Abstract
Inbred Lewis and Fisher 344 rat strains differ greatly in drug self-administration; Lewis rats operantly self-administer drugs of abuse including nicotine, whereas Fisher self-administer poorly. As shown herein, operant food self-administration is similar. Based on their pivotal role in drug reward, we hypothesized that differences in basal gene expression in GABAergic neurons projecting from nucleus accumbens (NAcc) to ventral pallidum (VP) play a role in vulnerability to drug taking behavior. The transcriptomes of NAcc shell-VP GABAergic neurons from these two strains were analyzed in adolescents, using a multidisciplinary approach that combined stereotaxic ionotophoretic brain microinjections, laser-capture microdissection (LCM) and microarray measurement of transcripts. LCM enriched the gene transcripts detected in GABA neurons compared to the residual NAcc tissue: a ratio of neuron/residual > 1 and false discovery rate (FDR) <5% yielded 6,623 transcripts, whereas a ratio of >3 yielded 3,514. Strain-dependent differences in gene expression within GABA neurons were identified; 322 vs. 60 transcripts showed 1.5-fold vs. 2-fold differences in expression (FDR<5%). Classification by gene ontology showed these 322 transcripts were widely distributed, without categorical enrichment. This is most consistent with a global change in GABA neuron function. Literature-mining by Chilibot found 38 genes related to synaptic plasticity, signaling and gene transcription, all of which determine drug-abuse; 33 genes have no known association with addiction or nicotine. In Lewis rats, upregulation of Mint-1, Cask, CamkIIδ, Ncam1, Vsnl1, Hpcal1 and Car8 indicates these transcripts likely contribute to altered signaling and synaptic function in NAcc GABA projection neurons to VP.
Keywords: GABA, nicotine, nucleus accumbens, addiction, ventral pallidum, synapse, transcriptome, Lewis rats, Fisher 344 rats, laser capture microdissection
Introduction
In rats, the inbred Lewis and Fisher 344 strains differ greatly in drug self-administration behavior; while Lewis rats operantly self-administer several drugs of abuse, including nicotine, Fisher rats self-administer poorly (Brower et al., 2002, Martin et al., 1999, Suzuki et al., 1988a, Suzuki et al., 1988b). The nucleus accumbens shell (NAcc shell), located in ventral striatum, and its primary output target, the ventral pallidum (VP), are both critical to the integration and discrimination of information encoding the rewarding dimension of motivated behavior (Carlezon & Thomas, 2009, Heimer & Wilson, 1975, Shirayama & Chaki, 2006, Smith et al., 2009). The vast majority of neurons in the NAcc are GABAergic medium spiny projection neurons (Meredith et al., 1993). The normal function of VP is essential for aspects of reward learning and to motivational “wanting” and hedonic “liking” (Smith et al., 2009). Based on their pivotal role in reward (Carlezon & Thomas, 2009, Meredith et al., 1993), we hypothesized that differences in basal gene expression by these GABAergic neurons in the Lewis versus Fisher 344 strains may contribute to drug-taking vulnerability, which for cigarette smoking is usually manifest during adolescence (Chassin et al., 1996, Dappen et al., 1996, S.A.M.H.S.A., 2001).
Differences in the gene expression profiles or transcriptomes between the NAcc shell-VP GABAergic neurons from these two strains were analyzed in adolescents [postnatal day 41 (PN41)]. We used a multidisciplinary approach, combining stereotaxic ionotophoretic brain microinjections, laser capture microdissection (LCM) and microarray detection of transcript expression levels. Neuroanatomical specificity was obtained by ionotophoretic delivery of fluorogold (FG) into VP (Pieribone & Aston-Jones, 1988) followed by capture of all FG+ neurons in sections from the NAcc shell by laser capture microdissection (LCM). The efficacy of this approach was evaluated by measuring the ratio of gene expression for neurons compared to the remaining NAcc tissue after removal of the neurons. RNA extracted from both neurons and remaining NAcc was amplified and then analyzed by microarray. Differential gene expression by rat strain was validated for a subset of genes by real-time PCR.
Methods
Animals and surgeries
Lewis rats were bred in the laboratory, using breeders purchased from Harlan Laboratories (Indianapolis, IN). Adolescent male Lewis rats (PN35) were anesthetized with ketamine/xylazine (90 and 10 mg/kg, respectively, i.m.) and then placed in a stereotaxic frame. Fluorogold (2% in sterile saline, Fluorochrome, Englewood, CO) was deposited into ventral pallidum (AP: −0.22, ML: ±2.2, DV: −8.0) bilaterally by ionotophoretic injection, using a Midgard high voltage current source (Stoelting Co., Wood Dale, IL, USA). The current was set at +5 μA (7s on, 7s off, for 5 min). Micropipettes were left in place for 15 min before the scalp incision was sutured. Five days later, rats were killed, brains removed, immediately frozen, and stored at −70°C. All procedures were conducted in accordance with the NIH Guidelines Concerning the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee of the University of Tennessee.
Chronic nicotine self-administration (SA)
Rats were acclimated to a reversed 12:12 h light:dark cycle. Standard rat chow and water was provided ad libitum. Nicotine (pH = 7.2; calculated as free base) was prepared weekly in 200 U heparinized saline to deliver 50 μl i.v. of 0.03 mg/kg b.wt. per injection. SA was performed according to our published protocol (Valentine et al., 1997). Briefly, PN38 adolescent rats received jugular cannulae under xylazine-ketamine anesthesia and were individually housed in operant chambers (Coulbourn Instruments, Whitehall, PA) inside a sound-attenuating chamber. Operant chambers were equipped with 2 levers positioned 4 cm above the floor and a green cue light 1 cm above each lever was illuminated when nicotine was available. Lever presses were recorded and syringe pumps controlled by computers and interfaces, using L2T2 or Graphic State software (Coulbourn Instruments). Rats were given 3 d to recover from jugular surgery, during which time they received antibiotic daily and hourly aliquots (50 μl) of 200 U heparin. At PN41, rats received 0.03 mg/kg nicotine in heparinized saline iv with each lever press.
One lever was randomly designated as the active lever. Pressing this lever elicited a computer-driven i.v. injection of nicotine over 0.81 s on a fix ratio 1 (FR1) schedule. Each injection was followed by a 7 s period during which the green cue light was extinguished and lever presses were recorded, but nicotine was not injected. Pressing the inactive lever had no programmed consequence. Rats were given access to nicotine or saline SA 23 h/d and learned to self-administer nicotine without prior training, priming or food deprivation. The final hour of the lights-on cycle was reserved for housekeeping tasks. During this interval, environmental enclosure doors were opened and green cue lights turned off; levers were not retracted, and lever press activity was not recorded nor rewarded during this period. Self-administration d 1 (SA d 1) is the 1st day nicotine was made available and the rats were given 10 d unlimited free access to acquire nicotine SA.
Food self-administration
Beginning on PN41, a separate cohort of adolescent male rats were trained on an FR1 schedule to self-administer food pellets (45 mg) during daily 2 h sessions in the same operant chambers used for nicotine SA. Animals were food deprived overnight prior to the first session. Thereafter, they received 16 g food per d.
Laser capture microdissection
Brains were sectioned in a Leica cryostat at 10μm, and sections were mounted onto uncharged glass slides and maintained at −20°C until dehydration. Slides were dehydrated by sequential immersion in the following: 100% methanol (3 min), 95% EtOH (2 min), 100% EtOH (1 min, twice) and xylene (5 min, twice), and then air dried (15 min). The Arcturus XT (Life Technologies, Carlsbad, CA) was used to capture Fluorogold-labeled neurons. The infrared laser produced spots approximately 15 μm in diameter, which allowed consistent capture of neurons onto CapSure LCM caps (Life Technologies). Approximately 750 neurons were captured from each brain.
RNA extraction
RNA trapped in the CapSure LCM caps was extracted using the PicoPure RNA isolation kit (Life Tech.); RNA was eluted with 13 μl nuclease-free H2O. RNA quality was analyzed using Bioanalyzer (Agilent Technologies, Foster City, CA). The mean value of RNA Integrity Number (RIN) was 7.9 for the 1.84 ± 2.4 ng of total RNA obtained from each rat.
RNA amplification and Affymetrix microarray
Total RNA were amplified by Nugen Ovation Pico WTA system, followed by cDNA synthesis using the WT Ovation Exon module. Ovation Biotin kits were then used to fragment and label the cDNA (Ovation reagents: NuGen Inc., San Carlos, CA). The labeled cDNA was then hybridized to the Rat Gene ST 1.0 array (Affymetrix, Santa Clara, CA), according to the manufacturer’s protocols for Whole Transcriptome arrays. Microarrays were then processed on a Fluidics Station 450 and scanned by the Affymetrix GeneChip 3000 7G Scanner. Expression Console software (ver 1.1.2) was used to normalize data. All microarray samples were processed by the same technician consecutively, interleaving the two strains. No batch effect was observed. The Rat Gene ST 1.0 array probe sets are generated from the Brown Norway rat genome. Therefore, false positive differences between Lewis and Fisher rat samples may occur due to the assymetical expression of single nucleotide polymorphisms (SNPs) by these 2 strains, differentially affecting cDNA hybridization to array probe sets.
Real-time PCR
Real-time quantitative PCR of 8 genes (and two reference genes Ywhaz and Sdha) was performed on the LightCycler 480 (Basel, Switzerland) using probes from the Universal Probe Library. The original unamplified RNA from each sample was used to prepare cDNA. The critical threshold (Ct) of genes of interest was normalized to the geometric mean of the two reference genes.
Data analysis
Microarray data were analyzed using the R-statistical package (Team, 2010). T-tests were used to compare the expression level of each gene from the two strains of rats and then all genes were analyzed for false discovery rate (FDR). Gene ontology analysis was conducted using DAVID Bioinformatics Resources (Huang et al. 2009). Nicotine and food self-administration data were analyzed using repeated measures ANOVA, with both lever and day treated as within subject variables. Strain was treated as a between subject variable.
Results
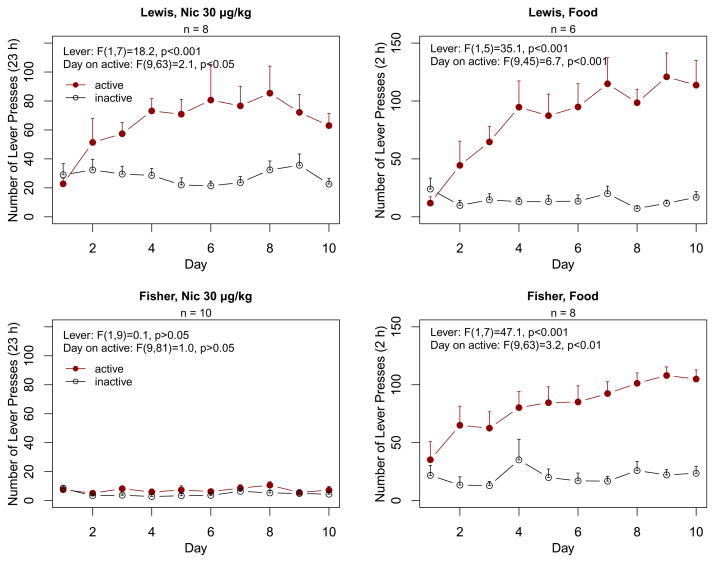
The capacity to acquire operant nicotine vs. food self-administration was evaluated in adolescent Lewis compared to Fisher rats. For nicotine SA, rats were housed in operant chambers and were not shaped, primed or food deprived. Three days after jugular vein surgery, nicotine was made available for 23 h per day. Figure 1 (data analyzed by repeated measures ANOVA) indicates that Lewis rats rapidly acquired nicotine self-administration, manifest by a significant increase in active lever presses in the absence of any change in the inactive lever [effect of lever on number of presses, F(1,7): 18.2, p<0.001; effect of day on active lever presses, F(9,63): 2.1, p<0.05; effect of day on inactive lever presses, F(9,63): 0.8, p>0.05]. In contrast, both active and inactive lever presses by Fisher rats failed to increase over time [F(9,81): 1.5, p>0.05]. The number of lever presses was different between strains [F(1,16): 111.8, p<0.001] and the interaction between lever and strain was significant [F(1,15): 17.5, p<0.001]. Thus, the number of active lever presses for nicotine was different between strains [F(1,16): 66.6, p<0.001], as was the number of inactive lever presses [F(1,16): 53.5, p<0.001)]. The higher number of inactive lever presses in Lewis rats is likely the result of stronger locomotor activation induced by a larger amount of nicotine intake. Both strains showed a specific increase in active lever presses reinforced by food [Fisher, F(1,7): 47.1, p<0.001; Lewis, F(1,5): 35.1, p<0.001). Neither active lever presses for food [F(1,12): 0.7, p=0.41] nor inactive lever presses [F(1,12): 0.7, p=0.41] differed by strain. The similar amount of inactive lever presses reflected similar level of arousal induced by food. Hence, both strains were capable of learning the operant task, but nicotine was only reinforcing in Lewis rats, under these conditions.
Figure 1.
Nicotine or food self-administration (SA) in Lewis vs. Fisher 344 adolescent male rats at postnatal day 41. Data shown in the left panels was from rats housed in operant chambers with nicotine available for 10d, beginning at PN41 (FR1 schedule), using our model of 23h drug access (Brower et al., 2002). Lewis rats rapidly acquired nicotine SA nicotine SA [effect of lever on number of presses, F(1,7): 18.2, p<0.001; effect of day on active lever presses, F(9,63): 2.1, p<0.05; effect of day on inactive lever presses, F(9,63): 0.8, p>0.05]. In contrast, Fisher rats failed to acquire nicotine SA. Right panels show results from both Lewis and Fisher rats that acquired operant food self-administration. Active lever presses for food did not differ by strain [F(1,12): 0.7, p=0.41].
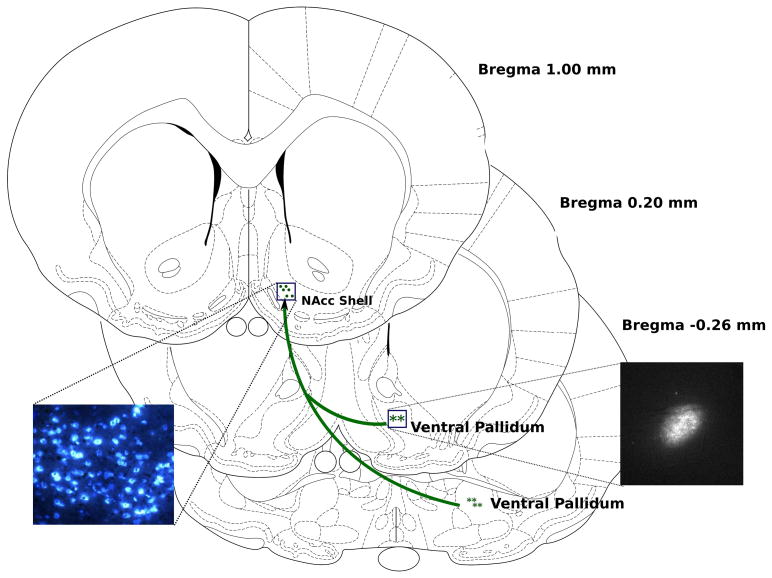
Neurons projecting from NAcc shell to VP were identified by retrograde uptake of fluorogold (FG) delivered by ionotophoresis. Figure 2 illustrates coronal sections of rat brain, identifying both NAcc shell and VP. Within VP, the central area of FG fluorescence for each of six rats is marked by an asterisk. A representative deposit of FG is shown in the inset. Within NAcc shell, the center of the distribution of retrograde-labeled FG+ neurons for each rat is shown by a green dot within the boxed area. The corresponding inset shows a representative fluorescence image of these FG+ neurons, located in ventral medial NAcc shell (20X). In this region of NAcc shell, virtually all neurons are FG+. All individual FG+ neurons were captured by LCM. Thus, this approach specifically captured a subpopulation of NAcc shell neurons that project from NAcc shell to VP, excluding neurons that project from NAcc shell to other brain regions.
Figure 2.
Identification of GABA neurons in nucleus accumbens shell (NAcc shell) that project to ventral pallidum (VP) at postnatal day 41. Neurons projecting from NAcc shell to VP were identified by retrograde uptake of fluorogold (FG) delivered by ionotophoresis bilaterally into VP (asterisks). Within VP, the central regions of FG fluorescence from six rats are marked by asterisks in a schematic coronal section. A representative deposit of FG is shown in the photomicrograph. Within NAcc shell, the center of the distribution of retrograde-labeled FG+ neurons is shown by green dots within the boxed area. The corresponding inset shows a representative fluorescence image of these FG+ neurons, located in ventral medial NAcc shell (20X). Individual FG+ neurons were captured by laser-capture microdissection.
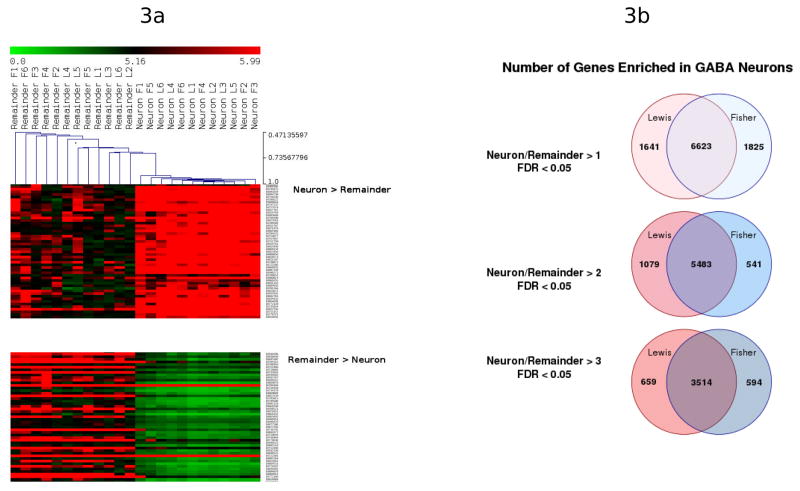
Microarray analysis was used to detect the level of specific gene transcripts expressed by the transcriptome from laser-captured FG+ neurons compared to the tissue remaining in the area of NAcc shell after removal of these fluorescent neurons. A heat map of all genes was generated; figure 3A shows representative areas that demonstrate clustering of genes expressed at significantly greater or lesser levels in neurons vs. tissue remainder. At a ratio of neuron/remainder > 1 and false discovery rate (FDR)<5%, 6,623 gene transcripts were identified in both Lewis and Fisher rats; at ratios > 2.0 and > 3.0, 5,483 and 3,514 genes, respectively, were enriched in the neuron populations from both inbred rat strains (Fig. 3b). Amongst the enriched transcripts, Table 1 shows those related to the neurotransmitter, GABA (e.g., GABA A receptor alpha 1, glutamate decarboxylase 1 and 2), and/or associated with regulation of GABA neuron-related functions (e.g., dopamine receptors D1A and D2; GABA vesicular transporter). These data indicate that laser capture microdissection of individual FG+ neurons successfully enriched transcripts from GABA neurons projecting to VP. At three incremental levels of enrichment (i.e., neuron/remainder >1 to >3), the majority of these transcripts were common to both strains of rat. Nonetheless, LCM unavoidably introduces glial cells, in intimate contact with FG+ neurons, within each sample captured. For example, myelin basic protein and glial fibrillary acidic protein, standard markers for oligodendrocytes and astrocytes, respectively, were detected at neuron/remainder ratios of 2.7 and 1.5. This enrichment of glial markers indicates that targeting most of the neuronal soma for capture, by using 15 μm diameter spot size, eliminates the ability to separate the neuronal vs. glial transcriptomes. This limitation of geometric capture by LCM of a high heterogeneous tissue was recently reported (Okaty et al., 2011).
Figure 3.
Microarray detection of transcripts enriched in NAcc shell GABA neurons, obtained by laser-capture microdissection, compared to the remainder of tissue. Unsupervised cluster analysis of all genes was used to generate the data shown in Fig. 3a; representative areas of the heat map show the clustering of genes expressed at significantly greater or lesser levels in neurons vs. tissue remainder. These differences in neuron/reminder ratios were consistent throughout all samples. Venn diagrams (Fig. 3b) show the number of gene transcripts that were significantly (false discovery rate < 0.05) enriched in GABA neurons at various ratios of neuron/remainder (i.e., 1, 2, 3). At each ratio of neuron/remainder, most of the enriched neurons were common to both Lewis and Fisher GABA neurons. Neuronal transcripts with very low mean fluorescence values (<3 on microarray) were excluded from the analysis.
Table 1.
Enrichment of GABA and GABA Neuron-Related Genes *
| Symbol | Gene Name | Neuron/Remainder (mean of both strains) | Neuron Fluorescence Intensity (mean of both strains) |
|---|---|---|---|
| Akap9 | A kinase (PRKA) anchor protein (yotiao) 9 | 5.18 | 7.28 |
| Dbi | diazepam binding inhibitor (GABA receptor modulator acyl-Coenzyme A binding protein) | 8.25 | 8.43 |
| Drd1a | dopamine receptor D1A | 9.83 | 9.20 |
| Drd2 | dopamine receptor D2 | 2.74 | 6.81 |
| Gabarap | GABA(A) receptor-associated protein | 10.55 | 9.41 |
| Gabarapl1 | GABA(A) receptor-associated protein like 1 | 9.18 | 8.73 |
| Gabarapl2 | GABA(A) receptor-associated protein like 2 | 6.01 | 8.03 |
| Gabarapl2 | GABA(A) receptor-associated protein like 2 | 5.32 | 7.78 |
| Gabra1 | gamma-aminobutyric acid (GABA) A receptor alpha 1 | 3.42 | 7.48 |
| Gabra2 | gamma-aminobutyric acid (GABA) A receptor alpha 2 | 3.90 | 6.69 |
| Gabra4 | gamma-aminobutyric acid (GABA) A receptor alpha 4 | 10.74 | 8.73 |
| Gabra5 | gamma-aminobutyric acid (GABA) A receptor alpha 5 | 1.72 | 5.97 |
| Gabrb1 | gamma-aminobutyric acid (GABA) A receptor beta 1 | 2.92 | 6.24 |
| Gabrb2 | gamma-aminobutyric acid (GABA) A receptor beta 2 | 6.04 | 7.45 |
| Gabrb3 | gamma-aminobutyric acid (GABA) A receptor beta 3 | 6.90 | 8.87 |
| Gabrg1 | gamma-aminobutyric acid (GABA) A receptor gamma 1 | 3.32 | 6.68 |
| Gabrg2 | gamma-aminobutyric acid (GABA) A receptor gamma 2 | 5.37 | 7.92 |
| Gabrg3 | gamma-aminobutyric acid (GABA) A receptor gamma 3 | 4.11 | 7.24 |
| Gabbr1 | gamma-aminobutyric acid (GABA) B receptor 1 | 3.93 | 7.42 |
| Gabbr2 | gamma-aminobutyric acid (GABA) B receptor 2 | 1.64 | 5.63 |
| Gad1 | glutamate decarboxylase 1 | 2.23 | 6.69 |
| Gad2 | glutamate decarboxylase 2 | 6.44 | 9.25 |
| Pdyn | prodynorphin | 3.73 | 6.84 |
| Penk1 | proenkephalin 1 | 4.01 | 6.50 |
| Slc32a1 | solute carrier family 32 (GABA vesicular transporter) member 1 | 6.41 | 8.38 |
| Slc6a1 | solute carrier family 6 (neurotransmitter transporter GABA) member 1 | 3.21 | 6.86 |
| Slc6a11 | solute carrier family 6 (neurotransmitter transporter GABA) member 11 | 2.90 | 6.59 |
| Snap25 | synaptosomal-associated protein 25 | 14.77 | 11.79 |
| Stxbp1 | syntaxin binding protein 1 | 9.05 | 9.15 |
Neuron > Remainder for all genes, FDR 5%
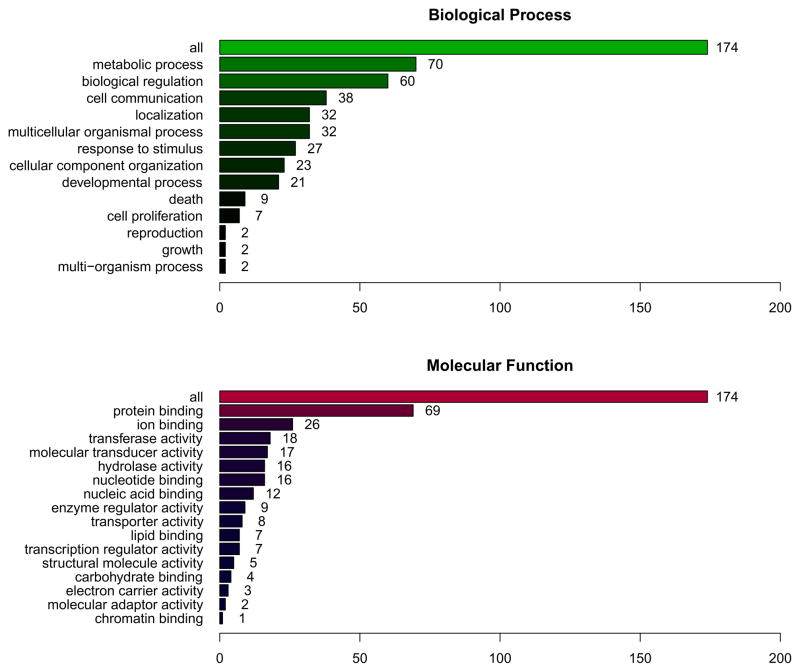
Genes that were differentially expressed in Lewis vs. Fisher NAcc shell GABA neurons projecting to VP were identified. One caveat about array analysis across strains concerns the potential for false positives due to assymetical expression of SNPs by the 2 strains, differentially affecting hybridization to the array probes. Table 2 shows the total number of gene transcripts as a function of the expression ratio in Lewis compared to Fisher rats (i.e., >1.5-fold and >2-fold); total transcript number declined with the application of increasingly stringent criteria to achieve statistical significance. A total of 322 genes were differentially expressed at a Lewis vs. Fisher ratio of >1.5-fold (p<0.05 and FDR<5%; see supplemental file). Of these, 174 gene transcripts could be mapped to biological processes, as shown in Figure 4. Transcripts mapped across a wide range of biological processes and molecular functions, showing no specific enrichment by either biological or molecular ontology.
Table 2.
Differential Gene Expression in Lewis vs. Fisher Rats
| Criteria | Number of genes |
|---|---|
| Lewis vs Fisher > 1.5 fold | 1210 |
| t-test p < 0.05 | 608 |
| FDR 10% | 573 |
| FDR 5% | 330 |
| Lewis vs Fisher > 2 fold | 267 |
| t-test p < 0.05 | 90 |
| FDR 10% | 71 |
| FDR 5% | 60 |
Figure 4.
Distribution of 322 genes showing differential expression in Lewis vs. Fisher NAcc shell GABA projection neurons to VP. 174/322 of these genes could be mapped by biological process and molecular function. Gene transcripts were distributed across all biological processes, showing no enrichment in any particular process. Widespread distribution and lack of enrichment were also shown for mapping by molecular ontology.
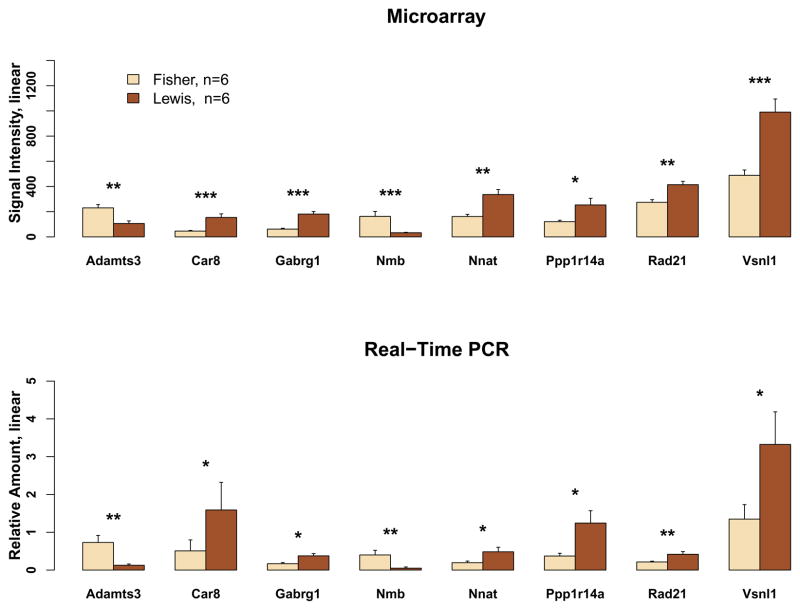
The differential gene expression detected by microarray analysis was validated by PCR using the same RNA samples. Eight gene transcripts were selected for validation. By microarray, the expression ratio of these transcripts in Lewis vs. Fisher rats varied by 1.5–2-fold (see Figure 5). Additionally, 6 of the 8 transcripts were expressed at higher levels in Lewis rats, whereas 2 (i.e., A disintegrin and metalloproteinase with thrombospondin motifs 3 and Neuromedin B) were greater in Fisher rats. Significant strain-dependent differences were detected by PCR analysis of all eight transcripts.
Figure 5.
Validation of differential gene expression in Lewis vs. Fisher rats by real-time PCR. Eight genes that varied in expression by 1.5–2-fold (by microarray) between rat strains were selected for measurement by PCR. Significant strain-dependent differences were found for all genes. The eight genes were: A disintegrin and metalloproteinase with thrombospondin motifs 3 (Adamts3), carbonic anhydrase 8 (Car8), GABAA receptor g-1 subunit (Gabrg1), neuromedin B (Nmb), neuronatin (Nnat), protein phosphatase 1 regulatory (inhibitor) subunit 14a (Ppp1r14a), cohesin subunit Rad21 (Rad21), Visinin-like protein 1 (Vsnl1). *, p<0.05; **, p<0.01, ***, p<0.001.
Gene interactions between the 322 differentially expressed transcripts were sought using Chilibot, an established data-mining software, to analyze relationships documented in all the abstracts compiled in PubMed (Chen & Sharp, 2004). We identified genes likely to determine drug abuse behavior based on their known relationships to cell signaling, synaptic plasticity, transcription, addiction and nicotine. Table 3 shows 22 genes that mapped to synaptic plasticity, 24 to signaling, 18 to transcription, 10 to addiction and 4 to nicotine. Twenty-one of 22 genes mapping to synaptic plasticity were also related to cell signaling. However, the strength of these gene relationships to signaling vs. synaptic plasticity varied in that gene × signaling interactions were based on a median of 14.5 unique publications, whereas gene × synaptic plasticity interaction were derived from a median of 3.5 publications. Only 4 of 18 genes mapped to transcription regulation were common to signaling and synaptic plasticity. Strong interactions between gene × transcription regulation were evident based on a median of 15.5 publications. Lastly, 7 of 10 genes mapped to addiction and all 4 genes mapped to nicotine were common to signaling and synaptic plasticity. Overall, 33 of the 41 genes in Table 3 have not previously been associated with addiction or nicotine. CASK (calcium/calmodulin-dependent serine protein kinase) exemplifies a gene strongly associated with all 3 biological processes, but not with addiction or nicotine.
Table 3.
Genes Related to Cell Signaling, Synaptic Plasticity and Transcription Regulation
| Gene Symb | Signaling | Synaptic plasticity | Transcription regulation | Addiction | Nicotine | Gene name |
|---|---|---|---|---|---|---|
| ADRBK1 | 345 | 14 | 5 | adrenergic, beta, receptor kinase 1 | ||
| AHI1 | 6 | 2 | Abelson helper integration site 1 | |||
| APBA1 | 15 | 30 | 15 | 1 | amyloid beta (A4) precursor protein-binding, family A, member 1 | |
| CAMK2D | 8 | 2 | 136 | 41 | 15 | calcium/calmodulin-dependent protein kinase II delta |
| CAR8** | 3 | 2 | carbonic anhydrase 8 | |||
| CASK | 30 | 51 | 16 | calcium/calmodulin-dependent serine protein kinase | ||
| DAAM1 | 25 | 2 | dishevelled associated activator of morphogenesis 1 | |||
| DSTN | 37 | 29 | destrin | |||
| EMD | 72 | emerin | ||||
| EPHA5 | 8 | 34 | 2 | EphA5 | ||
| ETFA | 1 | 3 | 1 | electron-transfer-flavoprotein, alpha polypeptide | ||
| GRIA4 | 6 | glutamate receptor, ionotrophic, AMPA 4 | ||||
| GRB10 | 78 | growth factor receptor-bound protein 10 | ||||
| GABRG1** | 1 | gamma-aminobutyric acid (GABA) A receptor, gamma 1 | ||||
| HBP1 | 48 | HMG-box transcription factor 1 | ||||
| HMBOX1 | 4 | homeobox containing 1 | ||||
| HPCAL1 | 6 | 2 | hippocalcin-like 1 | |||
| ILF3 | 35 | interleukin enhancer binding factor 3 | ||||
| MTSS1 | 14 | metastasis suppressor 1 | ||||
| NCAM1 | 277 | 675 | 11 | 5 | neural cell adhesion molecule 1 | |
| NFATC3 | 76 | nuclear factor of activated T-cells, calcineurin-dependent 3 | ||||
| NMI | 15 | N-myc (and STAT) interactor | ||||
| NNAT** | 6 | 1 | neuronatin | |||
| NXPH1 | 5 | 3 | neurexophilin 1 | |||
| POLE3 | 5 | polymerase (DNA directed), epsilon 3 (p17 subunit) | ||||
| PPP1R14A** | 26 | protein phosphatase 1, regulatory (inhibitor) subunit 14A | ||||
| PRDX1 | 44 | peroxiredoxin 1 | ||||
| PSMD7 | 1 | Psmd7 proteasome 26S subunit, non-ATPase, 7 | ||||
| PTAFR | 68 | 10 | platelet-activating factor receptor | |||
| RAD21 | 44 | RAD21 homolog (S. pombe) | ||||
| RICTOR | 90 | 2 | 1 | RPTOR independent companion of MTOR, complex 2 | ||
| RUFY2 | 6 | 4 | 2 | RUN and FYVE domain containing 2 | ||
| SSH2 | 2 | 2 | slingshot homolog 2 | |||
| SNAPC2 | 9 | small nuclear RNA activating complex, polypeptide 2 | ||||
| SPARC | 99 | 21 | secreted protein, acidic, cysteine-rich (osteonectin) | |||
| STXBP3 | 1 | syntaxin binding protein 3 | ||||
| TERF1 | 111 | telomeric repeat binding factor (NIMA-interacting) 1 | ||||
| VLDL-R | 17 | 4 | very low density lipoprotein receptor | |||
| VSNL1** | 14 | 10 | 1 | 2 | visinin-like 1 | |
| ZCCHC12 | 2 | 4 | zinc finger, CCHC domain containing 12 | |||
| ZFP410 | 6 | zinc finger protein 410 | ||||
| Number of genes | 24 | 22 | 18 | 10 | 4 | |
| Median PubMed hits | 14.5 | 3.5 | 15.5 | 1.5 | 3.5 |
denotes transcripts also measured by RT-PCR
Discussion
These studies demonstrate significant rat strain-dependent differences in the transcriptomes from neuroanatomically-defined sets of enriched GABA neurons that regulate the reinforcing dimension of motivated behavior. Differences in gene expression may impact the motivational function of these GABA neurons, since Lewis vs. Fisher rats differ greatly in their propensity to operantly self-administer nicotine and other drugs (Brower et al., 2002, Martin et al., 1999, Suzuki et al., 1988a, Suzuki et al., 1988b). In contrast, acquisition of operant food self-administration was similar between these strains. Therefore, differential gene expression is potentially related to drug reinforcement, rather than operant learning per se.
We combined neuroanatomical identification of specific neurons labeled with fluorogold and laser-capture of individual neurons. Fluorogold was transported from VP by GABA neurons in ventral medial NAcc shell. Despite RNA yield of only 1.8 ng/rat, requiring amplification before microarray, this protocol enriched >6,600 genes, including GABA-related transcripts. Strain-dependent effects on gene expression were identified; 322 vs. 60 transcripts showed 1.5-fold vs. 2-fold expression differences (FDR 5%). PCR validated the expression of selected gene transcripts. The LCM method unavoidably captures glial cells in close apposition to FG+ neurons, thus affecting the specificity of the transcriptome attributed to GABA neurons.
Classification by gene ontology showed these 322 strain-dependent transcripts lacked categorical enrichment. This wide-ranging alteration in the transcriptome from enriched NAcc GABA neurons is most consistent with a global change in neuronal function manifest throughout the cellular machinery. Since Chilibot literature-mining (Chen & Sharp, 2004) did not find networks amongst these genes, all 322 were analyzed for relationships to cellular processes pivotal to drug abuse (i.e., synaptic plasticity, signaling and regulation of gene transcription). Chilibot found genes with documented relationships to these 3 processes (Table 3). Almost all genes related to plasticity were common to signaling, whereas only 4 of 18 transcription genes were common to signaling/synaptic plasticity. Thirty-three of 38 genes related to plasticity, signaling and/or gene transcription have no association with addiction or nicotine. Based on novelty in the addiction literature, magnitude of the difference in strain-dependent expression, and prior knowledge of protein function, the following genes are highlighted.
Mint-1 (i.e., APBA1) has a relatively large number of associations with signaling, synaptic plasticity and transcription, but only 1 with addiction. Mint, an adaptor protein containing isoform-specific N-terminal sequences and common PTB and PDZ domains, is found in soma, axons and dendrites of excitatory and inhibitory neurons throughout the brain (Duclos & Koenig, 1995, Okamoto & Sudhof, 1997, Tanahashi & Tabira, 1999). Mints bind calcium channels, potentially affecting presynaptic calcium influx ((Maximov & Bezprozvanny, 2002, Maximov et al., 1999). Only the N-terminus of Mint-1 binds Cask (Borg et al., 1998, Butz et al., 1998), itself a synaptic membrane-associated guanylate kinase that interacts with membrane receptors affecting GABA release (Ho et al., 2003). Cask is associated with all 3 cellular processes, but not addiction/nicotine. In hippocampal interneurons, expressing high levels of Mint-1 vs. Mint-2, Mint-1 deficiency is associated with impaired regulation of GABA release, (Ho et al., 2003). Therefore, in Lewis vs. Fisher rats, the release of GABA by NAcc neurons regulating VP motivational functions might be affected by increased Mint-1-Cask heterodimers, reflecting increased basal levels of both transcripts (Lewis/Fisher ratios of Mint-1 and Cask: 1.7 and 1.6, respectively).
Cask associates with synaptic vesicles through interactions with neurexins, Mints and synaptotagmin (Biederer & Sudhof, 2000, Hata et al., 1993). Neurexins are presynaptic transmembrane cell-adhesion proteins (Nam & Chen, 2005) involved in initiating postsynaptic specializations at glutamate and GABA synapses (Graf et al., 2004). Cask binds and phosphorylates the cytoplasmic tail of neurexin-1, which may be essential for regulation of the presynaptic cytoskeleton (Mukherjee et al., 2008). Cask kinase activity is suppressed by neuronal activity and increased by silencing synapses. Two reports suggest that inhibitory synapses may undergo silencing (Bekkers, 2005, Kilman et al., 2002). In Lewis rats, the enhanced basal expression of Cask may promote phosphorylation of neurexin concomitant to the silencing of GABA synapses involving NAcc GABA projections.
In contrast to Mint-1 and Cask, Ca2+/calmodulin-dependent protein kinase IIδ (CamkIIδ) is related to all 3 cellular processes and addiction/nicotine. Its expression ratio in Lewis vs. Fisher NAcc GABA neurons was 1.76 (p=0.03, 5% FDR). CamkIIδ is present throughout rat brain (Takeuchi et al., 1999). Nuclear CamkIIδ is implicated in gene transcription, exemplified by mPer1 (Nomura et al., 2003). Dopamine D2 receptors (D2R) activate CamkIIδ to increase brain-derived neurotropic factor (BDNF) (Takeuchi et al., 2002), which is required for expression of activity-dependent inhibitory synapses (Hong et al., 2008, Yamada et al., 2002). These observations and the strain-dependent differences in basal CamkIIδ expression indicate that neuronal plasticity, involving BDNF and inhibitory synapses encoding aspects of behavioral reward, may be facilitated in Lewis rats.
Table 3 shows that neural cell adhesion molecule 1 (Ncam1) has the strongest associations with signaling and synaptic plasticity and is also associated with addiction/nicotine. Its expression ratio in Lewis vs. Fisher NAcc projection neurons was 1.54 (p=0.023, 5% FDR). Ncams mediate extracellular interactions with matrix and cells. Membrane clustering of Ncams initiate intracellular signaling cascades implicated in synaptic plasticity (Luthl et al., 1994, Maness & Schachner, 2007). By direct interaction with D2R, Ncam1 promotes internalization and degradation, regulating D2R signaling (Xiao et al., 2009). Increased Ncam1 expression appears to reduce surface D2R levels, which inhibit NAcc GABA neuron activity. Across 21 strains of recombinant inbred BXD mice, D2R and Ncam expression were associated with models of ethanol preference (Hitzemann et al., 2003). In Lewis rats, increased Ncam1 expression in NAcc GABA projection neurons is likely to enhance synaptic plasticity and alter surface expression of D2R, reducing dopamine-dependent inhibition.
Visinin-like 1 (Vsnl1; aka Vilip-1) and hippocalcin-like 1 (Hpcal1) are members of the neuronal Ca2+ sensor subfamily of visinin-like (Vsnl) proteins, which belong to the superfamily of EF-hand Ca2+ binding proteins (Braunewell & Gundelfinger, 1999). Vsnl interacts with multiple intracellular signaling cascades and affect exocytosis, modulation of adenylyl cyclase activity and regulation of both ligand- and voltage-gated ion channels (Burgoyne, 2007). Vsnl proteins are cytoplasmic at resting Ca2+ concentrations, and translocate to plasma or Golgi membrane with increased intracellular Ca2+ (Spilker et al., 2002). Vsnl1 has low to moderate associations with signaling and synaptic plasticity and minimal relationships to addiction/nicotine. Similarly, Hpcal1 has relatively few relationships with signaling and synaptic plasticity, and none with addiction/nicotine. The expression ratios for Vsnl1 and Hpcal1 in Lewis vs. Fisher were relatively high: 2.03 (p=0.023, FDR 5%) and 1.75 (p=0.043, FDR 5%), respectively. The Vsnl1 ratio was confirmed by RT-PCR.
Vsnl1 interacts directly with the alpha4 subunit of the most abundant brain nicotinic cholinergic receptor (nAChR), alpha4beta2, increasing the surface expression of functional receptors, depending on Ca2+ concentration (Zhao et al., 2009b). In contrast, alpha7-containing nAChRs elicit a calcium-dependent membrane localization of Vsnl1 (Zhao et al., 2009a). The inward Ca2+ current induced by nicotinic stimulation of alpha7 nAChRs (Berg et al., 2006) may drive the activation and membrane localization of Vsnl1 that is associated with upregulation of alpha4beta2 nAChRs. The brain region-specific upregulation of alpha4beta2 nAChRs has been reported in rats that chronically self-administer nicotine (Moretti et al., Parker et al., 2004). Upregulation of nAChRs would likely increase neuronal sensitivity and responsiveness to nicotine (Nguyen et al., 2004). However, in NAcc GABA projection neurons, the effect of chronic nicotine exposure on the level of nAChRs is not, to our knowledge, known. Nevertheless, the 2-fold increased expression of Vsnl1 in Lewis vs. Fisher GABA projection neurons may contribute to the motivational effects of nicotine in Lewis rats and to their ability to acquire nicotine self-administration.
Carbonic anhydrase 8 (Car8) displayed the greatest difference in expression between Lewis and Fisher rats (3.36-fold; p=.024, FDR 5%). This was confirmed by RT-PCR. Car8, an acatalytic carbonic anhydrase, binds inositol trisphosphate [IP(3)] receptors, inhibiting binding to inositol 1,4,5-trisphosphate (Hirota et al., 2003). Thus, Car8 modulates IP(3) receptor-dependent mobilization of Ca2+ from intracellular stores, which is critical to propagation of cytosolic Ca2+ signals (Taylor & Tovey, 2010). Car8 mutant mice demonstrate the essential role of Car8 in synaptogenesis and maintenance of synaptic function (Hirasawa et al., 2007). The difference in basal expression by Lewis vs. Fisher rats suggests that Car8 modulation of intracellular Ca2+ mobilization and signaling varies substantially between these inbred strains.
In summary, these studies demonstrate the feasibility of obtaining neuronal phenotype-enriched samples to ascertain strain-specific differences in gene transcript expression. Based on gene ontologies of 322 transcripts that differed between these 2 strains, there is a wide-ranging alteration in the transcriptome of GABA neurons projecting from NAcc shell to VP. This is most consistent with a global change in function. In Lewis rats, significant upregulation of Mint-1, Cask, CamkIIδ, Ncam1, Vsnl1, Hpcal1 and Car8, which participate in cellular signaling and synaptic plasticity, indicates that these gene transcripts may contribute to altered function of NAcc GABA neurons, potentially predisposing to nicotine self-administration in Lewis rats. However, data from a manifold of inbred strains and F1 crosses will be required to specify the potential role of individual genes.
Supplementary Material
Acknowledgments
These studies were supported by DA-028962 (B.M.S., H. Chen, S.G. Matta) from NIDA.
References
- Bekkers JM. Presynaptically silent GABA synapses in hippocampus. J Neurosci. 2005;25:4031–4039. doi: 10.1523/JNEUROSCI.4969-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DK, Conroy WG, Liu Z, Zago WM. Nicotinic signal transduction machinery. J Mol Neurosci. 2006;30:149–152. doi: 10.1385/JMN:30:1:149. [DOI] [PubMed] [Google Scholar]
- Biederer T, Sudhof TC. Mints as adaptors. Direct binding to neurexins and recruitment of munc18. J Biol Chem. 2000;275:39803–39806. doi: 10.1074/jbc.C000656200. [DOI] [PubMed] [Google Scholar]
- Borg JP, Straight SW, Kaech SM, de Taddeo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B. Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J Biol Chem. 1998;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- Braunewell KH, Gundelfinger ED. Intracellular neuronal calcium sensor proteins: a family of EF-hand calcium-binding proteins in search of a function. Cell Tissue Res. 1999;295:1–12. doi: 10.1007/s004410051207. [DOI] [PubMed] [Google Scholar]
- Brower VG, Fu Y, Matta SG, Sharp BM. Rat strain differences in nicotine self-administration using an unlimited access paradigm. Brain Res. 2002;930:12–20. doi: 10.1016/s0006-8993(01)03375-3. [DOI] [PubMed] [Google Scholar]
- Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz S, Okamoto M, Sudhof TC. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Thomas MJ. Biological substrates of reward and aversion: a nucleus accumbens activity hypothesis. Neuropharmacology. 2009;56(Suppl 1):122–132. doi: 10.1016/j.neuropharm.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassin L, Presson CC, Rose JS, Sherman SJ. The natural history of cigarette smoking from adolescence to adulthood: demographic predictors of continuity and change. Health Psychol. 1996;15:478–484. doi: 10.1037//0278-6133.15.6.478. [DOI] [PubMed] [Google Scholar]
- Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics. 2004;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dappen A, Schwartz RH, O’Donnell R. A survey of adolescent smoking patterns. J Am Board Fam Pract. 1996;9:7–13. [PubMed] [Google Scholar]
- Duclos F, Koenig M. Comparison of primary structure of a neuron-specific protein, X11, between human and mouse. Mamm Genome. 1995;6:57–58. doi: 10.1007/BF00350899. [DOI] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Davletov B, Petrenko AG, Jahn R, Sudhof TC. Interaction of synaptotagmin with the cytoplasmic domains of neurexins. Neuron. 1993;10:307–315. doi: 10.1016/0896-6273(93)90320-q. [DOI] [PubMed] [Google Scholar]
- Heimer L, Wilson RD. The subcortical projections of allocortex: similarities in the neural associations of the hippocampus, the periform cortex and the neocortex. In: MS, editor. Golgi centennial symposium proceedings. Raven Press; New York: 1975. pp. 173–193. [Google Scholar]
- Hirasawa M, Xu X, Trask RB, Maddatu TP, Johnson BA, Naggert JK, Nishina PM, Ikeda A. Carbonic anhydrase related protein 8 mutation results in aberrant synaptic morphology and excitatory synaptic function in the cerebellum. Mol Cell Neurosci. 2007;35:161–170. doi: 10.1016/j.mcn.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Ando H, Hamada K, Mikoshiba K. Carbonic anhydrase-related protein is a novel binding protein for inositol 1,4,5-trisphosphate receptor type 1. Biochem J. 2003;372:435–441. doi: 10.1042/BJ20030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Hitzemann B, Rivera S, Gatley J, Thanos P, Shou LL, Williams RW. Dopamine D2 receptor binding, Drd2 expression and the number of dopamine neurons in the BXD recombinant inbred series: genetic relationships to alcohol and other drug associated phenotypes. Alcohol Clin Exp Res. 2003;27:1–11. doi: 10.1097/01.ALC.0000047862.40562.27. [DOI] [PubMed] [Google Scholar]
- Ho A, Morishita W, Hammer RE, Malenka RC, Sudhof TC. A role for Mints in transmitter release: Mint 1 knockout mice exhibit impaired GABAergic synaptic transmission. Proc Natl Acad Sci U S A. 2003;100:1409–1414. doi: 10.1073/pnas.252774899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kilman V, van Rossum MC, Turrigiano GG. Activity deprivation reduces miniature IPSC amplitude by decreasing the number of postsynaptic GABA(A) receptors clustered at neocortical synapses. J Neurosci. 2002;22:1328–1337. doi: 10.1523/JNEUROSCI.22-04-01328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthl A, Laurent JP, Figurov A, Muller D, Schachner M. Hippocampal long-term potentiation and neural cell adhesion molecules L1 and NCAM. Nature. 1994;372:777–779. doi: 10.1038/372777a0. [DOI] [PubMed] [Google Scholar]
- Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- Martin S, Manzanares J, Corchero J, Garcia-Lecumberri C, Crespo JA, Fuentes JA, Ambrosio E. Differential basal proenkephalin gene expression in dorsal striatum and nucleus accumbens, and vulnerability to morphine self-administration in Fischer 344 and Lewis rats. Brain Res. 1999;821:350–355. doi: 10.1016/s0006-8993(99)01122-1. [DOI] [PubMed] [Google Scholar]
- Maximov A, Bezprozvanny I. Synaptic targeting of N-type calcium channels in hippocampal neurons. J Neurosci. 2002;22:6939–6952. doi: 10.1523/JNEUROSCI.22-16-06939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov A, Sudhof TC, Bezprozvanny I. Association of neuronal calcium channels with modular adaptor proteins. J Biol Chem. 1999;274:24453–24456. doi: 10.1074/jbc.274.35.24453. [DOI] [PubMed] [Google Scholar]
- Meredith GE, Pennartz CM, Groenewegen HJ. The cellular framework for chemical signalling in the nucleus accumbens. Prog Brain Res. 1993;99:3–24. doi: 10.1016/s0079-6123(08)61335-7. [DOI] [PubMed] [Google Scholar]
- Moretti M, Mugnaini M, Tessari M, Zoli M, Gaimarri A, Manfredi I, Pistillo F, Clementi F, Gotti C. A comparative study of the effects of the intravenous self-administration or subcutaneous minipump infusion of nicotine on the expression of brain neuronal nicotinic receptor subtypes. Mol Pharmacol. 78:287–296. doi: 10.1124/mol.110.064071. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Sudhof TC, Wahl MC. CASK Functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam CI, Chen L. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc Natl Acad Sci U S A. 2005;102:6137–6142. doi: 10.1073/pnas.0502038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Rasmussen BA, Perry DC. Binding and functional activity of nicotinic cholinergic receptors in selected rat brain regions are increased following long-term but not short-term nicotine treatment. J Neurochem. 2004;90:40–49. doi: 10.1111/j.1471-4159.2004.02482.x. [DOI] [PubMed] [Google Scholar]
- Nomura K, Takeuchi Y, Yamaguchi S, Okamura H, Fukunaga K. Involvement of calcium/calmodulin-dependent protein kinase II in the induction of mPer1. J Neurosci Res. 2003;72:384–392. doi: 10.1002/jnr.10581. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Sudhof TC. Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem. 1997;272:31459–31464. doi: 10.1074/jbc.272.50.31459. [DOI] [PubMed] [Google Scholar]
- Okaty BW, Sugino K, Nelson SB. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS One. 2011;6:e16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SL, Fu Y, McAllen K, Luo J, McIntosh JM, Lindstrom JM, Sharp BM. Up-regulation of brain nicotinic acetylcholine receptors in the rat during long-term self-administration of nicotine: disproportionate increase of the alpha6 subunit. Mol Pharmacol. 2004;65:611–622. doi: 10.1124/mol.65.3.611. [DOI] [PubMed] [Google Scholar]
- Pieribone VA, Aston-Jones G. The iontophoretic application of Fluoro-Gold for the study of afferents to deep brain nuclei. Brain Res. 1988;475:259–271. doi: 10.1016/0006-8993(88)90614-2. [DOI] [PubMed] [Google Scholar]
- SAMHSA; Services, DoHaH. Summary of Findings from the 2000 NHSDA. Substance Abuse and Mental Health Services Administration; Rockville: 2001. [Google Scholar]
- Shirayama Y, Chaki S. Neurochemistry of the nucleus accumbens and its relevance to depression and antidepressant action in rodents. Curr Neuropharmacol. 2006;4:277–291. doi: 10.2174/157015906778520773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KS, Tindell AJ, Aldridge JW, Berridge KC. Ventral pallidum roles in reward and motivation. Behav Brain Res. 2009;196:155–167. doi: 10.1016/j.bbr.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker C, Dresbach T, Braunewell KH. Reversible translocation and activity-dependent localization of the calcium-myristoyl switch protein VILIP-1 to different membrane compartments in living hippocampal neurons. J Neurosci. 2002;22:7331–7339. doi: 10.1523/JNEUROSCI.22-17-07331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, George FR, Meisch RA. Differential establishment and maintenance of oral ethanol reinforced behavior in Lewis and Fischer 344 inbred rat strains. J Pharmacol Exp Ther. 1988a;245:164–170. [PubMed] [Google Scholar]
- Suzuki T, Otani K, Koike Y, Misawa M. Genetic differences in preferences for morphine and codeine in Lewis and Fischer 344 inbred rat strains. Jpn J Pharmacol. 1988b;47:425–431. doi: 10.1254/jjp.47.425. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Fukunaga K, Miyamoto E. Activation of nuclear Ca(2+)/calmodulin-dependent protein kinase II and brain-derived neurotrophic factor gene expression by stimulation of dopamine D2 receptor in transfected NG108-15 cells. J Neurochem. 2002;82:316–328. doi: 10.1046/j.1471-4159.2002.00967.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Yamamoto H, Matsumoto K, Kimura T, Katsuragi S, Miyakawa T, Miyamoto E. Nuclear localization of the delta subunit of Ca2+/calmodulin-dependent protein kinase II in rat cerebellar granule cells. J Neurochem. 1999;72:815–825. doi: 10.1046/j.1471-4159.1999.0720815.x. [DOI] [PubMed] [Google Scholar]
- Tanahashi H, Tabira T. Genomic organization of the human X11L2 gene (APBA3), a third member of the X11 protein family interacting with Alzheimer’s beta-amyloid precursor protein. Neuroreport. 1999;10:2575–2578. doi: 10.1097/00001756-199908200-00025. [DOI] [PubMed] [Google Scholar]
- Taylor CW, Tovey SC. IP(3) receptors: toward understanding their activation. Cold Spring Harb Perspect Biol. 2010;2:a004010. doi: 10.1101/cshperspect.a004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, RDC. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2010. [Google Scholar]
- Valentine JD, Hokanson JS, Matta SG, Sharp BM. Self-administration in rats allowed unlimited access to nicotine. Psychopharmacol (Berl) 1997;133:300–304. doi: 10.1007/s002130050405. [DOI] [PubMed] [Google Scholar]
- Xiao MF, Xu JC, Tereshchenko Y, Novak D, Schachner M, Kleene R. Neural cell adhesion molecule modulates dopaminergic signaling and behavior by regulating dopamine D2 receptor internalization. J Neurosci. 2009;29:14752–14763. doi: 10.1523/JNEUROSCI.4860-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada MK, Nakanishi K, Ohba S, Nakamura T, Ikegaya Y, Nishiyama N, Matsuki N. Brain-derived neurotrophic factor promotes the maturation of GABAergic mechanisms in cultured hippocampal neurons. J Neurosci. 2002;22:7580–7585. doi: 10.1523/JNEUROSCI.22-17-07580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Anand R, Braunewell KH. Nicotine-induced Ca2+-myristoyl switch of neuronal Ca2+ sensor VILIP-1 in hippocampal neurons: a possible crosstalk mechanism for nicotinic receptors. Cell Mol Neurobiol. 2009a;29:273–286. doi: 10.1007/s10571-008-9320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CJ, Noack C, Brackmann M, Gloveli T, Maelicke A, Heinemann U, Anand R, Braunewell KH. Neuronal Ca2+ sensor VILIP-1 leads to the upregulation of functional alpha4beta2 nicotinic acetylcholine receptors in hippocampal neurons. Mol Cell Neurosci. 2009b;40:280–292. doi: 10.1016/j.mcn.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.