis isolates from patients in Sichuan on the Mycobacterial Interspersed Repetitive Units, and an epidemiological analysis of tuberculosis, especially drug-resistant tuberculosis in Sichuan, was also conducted.
Material & Methods
Strain selection and drug susceptibility testing: All isolated were obtained from patients suffering from pulmonary tuberculosis who were hospitalized from July 2006 to June 2007 in Chengdu Antituberculosis Hospital, the only professional anti-tuberculosis hospital of ethical clearance in Sichuan. Specimens were collected, and disposed by the guidelines of the World Health Organization20. All the tested isolates were characterized as M. tuberculosis by paranitrobenzoic acid and thiopnene 2 carborylic acid hydrazide20.
All the isolates were cultured on Löwenstein-Jensen slants medium at 37°C and identified by using the proportion method20 with the following drug (μg/ml) concentrations: INH, 0.2; RIF, 40; ethambutol (EMB), 2; and streptomycin (SM) 4. Resistance to any of these four drugs tested was defined when 1 per cent or more bacterial growth on drug-containing medium occurred compared with a control medium. The isolates were also identified as M. tuberculosis on the basis of a positive niacin test. The medical records were examined to make sure that the corresponding 89 patients who provided isolates had no familial ties: no TB patients in the same family.
A total of 80 drug-resistant and nine susceptible M. tuberculosis isolates were selected randomly over a period of one year between July 2006 and June 2007.
DNA extract, PCR amplification and electrophoresis: Genomic DNA was prepared by chloroform-isoamyl alcohol extraction as previously described21. The primers for 12 MIRU loci were originally described by Supplly et al17 and synthesized by Invitrogen (Shanghai). Each MIRU locus were amplified individually in 50 μl reaction mixture containing 5 μl of extracted DNA, 50 mM KCl, 10 mM Tris (p H 8.3), 1.5 mM MgCl2, 0.6 μM (each) primer, 200 μM (each) deoxynucleotide triphosphates and 1.5 U Taq polymerase. The cycling parameters included initial denaturation step at 95°C for 5 min, 36 cycles of denaturation at 94 °C for 1 min, annealing 1 min, extension at 72 °C for 1 min, and then a final extension at 72 °C for 10 min.
The PCR products were run on 3 per cent agarose gel. Molecular weight standard ladder (Marker D2000, GE Healthcare) was used every 19 lanes. The reference strain H37Rv, provided by Sichuan c0 entre for Disease and Prevention, with known MIRU pattern, was run as an additional positive control.
MIRU genotyping: MIRU-VNTR analysis was performed essentially as described previously13,17. To test the reproducibility of the results obtained for MIRU loci, the PCRs were repeated and initial results were confirmed. The size analysis of the PCR fragments in agarose gels and the assignment of the MIRU alleles were performed using TotalLab TL100 software (Nonlinear Dynamics Ltd., United Kingdom). The number of MIRU copies for each isolate was determined according to length of the PCR fragments, which was calculated according to a scheme22.
A strain cluster was defined when more than two patients were infected by strains of the same MIRU pattern. To assess the diversity of these types, the electrophoresis bands were converted to “0” or “1” and put into computer. A dendrogram based on 12 MIRU loci was constructed using UPGMA (Unweighted Pair Group Method with Arithmetic Mean) after pair-wise comparison of strains by calculating Jaccard Index through the NTSYS-pc version 2.00 software package (Exeter Software, Setauket, NY, USA).
Statistical analysis: Chi-squared tests were used to compare proportions with unique MIRU patterns, with P<0.05 considered as significant.
The allelic diversity (h) at a given MIRU locus was also calculated as described previously23 and was used to evaluate the discriminatory power of the typing method utilized.
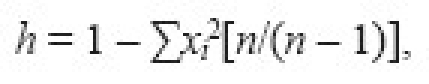
where, xi is the frequency of the ith allele at the locus and the n the number of isolates in the sample23,24. In this study the n was 89.
The discrimination power, evaluated by Hunter-Gaston index (HGI), was calculated by using the following equation24:
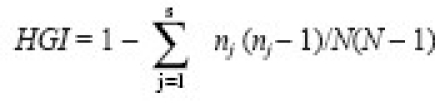
where, N is the total number of isolates in the sample, s the number of different MIRU types described, and nj the number of strains belonging to the jth MIRU pattern.
Results
Drug-resistant phenotypic profile: Among all 198 culture-positive isolates from 198 patients, 6 (3%) were non-tuberculous mycobacteria, one M. bovis, and the others M. tuberculosis. Among all 191 M. tuberculosis isolates from Sichuan province, 80 were resistant to at least one of the tested anti-TB drug, and 112 were sensitive to all of the four drugs tested. Of the 80 drug-resistant isolates, 61 were RIF-resistant, 51 INH-resistant, 46 EMB-resistant, and 38 SM-resistant. Thirty-nine drug-resistant isolates were MDR-TB.
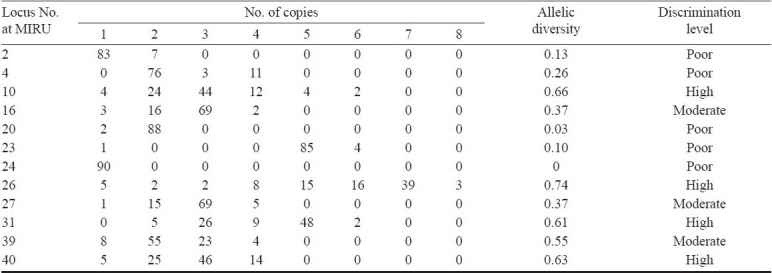
Discriminatory power of MIRU typing: A total of 89 isolates of M. tuberculosis were typed by the 12 MIRU-VNTR loci. All the primers could amplify various lengths of segments since different isolates possess distinctive MIRU except locus 24. A representative diversity of amplified bands at locus 10 for some tested isolates is shown in Fig. 1. The primer 26a/26b produced the most abundant electrophoretic bands, and its PCR products gave 8 different lengths of bands, with 1, 2, 3, 4, 5, 6, 7, or 8 repeat units, respectively. The allelic diversity (h) of the sample studied by every locus, an indirect index of both the heterogeneity of the sample and the discriminatory power provided by the loci, and the distribution of MIRU allele's number are summarized in Table I. The 12 loci investigated showed a remarkable difference in their allelic diversity. Based on this index, 4 MIRU loci in our samples (MIRU nos. 10, 26, 31 and 40) were designated as being “highly discriminant” (h > 0.6), three (MIRU nos. 16, 27 and 39) as “moderately discriminant” (0.3 ≤ h ≤ 0.6), and 4 (MIRU nos. 2, 4, 20 and 23) “poorly discriminant” (h < 0.3). Lastly, one MIRU loci (Locus 24) could not generate polymorphism bands and the allelic diversity could not be detected.
Fig. 1.
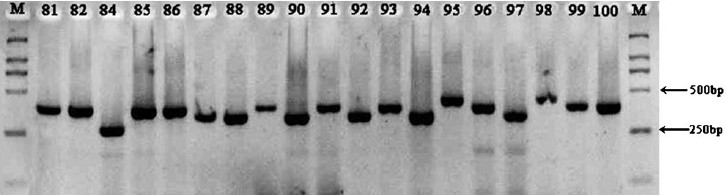
Length polymorphisms with the primers 10a/10b (M:Marker D2000).
Table I.
Allelic diversity of each MIRU-VNTR locus
Cluster analysis and isolate genotype: A single combined numerical tree was constructed based on the MIRU typing. The genetic distances among the isolates based on their MIRU patterns are shown in Fig. 2. Because none of the isolates contained a MIRU locus of more than 9 MIRU units, 12 digital numbers were used to represent the MIRU patterns in isolates. These 12 digital numbers represented the number of repeated copy of each corresponding MIRU loci, e.g., MIRU locus 2-4-10-16-20-23-24-26-27-31-39-40. The numerical MIRU typing gained in this study is listed in Fig. 2.
Fig. 2.
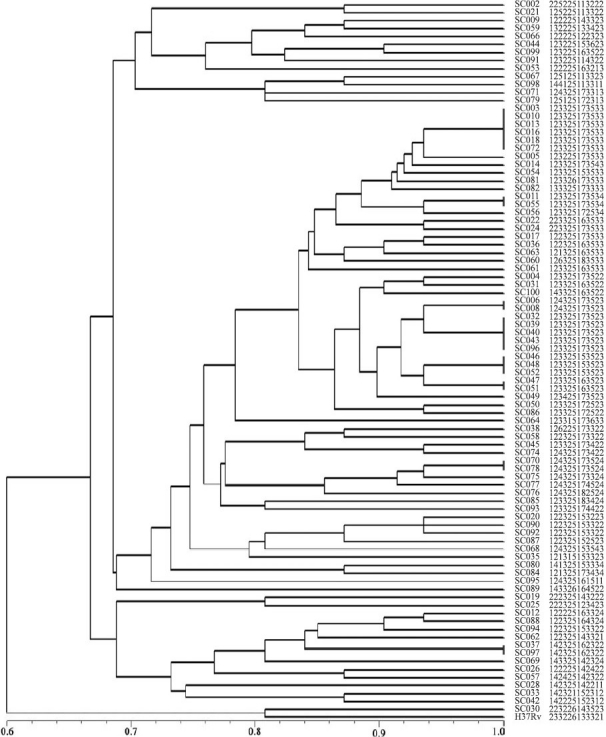
Dendrogram deduced from the clustering analysis of 90 strains.
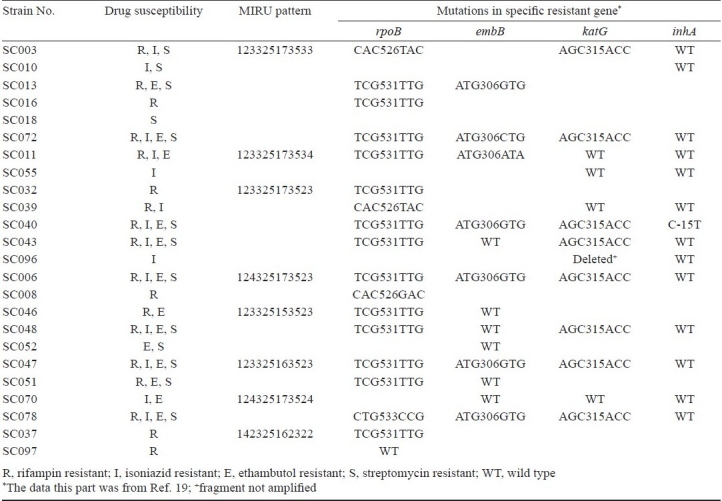
Isolates SC003, SC010, SC013, SC016, SC018, and SC072 shared the same MIRU pattern, but their drug-resistance was varied. SC003 was resistant to INH, EMB, and SM but susceptible to RIF, while isolate SC010 was resistant to SM and INH but susceptible to RIF and EMB. To assess the correlation between MIRU pattern and mutation pattern on specific resistant gene, the drug susceptibility, MIRU pattern and mutation type in specific resistant gene of isolates that clustered together were listed in Table II. Some isolates, such as SC037 and SC097, shared the same MIRU pattern (142325162322) and drug-resistant pattern (resistance to RIF), but these had different mutation pattern on rpoB gene. SC037 contained TCG→TTG mutation on loci 531 while SC097 had a wide type of rpoB gene. Among 65 isolates with distinct MIRU patterns, 41 were resistant to more than two anti-TB drugs, nine to one drug. All the nine drug susceptible isolates have distinct MIRU patterns.
Table II.
Drug susceptibility, MIRU pattern and mutations pattern in specific resistant gene of clustered isolates (n=24) from Sichuan
A dendrogram based on 12 MIRU loci was constructed using UPGMA and shown in Fig. 2. Sixty five of the isolates had unique MIRU patterns and 24 were grouped into 8 clusters (n = 2 for 6 isolates). The isolates within the same group had the same MIRU numerical results. The largest cluster, with six isolates composed a group with the same MIRU pattern, had the numerical order of 123325173533, followed by five isolates composed another cluster with the same MIRU numerical order of 123325173523. Altogether 73 MIRU profiles were obtained. Another group isolates had three isolates with MIRU profiles 123325153523 while the remaining clustered groups had two isolates.
Discussion
In this study, application of MIRU typing on a set of 89 clinical isolates of M. tuberculosis from Sichuan province in China demonstrated that, with a good reproducibility, MIRU typing was a discriminate alternative to IS6110-RFLP, a well-known gold standard method for M. tuberculosis DNA fingertyping, and thus it could be used to type a large amount of isolates due to its fast speed and simplicity.
To evaluate the discrimination power of 12 MIRU typing on the M. tuberculosis isolates, Hunter-Gaston index was utilized. In the current study, we had N = 100, s = 73. n1 = 6, n2 = 5, n3 = 3, n4 to n8 = 2, n9 to n73 = 1, and then HGI = 0.991, which was higher than that from Beijing isolates16, indicating a great discrimination power of 12 MIRU loci on the M. tuberculosis isolates. Further, 11 MIRU loci could generate polymorphic bands, with locus 26 giving the most polymorphic bands, while the locus 24 only produced the same band for all the isolates. Whether the isolates from Sichuan have polymorphism or not at locus 24 deserves further investigation, our result was in agreement with those on isolates from Shanghai and Beijing in China, where polymorphic bands were not detected at locus 2415,16. The isolates from Russia5 and England25 have shown polymorphism at locus 24. Polymorphic MIRU repeats at locus 2 were not detected in both Beijing and Shanghai isolates, whereas in our study, a low polymorphism with the allelic diversity of 0.13 at the locus 2 was detected.
Our results showed that 73.0 per cent isolates (65 in 89) had unique MIRU patterns, which was significantly higher than those from previous studies from Beijing16and Taipei26, while the remaining 24 isolates belonged to 8 MIRU-pattern clusters (each cluster has 2 to 6 isolates). The isolates with the same MIRU pattern possibly have the common epidemic origin, however, these could have different drug-resistant type.
In our study, the isolates with the same MIRU patterns were about 27.0 per cent of total isolates (24 in 89), while the others contained unique MIRU pattern, with the latter making up the majority of M. tuberculosis isolates, especially the drug-resistant ones in Sichuan province of China. Whether the isolates belong to endogenous or exogenous infection deserve further investigation. The exiguous molecular epidemiology data in west China such as Sichuan province could provide information on the TB transmission status in this region. Establishment of MIRU digital profiles at the nearest future is essential for the control of tuberculosis in China, especially in the region of high occurrence of TB like Sichuan. The 12 loci based MIRU typing method, which is fast and cost- efficient with satisfactory discrimination power, may also be regarded as a valuable tool for phylogenetic studies in M. tuberculosis isolates. Another study from China showed that the 15 loci could give better differentiation power of M. tuberculosis27. For this reason, more loci should be tested on the M. tuberculosis isolates from Sichuan region.
Acknowledgments
The authors acknowledge the financial support received from National Nature Science Foundation of China, and the Department of Science and Technology of Sichuan Province of China.
References
- 1.Frieden TR, Sterling TR, Munsiff SS, Watt CJ, Dye C. Tuberculosis. Lancet. 2003;362:887–99. doi: 10.1016/S0140-6736(03)14333-4. [DOI] [PubMed] [Google Scholar]
- 2.Global tuberculosis control: surveillance, planning and financing. Geneva, Switzerland: WHO; 2008. World Health Organization. WHO/HTM/TB/2008.397. [Google Scholar]
- 3.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349:1149–56. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 4.Middelkoop K, Bekker LG, Mathema B, Shashkina E, Kurepina N, Whitelaw A, et al. Molecular epidemiology of Mycobacterium tuberculosis in a South African community with high HIV prevalence. J Infect Dis. 2009;200:1207–11. doi: 10.1086/605930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baranov AA, Mariandyshev AO, Mannsaker T, Dahle UR, Bjune GA. Molecular epidemiology and drug resistance of widespread genotypes of Mycobacterium tuberculosis in northwestern Russia. Int J Tuberc Lung Dis. 2009;13:1288–93. [PubMed] [Google Scholar]
- 6.Doustdar F, Khosravi AD, Farnia P, Masjedi MR, Velayati AA. Molecular analysis of isoniazid resistance in different genotypes of Mycobacterium tuberculosis isolates from Iran. Microb Drug Resist. 2008;14:273–9. doi: 10.1089/mdr.2008.0842. [DOI] [PubMed] [Google Scholar]
- 7.Affolabi D, Anyo G, Faihun F, Sanoussi N, Shamputa IC, Rigouts L, et al. First molecular epidemiological study of tuberculosis in Benin. Int J Tuberc Lung Dis. 2009;13:317–22. [PubMed] [Google Scholar]
- 8.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolotin S, Alexander DC, Chedore P, Drews SJ, Jamieson F. Molecular characterization of drug-resistant Mycobacterium tuberculosis isolates from Ontario, Canada. J Antimicrob Chemother. 2009;64:263–6. doi: 10.1093/jac/dkp183. [DOI] [PubMed] [Google Scholar]
- 10.Martin A, Inigo J, Chaves F, Herranz M, Ruiz-Serrano MJ, Palenque E, et al. Re-analysis of epidemiologically linked tuberculosis cases not supported by IS6110-RFLP-based genotyping. Clin Microbiol Infect. 2009;15:763–9. doi: 10.1111/j.1469-0691.2009.02839.x. [DOI] [PubMed] [Google Scholar]
- 11.Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J Clin Microbiol. 2002;40:1592–602. doi: 10.1128/JCM.40.5.1592-1602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer J, Andersen AB, Kremer K, Miorner H. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microb. 1999;37:2602–6. doi: 10.1128/jcm.37.8.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gendes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan LS, Diem L, Monson T, Wand P, Temporado D, Oemig TV, et al. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 2005;43:688–95. doi: 10.1128/JCM.43.2.688-695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Chen J, Shen X, Gui X, Mei J, Deriemer K, et al. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol Lett. 2008;282:22–31. doi: 10.1111/j.1574-6968.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- 16.Jiao WW, Mokrousov I, Sun GZ, Guo YJ, Vyazovaya A, Narvskaya O, et al. Evaluation of new variable-number tandem-repeat typing system of Mycobacterium tuberculosis with Beijing genotype isolates from Beijing, China. J Clin Microbiol. 2008;46:1045–9. doi: 10.1128/JCM.01869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–71. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 18.Duanmu HJ. Report on fourth national epidemiological sampling survey of tuberculosis in China. Chin J Tuberc Respir Dis. 2002;25:3–7. [PubMed] [Google Scholar]
- 19.Guo JH, Xiang WL, Zhao QR, Luo T, Huang M, Zhang J, et al. Molecular characterization of drug-resistant Mycobacterium tuberculosis isolates from Sichuan Province in China. Jpn J Infect Dis. 2008;61:264–8. [PubMed] [Google Scholar]
- 20.Guidelines for surveillance of drug resistance in tuberculosis. Geneva, Switzerland: WHO; 1997. WHO/ IUATLD; p. 216. WHO/TB/96. [Google Scholar]
- 21.Parra A, Larrasa J, Garcia A, Alonso JM, de Mendoza JH. Molecular epidemiology of bovine tuberculosis in wild animals in Spain: A first approach to risk factor analysis. Vet Microbiol. 2005;110:293–300. doi: 10.1016/j.vetmic.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Frothingham R, Meeker-O’Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–96. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 23.Selander RK, Caugant DA, Ochman H, Musser JM, Gilmour MN, Whittam TS. Methods of multilocus enzyme electrophoresis for bacterial population-genetics and systematics. Appl Environ Microbiol. 1986;51:873–84. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love J, Sonnenberg P, Glynn JR, Gibson A, Gopaul K, Fang Z, et al. Molecular epidemiology of tuberculosis in England, 1998. Int J Tuberc Lung Dis. 2009;13:201–7. [PubMed] [Google Scholar]
- 26.Dou HY, Tseng FC, Lin CW, Chang JR, Sun JR, Tsai WS, et al. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infect Dis. 2008;8:170–81. doi: 10.1186/1471-2334-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama E, Kishida K, Uchimura M, Ichinohe S. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect Genet Evol. 2007;7:499–508. doi: 10.1016/j.meegid.2007.02.006. [DOI] [PubMed] [Google Scholar]


