Abstract
Protein pKa calculation methods are developed partly to provide fast non-experimental estimates of the ionization constants of protein side chains. However, the most significant reason for developing such methods is that a good pKa calculation method is presumed to provide an accurate physical model of protein electrostatics, which can be applied in methods for drug design, protein design and other structure-based energy calculation methods. We explore the validity of this presumption by simulating the development of a pKa calculation method using artificial experimental data derived from a human-defined physical reality. We examine the ability of an RMSD-guided development protocol to retrieve the correct (artificial) physical reality and find that a rugged optimization landscape and a huge parameter space prevent the identification of the correct physical reality. We examine the importance of the training set in developing pKa calculation methods and investigate the effect of experimental noise on our ability to identify the correct physical reality, and find that both effects have a significant and detrimental impact on the physical reality of the optimal model identified. Our findings are of relevance to all structure-based methods for protein energy calculations and simulation, and have large implications for all types of current pKa calculation methods. Our analysis furthermore suggests that careful and extensive validation on many types of experimental data can go some way in making current models more realistic.
Keywords: pKa prediction, Poisson-Boltzmann Equation, F-test
Introduction
Our understanding of protein biophysical characteristics relies critically on the ability to construct quantitative theoretical models that can reproduce and predict experimental data. Measurements of residue-specific protein ionization constants (protein pKa values) via NMR, isothermal titration calorimetry, enzymatic pH-activity profiles and pH-dependent stability changes play a central role in the development of models for predicting electrostatic field effects in and around proteins, and in the last decade the number of experimental data points have more than doubled. The increase in the amount of experimental data has spurred the development of novel algorithms1; 2; 3; 4; 5; 6 that encompass a host of new features and have yielded new insights in aspects protein evolution, structure and function. There is thus much cause for optimism with regard to our understanding of protein structure-function relationships from an electrostatic viewpoint. However, if one examines the improvement in the accuracy of the theoretical models for predicting pKa values, then it is questionable if the increasing amount of experimental data has been paralleled by an improvement in the prediction accuracy. For example, the RMSD achieved by the best pKa calculation packages in 2001 was around 0.8–1.17 pKa units for a dataset of around 140 pKa values, while the present accuracy is similar8 although current datasets are an order of magnitude larger.
We are faced with the question of how to improve the accuracy of current pKa calculation models and thus achieve a better understanding of protein electrostatics. A central issue in establishing the direction of future work is to identify and prioritize applications for a highly accurate pKa calculation package. Applications of pKa calculation packages range from predicting and re-designing enzymatic pH-activity profiles4 to quick evaluations of the pI of a protein, and a large number of applications in protein engineering and protein structure analysis are found between these two extremes. The accuracy and speed requirements for each application of pKa calculations differs, and while very approximate methods are inappropriate for certain types of problems, more time-consuming methods, such as the MD-based algorithms, will be difficult to apply in protein design applications due to the combinatorial explosion encountered in this type of problems. There is thus room for a variety of pKa calculation algorithms due to the many types of problems that we wish to address, and when developing a pKa calculation algorithm we must carefully specify the type of problem we are developing our algorithm to solve and explicitly state what the algorithm can and cannot do. In general, a pKa calculation algorithm benchmarked on pKa values cannot be expected to be able to predict other quantities such as desolvation energies, ΔpKa values or electrostatic site-site interaction energies without having been benchmarked directly on these values.
In spite of this somewhat obvious fact, pKa calculation algorithms benchmarked only on pKa values are often used to study and analyze other biophysical quantities and phenomena because we assume that the rigor of the underlying physical models (e.g. an MD force field, a semi-empirical energy calculation method or the Poisson-Boltzmann Equation) ensures that the adjustable parameters of the method have been calibrated to give a model that describes the physics accurately. In the present paper we examine the validity of this assumption by simulating the development, calibration and testing of a pKa calculation algorithm on a set of simulated experimental data. We simulate the development of the pKa calculation algorithm by combining 10 descriptors, and we then search for the optimal combination of these descriptors using the RMSD between experimental and predicted pKa values as a guide. Since we define the physics underlying the simulated experimental data we can quantify the physical realism of the best pKa calculation method in a given development simulation.
We examine the physical realism that we can expect to achieve in a pKa calculation algorithm using simulated experimental datasets that are similar to the currently available experimental data. We use these simulations to make predictions on how we should optimize and test pKa calculation algorithms and which experimental datasets we would like to have access to.
Assessing the physical realism of pKa calculation algorithms
Protein pKa calculation algorithms come in many flavors that differ in their physical realism. QM/MM and MD-based methods are relatively close to first principles, whereas continuum-based methods using Poisson-Boltzmann solvers are somewhat more removed from first principles. Similarly it is clear that methods consisting mostly of adjustable parameters connected to simplified physical or empirical equations are far removed from physical reality, but it is unclear how we quantitatively assess the physical realism of pKa calculation algorithms in general. We start by defining a fully physically realistic pKa calculation algorithm as one that includes a full quantum mechanical description of an entire protein conformational ensemble and its surrounding solvent using a perfect basis set. The pKa values of all ionizable residues are found by performing simulations at every 0.1 pH value and using these to determine the charge vs. pH titration curves. The resulting titration curves are then fit using the Henderson-Hasselbalch equation to extract the final pKa value. In the following we refer to this model as a “full, true microscopic model”. Such a model is presently computationally unfeasible and will probably never be achieved, but it serves as a useful reference point for the development of our framework.
We now envisage a situation where we gradually simplify the true microscopic model by substituting parts of the algorithm with a more approximate physical description, by reducing simulation time to capture only a portion of the conformational ensemble, by calculating pKa values from a single simulation at a given pH, by using classical force fields rather than quantum mechanical descriptions, or by making some other adjustment that makes the procedure less physically realistic. The performance of our method will now have deteriorated, and additionally we now have one or more parameters that we can adjust to optimize prediction accuracy. These parameters can be part of the approximate description of the physics (e.g. a dielectric constant, a scaling factor or a simplified force field), they can be technical simulation parameters (simulation time, time step, integration scheme) or they can be part of the sampling algorithm (e.g. they can be contained in the inner workings of a rotamer sampling scheme or in a hydrogen-building algorithm). We can repeat the simplification procedure until we have replaced the full, true microscopic model with fully empirical equations based on principles similar to those of the Hammett equation9 and other forms used in QSAR methods. At some point during this simplification process we reach a situation where the adjustable parameters lose their physical meaning and start becoming variables that simply are adjusted to give the best agreement with the experimental data used in the calibration/training process. We believe that this transition from physical to empirical parameters happens quite early and that the parameters/methods used in constant pH MD simulations to a certain extent are empirical. This is corroborated by the large variation observed in the performance of different constant pH MD simulations2; 10; 11, which suggest that many parameters/choices reflect the performance of these algorithms. We are therefore concerned with examining the consequences of the presence of empirical parameters pKa calculation methods. In the present article we simulate the development of pKa calculation methods using calculated experimental data to examine our ability to identify the most physically realistic method when being guided only by the agreement between experimental and predicted pKa values.
Materials and methods
WHAT IF pKa calculations
pKa calculations with the WHAT IF pKa calculation package (WIpKa) were carried out as described previously7 with the exception that a uniform protein dielectric constant of 8 was used unless stated otherwise. WIpKa employs DelPhi II12 for solving the linear form of the Poisson-Boltzmann Equation and uses a hydrogen bond optimization scheme13 to arrive at the best possible hydrogen bond network for a given protonation state. This pKa calculation method was initially calibrated on a set of 120 experimental pKa values and optimized to predict pKa values for active site residues.
pdb2pka pKa calculations
The open source pdb2pka package utilizes pdb2pqr14 to construct hydrogen positions and assign charges/radii. pdb2pka employs a hydrogen bond sampling network similar to that used in initial versions of MCCE15 and uses APBS16 to solve the PBE.
Null-model calculations
The null model is defined as the situation where all titratable groups have a pKa value similar to their model pKa value. In the current study we use model pKa values if N-terminal: 8.0, Lys: 10.4, Glu:4.4, His:6.3, Asp:4.0, Tyr: 9.6, Arg: 13.0, C-terminus: 3.8, Cys: 8.7.
Simulated experimental datasets
Experimental datasets were simulated by randomly generating artificial environments for the titratable groups in a protein. A given protein (P) with N titratable groups was generated as follows:
Each titratable group is randomly chosen to be an Asp, Glu, His, Lys, or Arg. The distribution is skewed to ensure equal amounts of acidic and basic titratable groups. In addition each protein contains an N-terminal titratable group and a C-terminal titratable group.
Each titratable group interacts with 0–15 non-titratable atoms (uniform distribution) that each have the following randomly chosen characteristics: charge (−1.0 → 1.0) and distance (3 → 15 Å). Both the charge and the distance are chosen from uniform distributions.
The distances between titratable groups in the protein are chosen from a Gaussian distribution (mean 21.0 Å and std. deviation 8.0 Å) chosen to mirror the distances between titratable groups observed in SNAse. A notable feature of the experimentally observed distribution is an increased probability that a favorable interaction is observed at distances < 10Å. We reproduce this observation by picking 10 % of favorable interactions from a Gaussian distribution with mean 8.0 Å and std. dev. 2.0 Å.
pKa values are calculated from the above characteristics through the calculation of intrinsic pKa values (second term in Eq. 1) and site-site interaction energies (third term in Eq. 2) that are combined linearly to facilitate the use of linear regression.
| (Eq. 1) |
In Eq. 1 pKa,mod,i is the model pKa value of that residue, γi is −1 for acids and +1 for bases, rij are distances in Ångstrom between groups i and j. The first sum is over all atoms that influence the intrinsic pKa value, whereas the second sum is over all titratable groups.
The above method was calibrated to provide pKa value distributions similarly to those observed in currently available experimental datasets (Figure 1). Table 1 provides a summary of the characteristics of the four types of datasets used in this work. We refer to theses datasets at Set<name><number of pKa values>, so that SetA1000 refers to 1000 simulated pKa values generated according to the settings for set A in Table 1.
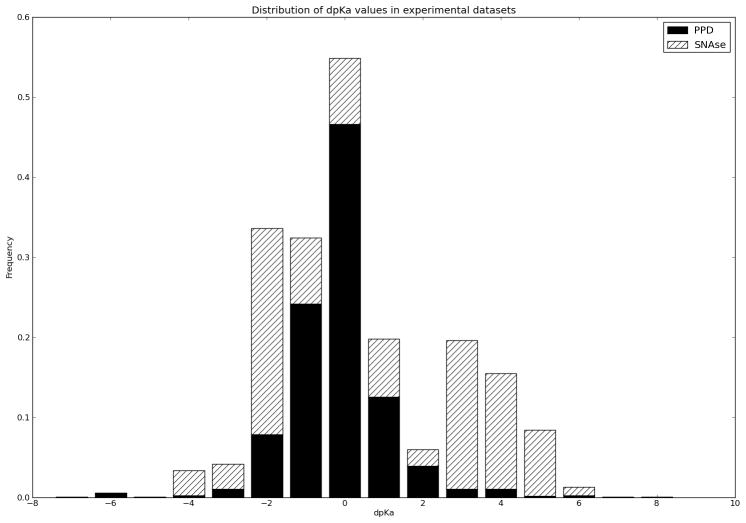
Figure 1.
The distribution of ΔpKa values relative to amino acid model pKa values in the PPD33 and in the SNAse dataset24. The majority of ΔpKa values are within +/−1.0 units for most proteins, whereas the SNAse dataset stands out in containing a large number of highly perturbed pKa values.
Table 1.
Characteristics of simulated datasets used in this work. Sets Default and A model ‘average’ proteins, set B models effect pKa values that are affected only by desolvation and background interaction energies, whereas set C models pKa values that are perturbed only through charge-charge interactions.
| Dataset | max. # of near groups | Near group distance (Å) | # of groups per protein | Favorable charge interactions | Unfavorable charge interactions |
|---|---|---|---|---|---|
| Default | 10 | 3–15 | 50 | 0.1 | 0.0 |
| Set A | 5 | 3–15 | 50 | 0.0 | 0.0 |
| Set B | 10 | 3–15 | 1 | 0.1 | 0.0 |
| Set C | 0 | - | 50 | 0.5 | 0.4 |
Near groups: Groups influencing the intrinsic pKa value.
Favorable charge interaction: Fraction of charge-charge interactions that are strongly favorable.
Unfavorable charge interactions: Fraction of charge-charge interactions that are strongly unfavorable.
Simulating the development of a pKa calculation method
We examine the ability of an automatic RMSD-guided optimization procedure to find expressions of the type provided by Eq. 1. We do this by randomly combining fragments of this and other equations and evaluating the ability of these combined equations to reproduce and predict a simulated set of pKa values.
Specifically we use the following fragments:
| (Eq. 2) |
for calculating the intrinsic pKa value, and the fragments
| (Eq. 3) |
for the calculation of site-site interaction energies. Each pair of fragments can be combined as a product or as a sum. In the latter case each term in the sum is scaled by a constant, which increases the number of freely adjustable parameters.
The final combined model is fitted to the training set using linear regression to optimize the free adjustable parameters in the specific model. The physical reality of each model is calculated by representing each model as a string of 28 bits, where odd bits represent the absence/presence of a specific term and the even bits decide whether two terms are combined as a product or as a sum. The physically correct model (Eq. 1) is also represented by a string of bits, and the physical reality is calculated as the percentage of bits that are identical when comparing a given model to the correct model. It is important to note that this quantification of physical realism is not linear, and that each term does not contribute equally to the final pKa value. The physical realism should therefore be taken only as a preliminary attempt at defining the concept of physical realism in biophysical models.
Finding the best model
The model describing a simulated dataset best is found by a simple RMSD minimization procedure or by a Monte Carlo optimization procedure. In the minimization procedure terms are added sequentially in the order of the most beneficial as judged by the decrease in RMSD to the training set. Terms are added as long as they give at least an 0.01 improvement in the RMSD.
In the Monte Carlo optimization procedure a new model is chosen by randomly switching 1 to 3 bits in the model definition thus allowing fairly large jumps in model space. A new model is chosen if it gives an improvement in the RMSD to the training set.
F-test
The F-test compares the sum of squares (SS) of two models to the number of parameters used to achieve that variance.
| (Eq. 4) |
NDF1 and NDF2 is the number of degrees of freedom for models 1 and 2 respectively, and is calculated as the number of parameters subtracted from the number of data points. The p-value of the F-statistic is calculated from the incomplete beta function, I, by numeric methods [Numerical Recipes, 3rd ed.].
| (Eq. 5) |
If there is no significant improvement in the variance upon switching to a model with more degrees of freedom (p (F, NDF1, NDF2) > 0.05), then that fitting model is discarded. Otherwise the more advanced model is accepted and the procedure is continued with another parameter added.
Results and discussion
We start by examining how likely we are to arrive at a physically interpretable pKa calculation algorithm as a function of the size and characteristics of the experimental dataset that is available to us for training and benchmarking procedures. We perform this analysis using simulated datasets of varying sizes and characteristics, and use these to train, select and benchmark a set of automatically constructed pKa calculation algorithms. Finally we examine a very simple but quite accurate model for reproducing the SNAse mutant pKa values used in the pKa cooperative blind test exercise, and ask what this tell us about the development of more sophisticated pKa calculation procedures.
We simulate the development of protein pKa calculation using four simulated datasets (see Table 1 for dataset simulation parameters): An unbiased dataset (default dataset), a dataset similar to what is available in databases and literature (dataset A), a dataset containing highly shifted pKa values due to high desolvation penalties (dataset B), a dataset with highly shifted pKa values due to strong electrostatic interactions (dataset C).
Does optimization guided by RMSD yield the most physically realistic model?
The development of a protein pKa calculation algorithm is a tedious process that is dominated by a process of programming, scripting, bug fixing and benchmarking to identify an algorithm that gives good results with a model that is judged to be physically sound. Often improvements/changes to the algorithm initiated by the study of cases where the algorithm does poorly. E.g. being able to correctly predict the pKa values of Glu35 (buried, pKa = 6.2) and Asp66 (buried, pKa < 2.0) in HEWL requires that the charge-stabilizing hydrogen bonds to Asp66 offset the desolvation penalty for this residue. Attempting to get these two pKa values right will thus often lead to changes in the hydrogen bond model used in the pKa calculation method. Following such an algorithmic change, the algorithm is re-benchmarked on the training dataset and the next problem is identified and corrected. Typically this process continues until the algorithm reaches an RMSD on a benchmarking dataset that is ‘low enough to publish’, and the pKa calculation model is thus built up in a piece-meal fashion.
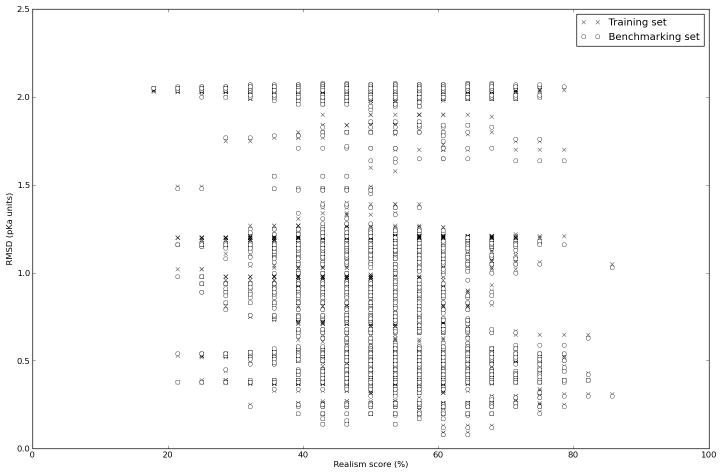
In this process the scientist is guided by a measure of accuracy of the pKa calculation algorithm, typically the RMSD between calculated and experimental values, and the accuracy thus drives the development of the pKa calculation algorithm; what improves the RMSD stays, what makes it worse is left out. We investigate if such an optimization process is well suited for identifying the physically most realistic model, by simulating the development of a pKa calculation algorithm using RMSD as a guide. Figure 2 shows the RMSD of all models evaluated in a Monte Carlo optimization run plotted as a function of their realism score. The best models evaluated in this simulation achieve an RMSD around 0.2 units, which compares very favorably to the RMSD 2.1 achieved by the null model. However, the same models only achieve a physical realism score of around 60%. The best performing model has a realism score of 61% and takes the following form:
| (Eq. 6) |
with k1 to k5 taking values of 2.5, 0.13, −3.4*10−3, 1.8*10−3, and 2.82 respectively. While this equation certainly is very close to Eq. 1, it is not physically correct. Nevertheless, the presence of 5 free parameters enables this model to achieve a low RMSD for the simulated experimental dataset.
Figure 2.
The realism score plotted vs. the RMSD between experimental and calculated pKa values for 1000 step Monte Carlo optimization run using the automatically generated pKa calculation models. It is evident that the correlation between RMSD and realism score is very weak.
Thus, the simulations are not able to identify the physically correct model, but the results of the models are virtually indistinguishable from the physically true model on the current datasets. For the datasets of 1000 pKa values used above, the difference in RMSD between the training and benchmarking datasets is insignificant, and the model is thus not over fitted to the training set. However, if we use smaller datasets this changes significantly with datasets of size 100 giving and average difference of 0.03, and datasets of size 10 giving a difference of 0.21. (Table 2)
Table 2.
Effect of the size of the experimental dataset and sampling protocol on the RMSDs achievable and the physical realism of the identified models when trained on the standard dataset. Larger experimental datasets give a smaller RMSD difference between the training dataset (RMSDtrain) and the benchmarking dataset (RMSDtest). The best models typically achieve a physical realism of only 55–65%.
| # of MC runs | MCsteps per run | # pKa values | RMSDtrain (stddev) | RMSDbench (stddev) | Avg. physical realism (std. dev) | Best model RMSD (realism) |
|---|---|---|---|---|---|---|
| 10 | 100 | 10 | 0.21 (0.04) | 0.42 (0.13) | 57% (9.3%) | 0.04 (66%) |
| 50 | 100 | 10 | 0.22 (0.21) | 0.47 (0.88) | 56% (8.3%) | 0.05 (64%) |
| 100 | 100 | 10 | 0.19 (0.14) | 0.36 (0.30) | 57% (7.8%) | 0.00 (57%) |
| 10 | 100 | 100 | 0.24 (0.20) | 0.27 (0.25) | 60% (7.4%) | 0.06 (64%) |
| 50 | 100 | 100 | 0.15 (0.05) | 0.20 (0.08) | 62% (7.3%) | 0.00 (57%) |
| 100 | 100 | 100 | 0.19 (0.10) | 0.20 (0.11) | 61% (7.8%) | 0.00 (79%) |
The model development space produced by the combination of the 10 expressions (Eqs. 2 and 3) is very large and is therefore not sampled completely during the Monte Carlo procedure. The identification of the most physically realistic model is therefore also dependent on the ability of the Monte Carlo procedure to make big enough jumps in model space and on an RMSD gradient leading to the physically correct model. Figure 2 gives a good impression of the ruggedness of the model RMSD landscape and convincingly shows that there is no general RMSD gradient leading to Eq. 1. Instead the RMSD gradient favors models with a large number of adjustable parameters (See figure SI3) since these models can be fitted to a wide range of experimental data sets.
Different types of data
If a given model, although not perfect, is gives results very close to the true model, then what are the consequences of using such a model? The answer to this question is twofold; first and foremost it means that the pKa calculation model can only be used to predict pKa values from a distribution similar to the one used for training and benchmarking. It is important to note that it is not enough for the distribution of pKa values to be the same as that of the training set – the underlying physical effects that produce the pKa values must also be the same.
Secondly it means that any physical quantities other than pKa values can be incorrect. These first of these effects is demonstrated in table 3, where the performance of four models optimized on each their dataset (setA100, setB100, setC100 or setC1000) is compared using the same four datasets. The performance of each model is clearly skewed depending on the dataset used to optimize the model, with each model performing best on the dataset it was trained on. In the case of the models trained on setC100 and setC1000, the model is not able to predict any pKa changes originating from changes in the intrinsic pKa value, thus clearly indicating that this model is physically unrealistic as a general method for predicting protein pKa values. The function form of each model closely reflects the data used for training the model as illustrated by Table 4.
Table 3.
Training set dependence of pKa calculation models as shown by the RMSD values between calculated and simulated experimental pKa values for three datasets. Each model was optimized using a greedy optimization algorithm using different training datasets. The results show that the training set used has a big impact on the ability of each model to accurately predict pKa values for a given set. The relatively small size of the training set (100 pKa values – 1000 pKa values for GreedyC1000) furthermore results in significant differences between the training set RMSD (values in parentheses) and the benchmark test set RMSD. The lowest RMSD for each method is shown in bold and is found for the training set for 3 of the 4 models.
| Model/Dataset | RMSD SetA100 | RMSD SetB100 (intpKa) | RMSD SetC100 (Charge) |
|---|---|---|---|
| GreedyA100 | 0.37 (0.34) | 0.42 | 0.54 |
| GreedyB100 | 0.51 | 0.42 (0.43) | 0.85 |
| GreedyC100 | 0.76 | 2.02 | 1.11 (0.33) |
| GreedyC1000 | 0.92 | 2.02 | 0.43 (0.76) |
| null model | 0.90 | 2.02 | 0.76 |
Table 4.
Functional forms of the models found during the optimization shown in Table 2.
| Model | pKaint term | Charge-charge interaction term | |
|---|---|---|---|
| GreedyA100 | −K*γi | K+K*γi | |
| GreedyB100 | −K*γi | K*γi | |
| GreedyC100 | - | K+K*γi +K*γk | |
| GreedyC1000 | - |
|
The effect of noise in experimental data
Until now we have used simulated experimental datasets that are infinitely accurate. However NMR-measured experimental pKa values are only accurate to within 0.2–0.3 units in best cases and significantly more inaccurate in the worst cases17. Similarly structural data has a finite accuracy, which is highly dependent on the method used structure determination. In the case of X-ray structures the space group has been found to have an effect on the calculated pKa values18, and other experimental conditions presumably influence the structural details for a large number of proteins. pKa calculation methods that use a fixed representation of the protein structure are more sensitive to the structural errors than those who employ some element of structural relaxation. However, using structural relaxation also carries the risk of making parts of the protein structure unrealistic, and thus increases the risk that a physically unrealistic model is chosen during the optimization/calibration of the pKa calculation method. Table 5 shows the effect of adding various levels of noise to the simulated experimental data and demonstrates that the noise has a large effect on the RMSD of the best model. Surprisingly, the physical realism score of the best method does not change with increasing levels of noise. The explanation for this result is shown in figure SI3, which plots the average number of parameters vs. the physical realism of each method. The plot shows a broad optimum ranging from 40 – 60% and further supports the conclusion that an RMSD-based optimization of a pKa calculation algorithm tends to gravitate towards models with a large number of adjustable parameters. The functional forms of the models identified and the associated parameters differ substantially as would be expected. Finally it is worth noticing that for several of the datasets, the RMSD found during minimization is as low as the RMSD for the correct model. This result is significant since it shows that physically incorrect models with many free parameters are able to exploit patterns in random noise to achieve very low RMSDs. Indeed for higher levels of noise and smaller experimental datasets we have observed models with a little physical realism achieve RMSDs lower than that of the physically correct model.
Table 5.
The effect of including experimental noise for pKa calculation model optimizations using the minimization protocol. A single minimization was performed on Set Default dataset with 1000 pKa values. All RMSD values are reported for the training set only. pKa noise: standard deviation of Gaussian noise added to experimental pKa values, Struct noise, standard deviation of noise (in Å) added to interatomic distances, RMSDsearch: RMSD of best model found during minimization, RMSDcorr: the RMSD of the physically correct model, RMSDpath: the highest RMSD encountered on the best path from the best model found during the search to the physically correct model, numpar: number of free parameters for best model found during minimization.
| pKa Noise | Struct noise (Å) | RMSDsearch | numpar | Realism (%) | RMSDcorr | RMSDpath |
|---|---|---|---|---|---|---|
| 0.0 | 0.0 | 0.08 | 10 | 60% | 0.0 | 0.0 |
| 0.2 | 0.2 | 0.21 | 10 | 61% | 0.20 | 0.44 |
| 0.5 | 0.5 | 0.51 | 7 | 64% | 0.51 | 0.62 |
| 0.5 | 0.0 | 0.50 | 10 | 61% | 0.50 | 0.61 |
| 0.0 | 0.5 | 0.12 | 10 | 68% | 0.11 | 0.40 |
pKa calculation performance as a function of parameters and calculation methods
Having established that the development of our simulated pKa calculation methods are uniquely sensitive to the characteristics of the experimental datasets used, it prudent to ask if real-world pKa calculation methods share this characteristic. It is well known that the value of the protein dielectric constant influences pKa calculation results significantly, but also details of the calculational method such as the explicit modeling of protein dynamics has the ability to change calculational results significantly. This is evident by papers that describe improvements due to hydrogenbond network optimization15; 19; 20, rotamer optimization5, general conformational sampling21 and constant-pH MD simulations2; 10; 22; 23. The choice of X-ray structure and the protocol used for preparing/filtering the used X-ray structures similarly has a large effect, and taken together this data strongly suggest that real pKa calculation algorithms and protocols possess many of the characteristics displayed by the simple pKa calculation models investigated here.
SNase mutant pKa values
We now turn our attention to the issue of validating pKa calculation algorithms on the set of SNAse pKa values produced by Garcia-Moreno and co-workers24. This issue of Proteins contains a number of papers [refs] reporting the performance of various pKa calculation methods on this set of mutant pKa values. We have indeed also performed a validation of our own methods; the WHAT IF pKa calculation package (WipKa 7) and pdb2pka14 and found the performance to be similar in accuracy to most other methods. However, instead of providing a navel-gazing account of where we go wrong with our models, and where we succeed, we are interested in examining what a good or a poor performance on this dataset means when attempting to assess the physical realism/goodness of a specific pKa calculation algorithm. As shown earlier, a low RMSD of a pKa calculation algorithm does not guarantee that the model is physically realistic, and the question is thus what we can learn from a blind prediction exercise such as the one undertaken by the members of the pKa cooperative.
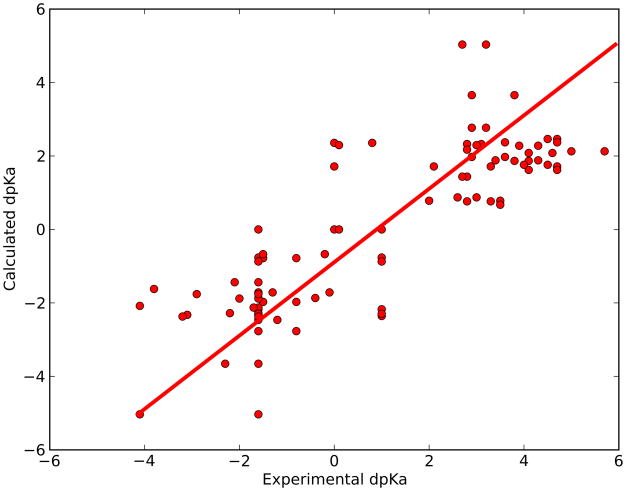
Figure SI2 shows a histogram of the RMSD values obtained by members of the pKa cooperative. The average RMSD for all methods is 2.2 pKa units, and display a large variability in RMSD from 0.9 to 3.8 pKa units. Only 7 of 35 submissions achieved an RMSD lower than 1.7. Three of these were predictions for a limited number of groups (17, 9 and 21 pKa values, respectively), and a further two predictions were 2nd round prediction which allowed calibration of the algorithms on a subset of the SNAse pKa values. We had previously speculated that a very simple model (Eq. 7) could produce such a correlation. Figure 3 displays the correlation between experimental and predicted pKa values for the SNAse dataset, then predicted with equation 7:
| equation 7 |
where α is a constant, acc (res) is the number of heavy atoms within 8.0 Å of the wild type residue and γ is −1 for acids and +1 for bases. A simple linear fit of this equation to the observed pKa values gives an optimal value of α = −0.153 and produces an RMSD of 1.7 and a correlation coefficient of 0.84. This model is therefore better at reproducing the pKa values of the SNAse mutant dataset than the vast majority of the pKa calculation methods examined in the pKa cooperative blind prediction exercise.
Figure 3.
The pKa value shifts displayed by the 100 inserted titratable groups in the SNAse dataset can be reproduced using the model ΔpKa = αγ*acc(res), where α is a freely adjustable parameter, acc(res) is the number of heavy atoms surrounding the wild type residue, and γ is a parameter indicating if the inserted residue is an acid (γ= −1) or a base (γ=+1). A simple linear fit gives an optimal value of α = −0.153 and produces an RMSD of 1.70 and a correlation coefficient of 0.84.
The reasons for this difference in accuracy are many. First and foremost Eq. 7 is calibrated on the SNAse mutant dataset and is therefore not truly predictive. Furthermore the SNase dataset is unique because it (presumably) predominantly describes desolvation-induced pKa shifts. Since all other pKa calculation methods have been benchmarked and trained on previously available collections of pKa values that consist mostly of pKa values for solvent accessible residues, there is the possibility that these methods have been over fitted to the more natural distributions of pKa values. Secondly, Equation 7 does not utilize 3D structures or models of the mutants of SNAse, but calculates acc (res) from the wild type SNAse structure. The performance of Equation 7 is therefore not influenced by uncertainties introduced by the modeling of mutant structures to the same extent as methods that use explicitly modeled 3D representations of the SNase mutants.
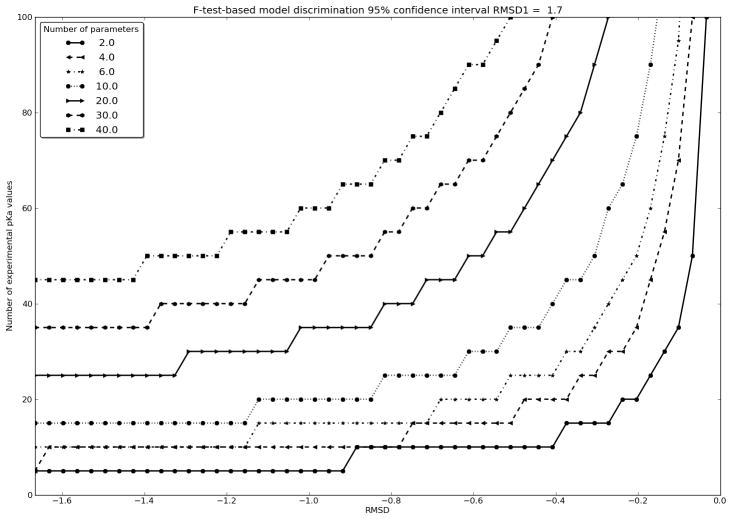
We now seek to examine how we can establish that a full-fledged pKa calculation algorithm provides a more realistic physical description of protein pKa values than that provides by Equation 7. The first requirement is of course that the other method has an RMSD lower than that of Equation 7. It must furthermore be a requirement that an F-test demonstrates that the better agreement obtained with the pKa calculation algorithm is not simply due to more parameters. An F-test is dependent on the residual sum of squares of the two models, the number of parameters in each model, and the number of data points in the dataset. Eq. 7 has 1 free parameter and has been trained on 100 pKa values. To establish which RMSD we should expect for a physically more realistic pKa calculation algorithm we plot the required RMSD improve relative to Eq. 7 vs. the number of pKa values in the dataset for pKa calculation models with different numbers of parameters (Figure 4). It is difficult to estimate the number of free parameters in PBE-based or MD-based pKa calculation models, especially since procedures and algorithmic choices in both structure preparation, electric field calculation and pKa value determination to a good approximation can be described as partially free parameters. If one adds force field values, dielectric constants, GB model parameters etc., one can achieve very high estimates for the number of free parameters in these methods, and it is therefore quite likely that the improved descriptions of the SNase dataset achieved by some methods, indeed is statistically insignificant if the full parameter set has been validated only on a limited set of pKa values.
Figure 4.
Required ΔRMSD for a given pKa calculation model (model2) relative to the performance of Eq. 7 vs. number of pKa values in experimental dataset. The different lines refer to number of free parameters in model2. It is seen that a pKa calculation model with 10 free parameters requires a ΔRMSD of at least −0.2 for 100 pKa values for it to provide a better description of the SNase dataset than Eq. 7.
Discussion
In the present paper we have simulated the development of a pKa calculation algorithm in an artificial physical reality. We acknowledge that the artificial reality employed here is simplistic and that significant differences exist between an automatic pKa calculation model optimization procedure and human-based careful construction of predictive models. However, while the details of the conclusions might differ due to differences between the artificial reality and real life, the following main conclusions will hold true:
The optimization of a pKa calculation algorithm guided by RMSD is not guaranteed to arrive at a physically realistic model. Moreover, the characteristics of the most accurate model depend greatly on the characteristics of the dataset used for calibrating the pKa calculation method.
The the inclusion of experimental noise produces significantly altered models and obscures the difference in performance between unrealistic pKa calculation methods and the physically correct method.
A simple one-parameter model can explain the pKa values in the SNAse mutant dataset to a high degree of accuracy, thus raising questions on the significance of the RMSDs reported by other methods in this volume.
Together these three observations suggest that pKa calculation model optimization is a complex procedure that can lead to the construction of models with limited validity.
The importance of the experimental dataset
The experimental dataset plays a very important role when developing statistical methods and parameterized semi-empirical methods because the dataset determines the subsection of physical reality that the method can hope to describe. It is non-trivial to assess the bias that the experimental dataset introduces when developing pKa calculation methods based on more rigorous physical descriptions such as those employed in Poisson-Boltzmann Equation solvers, constant pH MD simulations and QM/MM methods, since both human physical intuition and the dataset determines which model we will arrive at. Here we make the point that human physical intuition can be deceived into thinking that a parameter carries a physical meaning in cases where it behaves more like a free parameter.
It is well known that PBE-based pKa calculation methods can be tuned to give a very wide range of answers depending on the protein dielectric constant, the PBE boundary conditions, the force field and several calculational details. One might argue that this is insignificant since good physical reasons exist for choosing the ‘right’ parameter combinations, parameter values and the correct way of modeling dynamics, boundary conditions etc. However, a quick scan of the literature reveals that good arguments for almost any value of the dielectric constant 7; 25; 26, choice of force field or choice of calculational procedure can be produced, thus effectively nullifying the supposedly positive contribution of human physical intuition.
The parameters, calculational details and structure preparation procedures for PBE-based pKa calculation methods can therefore arguably be we viewed as parameters that have been chosen to give the best agreement with experimental results. This might be perfectly acceptable provided that PBE-based pKa calculation methods are used only to calculate pKa values. However any conclusions regarding the magnitude of energetic terms not derived from pKa values must be interpreted very carefully if the method in question has not been benchmarked on this type of energy. A solution to this problem is to benchmark protein pKa calculation methods on other types of experimental data such as electrostatic interaction energies27, ΔpKa values4; 28, measurements of electric field differences29 and measurements of the intrinsic pKa value. In addition the field still has to develop an appreciation for the impact of experimental uncertainty30; 31 on the performance of pKa calculation algorithms.
An argument along the same lines can be constructed for MD-based methods for pKa calculations (and indeed for any other MD-based application). Since MD force fields and MD simulation setups contain many more parameters than PBE solvers, the comparison between fundamental energetic terms and experimentally observed quantities is even more indirect. Similarly to PBE solvers, we believe that MD-based methods should be carefully benchmarked on a large body of experimental data that probes individual energies and forces to ensure that all terms and simulation procedures are physically realistic.
Producing a balanced dataset
Until we have accumulated enough additional experimental data it is important to train and benchmark protein pKa calculation methods on diverse sets of experimental data as demonstrated above, since this is likely to give more physically realistic models. Initial progress in this area has been provided by the publication of the present SNAse dataset24 and the Houk32 dataset of highly perturbed pKa values, and these will hopefully be used extensively for future benchmarking purposes. Nevertheless, the number of relatively unperturbed pKa values is orders of magnitudes larger than the number of highly perturbed pKa values33; 34, and training on the full dataset of pKa values will therefore naturally produce a pKa calculation model that is better at predicting relatively unperturbed pKa values. Researchers should consequently take care to specify the conditions that their model performs well under, and we encourage the development of automatic methods that will determine if the energetic environment of a specific titratable group falls within those conditions. Finally we believe that it is important that the protein electrostatics community continues to hold blind prediction exercises to provide independent evaluation of model performance since this is the only way in which we can obtain independent confirmation of the performance of a model.
Assessing the state of the pKa calculation field
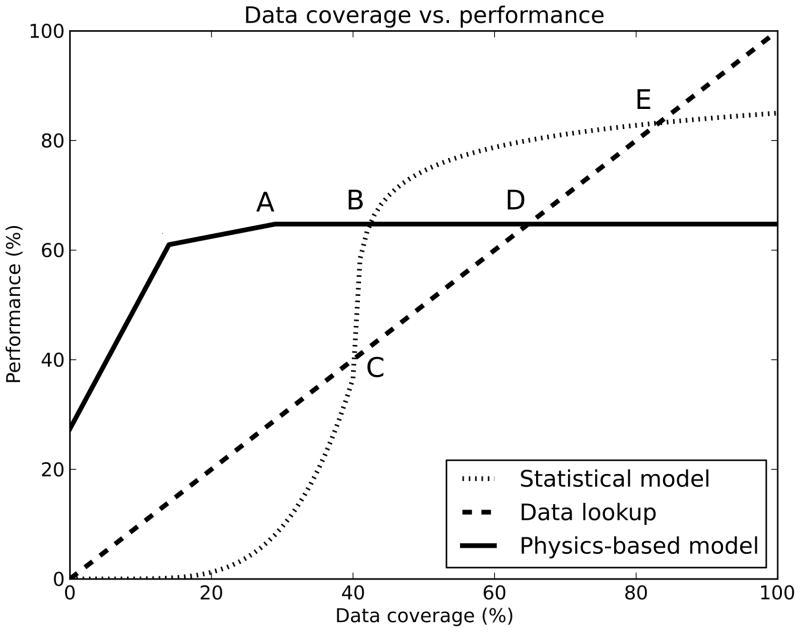
Presently pKa calculation algorithms can provide estimates of protein pKa values that are accurate to within 1.0 units for most experimental datasets whereas more challenging cases, such as the ones provided by the SNase dataset, are predicted with a significantly lower accuracy. To determine how to focus our future efforts, we find it instructive to examine a diagram such as the one provided in figure 5. In this figure three hypothetical pKa calculation algorithms are compared. A simple data look-up model is compared with a statistical/empirical model and a physics-based model, and whereas the performance of the data look-up model is fixed, the relative performance of the two other models will depend on their characteristics. The general features of the plot should, however, be universally applicable.
Figure 5.
Theoretical performance of an approximate physics-based model, a statistical model and a simple data look-up as a function of data coverage. We can define four important points: A: the point where the approximate physical model cannot be improved further, B: The point where enough experimental data is available for the statistical model to surpass the physics-based model, C: the point where the statistical model surpasses a simple data look-up, and D: the point where data look-up is more accurate than using a. physics-based model. A final point (E) identifies the point where data lookup is more accurate than using the statistical model.
An unrestrained statistical model becomes increasingly better as the data coverage increases surpassing data lookup a given level (C in figure 5). However, the physical model only improves marginally with increasing amounts of experimental data and levels off at a given performance level (A). This is due to the restraints of the basic physical equations and the approximations implicit in the model. At low levels of data coverage the physical model should prevail, whereas we expect the statistical model to be more accurate than the physical model at high levels of data coverage (B in Figure 5). This situation is similar to the current state in protein structure prediction, where homology-based models outperform de novo structure prediction algorithms if a homologous protein structure is known. The question is now where the field of protein pKa predictions is located in this figure. Levels of overall data coverage are quite low, but we have a large mount of data for solvent exposed residues with relatively unshifted pKa values. We therefore expect statistics-based and empirical models to outperform physics-based models for this type of groups, whereas physics-based models should be able perform better on buried groups and titratable groups in highly charged environments. However, the success of the physics-based models in studying such exceptional groups relies on proper modeling of the structural ensemble, which is of crucial importance for calculating desolvation energies correctly. Small errors in coordinates can give rise to large changes in desolvation energies when applying PBE-based or GB-based desolvation models, and dealing correctly with this high sensitivity to structural detail is a formidable challenge to physically realistic structure-based pKa calculation algorithms.
The role of pKa calculation methods
Finally it is worth contemplating the role of pKa calculation methods in modern biology/biochemistry. In our opinion pKa calculation methods are interesting because
They can be used to understand the physics of protein electrostatics.
They can be used to understand and redesign proteins, their characteristics and function.
We believe that both of these goals should be pursued, but note that present efforts, in our opinion, focus too narrowly on pKa values for benchmarking purposes. Methods that aim to understand the underlying physics should be benchmarked on multiple types of data as argued earlier, and similarly methods that primarily attempt to analyze and redesign proteins should be benchmarked on the characteristics that they seek to change. Thus methods for predicting enzymatic pH-activity profiles should calculate pH-dependent population profiles of the catalytically competent protonation state, and methods for calculating changes in pKa values should be benchmarked in experimental ΔpKa values. Such efforts are currently hampered by the fact that little or no data is available in electronic form, and we therefore believe that future efforts should contain efforts aimed at getting experimentalists and theoreticians collaborating through online sharing of experimental and computational data35.
Conclusion
We have shown that the experimental dataset, the experimental error and the number of free parameters to be of high importance when developing pKa calculation models in a simulated environment. While we have no hard proof that real pKa calculation algorithms suffer from the same artifacts as our simulated models, we believe that it is prudent for scientists in the field to assess the parameter dependence of their methods and critically examine their development procedures in particular with respect to the composition and accuracy of their experimental datasets. In our opinion, a better understanding of protein electrostatics relies critically on a sober assessment of the abilities and deficiencies of current theoretical models through testing on novel experimental data in the form of blind predictions.
Acknowledgments
This work was sponsored by NIH grants R01 GM069702 and P41 RR0860516 to NAB, and by a Science Foundation Ireland PIYRA grant (04/YI1/M537), a project grant from the Irish Health Research Board (RP/2004/140), a CSCB (HEA) grant, and a Science Foundation Ireland Research Frontiers Program Award (08/RFP/BIC1140)to JN.
Abbreviations
- SNase
Staphylococcal nuclease
References
- 1.Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 2008;73:765–83. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 2.Khandogin J, Brooks CL., III Toward the Accurate First-Principles Prediction of Ionization Equilibria in Proteins. Biochemistry. 2006;45:9363–9373. doi: 10.1021/bi060706r. [DOI] [PubMed] [Google Scholar]
- 3.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–71. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tynan-Connolly BM, Nielsen JE. Re-Designing protein pKa values. Protein Sci. 2007;16:239–249. doi: 10.1110/ps.062538707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Y, Mao J, Gunner MR. MCCE2: improving protein pKa calculations with extensive side chain rotamer sampling. J Comput Chem. 2009;30:2231–47. doi: 10.1002/jcc.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aleksandrov A, Polydorides S, Archontis G, Simonson T. Predicting the acid/base behavior of proteins: a constant-pH Monte Carlo approach with generalized born solvent. J Phys Chem B. 114:10634–48. doi: 10.1021/jp104406x. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen JE, Vriend G. Optimizing the hydrogen-bond network in Poisson-Boltzmann equation-based pK(a) calculations. Proteins. 2001;43:403–12. doi: 10.1002/prot.1053. [DOI] [PubMed] [Google Scholar]
- 8.Davies MN, Toseland CP, Moss DS, Flower DR. Benchmarking pK(a) prediction. BMC Biochem. 2006;7:18. doi: 10.1186/1471-2091-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammett LP. The Effect of Structure upon the Reactions of Organic Compounds, Benzene Derivaties. J Am Chem Soc. 1937;59:96–103. [Google Scholar]
- 10.Mongan J, Case DA, McCammon JA. Constant pH molecular dynamics in generalized Born implicit solvent. J Comput Chem. 2004;25:2038–48. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- 11.Machuqueiro M, Baptista AM. Acidic range titration of HEWL using a constant-pH molecular dynamics method. Proteins. 2008;72:289–98. doi: 10.1002/prot.21923. [DOI] [PubMed] [Google Scholar]
- 12.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23:128–37. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 13.Hooft RW, Sander C, Vriend G. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins. 1996;26:363–76. doi: 10.1002/(SICI)1097-0134(199612)26:4<363::AID-PROT1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 14.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–5. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexov EG, Gunner MR. Incorporating protein conformational flexibility into the calculation of pH-dependent protein properties. Biophys J. 1997;72:2075–93. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb H, Tynan-Connolly BM, Lee GM, Farrell D, O’Meara F, Sondergaard CR, Teilum K, Hewage C, McIntosh LP, Nielsen JE. Remeasuring HEWL pK(a) values by NMR spectroscopy: Methods, analysis, accuracy, and implications for theoretical pK(a) calculations. Proteins. doi: 10.1002/prot.22886. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen JE, McCammon JA. On the evaluation and optimisation of protein X-ray structures for pKa calculations. Protein Science. 2003;12:313–326. doi: 10.1110/ps.0229903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen JE, Borchert TV, Vriend G. The determinants of alpha-amylase pH-activity profiles. Protein Eng. 2001;14:505–12. doi: 10.1093/protein/14.7.505. [DOI] [PubMed] [Google Scholar]
- 20.Alexov EG, Gunner MR. Calculated protein and proton motions coupled to electron transfer: electron transfer from QA-to QB in bacterial photosynthetic reaction centers. Biochemistry. 1999;38:8253–70. doi: 10.1021/bi982700a. [DOI] [PubMed] [Google Scholar]
- 21.van Vlijmen HW, Schaefer M, Karplus M. Improving the accuracy of proteinpKa calculations: conformational averaging versus the average structure. Proteins. 1998;33:145–58. doi: 10.1002/(sici)1097-0134(19981101)33:2<145::aid-prot1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 22.Baptista AM, Martel PJ, Petersen SB. Simulation of protein conformational freedom as a function of pH: constant-pH molecular dynamics using implicit titration. Proteins. 1997;27:523–44. [PubMed] [Google Scholar]
- 23.Khandogin J, Brooks CL., 3rd Constant pH molecular dynamics with proton tautomerism. Biophys J. 2005;89:141–57. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isom D, Castaneda CA, Cannon BR, Velu P, Garcia-Moreno B. Charges in the hydrophobic interior of proteins. Proc Natl Acad Sci U S A. 2010;107:16096–16100. doi: 10.1073/pnas.1004213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang AS, Gunner MR, Sampogna R, Sharp K, Honig B. On the calculation of pKas in proteins. Proteins. 1993;15:252–65. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- 26.Antosiewicz J, McCammon JA, Gilson MK. The determinants of pKas in proteins. Biochemistry. 1996;35:7819–33. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]
- 27.Sondergaard CR, McIntosh LP, Pollastri G, Nielsen JE. Determination of electrostatic interaction energies and protonation state populations in enzyme active sites. J Mol Biol. 2008;376:269–87. doi: 10.1016/j.jmb.2007.09.070. [DOI] [PubMed] [Google Scholar]
- 28.Russell AJ, Fersht AR. Rational modification of enzyme catalysis by engineering surface charge. Nature. 1987;328:496–500. doi: 10.1038/328496a0. [DOI] [PubMed] [Google Scholar]
- 29.Suydam IT, Snow CD, Pande VS, Boxer SG. Electric fields at the active site of an enzyme: direct comparison of experiment with theory. Science. 2006;313:200–4. doi: 10.1126/science.1127159. [DOI] [PubMed] [Google Scholar]
- 30.Webb H, Tynan-Connolly BM, Lee GL, Farrell D, O’Meara F, Sondergaard CR, Teilum D, Hewage CH, McIntosh LP, Nielsen JE. Remeasuring HEWL pKa values by NMR spectroscopy: methods, analysis, accuracy and implications for theoretical pKa calculations. Proteins. doi: 10.1002/prot.22886. in press. [DOI] [PubMed] [Google Scholar]
- 31.Farrell D, Miranda ES, Webb H, Georgi N, Crowley PB, McIntosh LP, Nielsen JE. Titration_DB: Storage and analysis of NMR-monitored protein pH titration curves. Proteins. 2010;78:843–857. doi: 10.1002/prot.22611. [DOI] [PubMed] [Google Scholar]
- 32.Stanton CL, Houk KN. Benchmarking pKa prediction Methods for Residues in Proteins. J Chem Theory Comput. 2008;4:951–966. doi: 10.1021/ct8000014. [DOI] [PubMed] [Google Scholar]
- 33.Toseland CP, McSparron H, Davies MN, Flower DR. PPD v1.0--an integrated, web-accessible database of experimentally determined protein pKa values. Nucleic Acids Res. 2006;34:D199–203. doi: 10.1093/nar/gkj035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18:247–51. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrell D, Georgi N, O’Meara F, Bradley J, Søndergaard CR, Webb H, Tynan-Connolly BM, Bjarnadottir U, Carstensen T, Nielsen JE. Capturing, sharing and analyzing experimental data form protein engineering and directed evolution studies. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq726. [DOI] [PMC free article] [PubMed] [Google Scholar]