Abstract
Proteins requiring post-translational modifications such as N-linked glycosylation are processed in the endoplasmic reticulum (ER). A diverse array of cellular stresses can lead to dysfunction of the ER and ultimately to an imbalance between protein-folding capacity and protein-folding load. Cells monitor protein folding by an inbuilt quality control system involving both the ER and the Golgi apparatus. Unfolded or misfolded proteins are tagged for degradation via ER associated degradation (ERAD) or sent back through the folding cycle. Continued accumulation of incorrectly folded proteins can also trigger the Unfolded Protein Response (UPR). In mammalian cells, UPR is a complex signaling program mediated by three ER transmembrane receptors: activating transcription factor 6 (ATF6), inositol requiring kinase 1 (IRE1) and double-stranded RNA-activated protein kinase (PKR)-like endoplasmic reticulum kinase (PERK). UPR performs three functions, adaptation, alarm and apoptosis. During adaptation, the UPR tries to reestablish folding homeostasis by inducing the expression of chaperones that enhance protein folding. Simultaneously, global translation is attenuated to reduce the ER folding load while the degradation rate of unfolded proteins is increased. If these steps fail, the UPR induces a cellular alarm and mitochondrial mediated apoptosis program. UPR malfunctions have been associated with a wide range of disease states including tumor progression, diabetes, as well as immune and inflammatory disorders. This review describes recent advances in understanding the molecular structure of UPR in mammalian cells, its functional role in cellular stress, and its pathophysiology.
Keywords: Protein Folding, UPR, PERK, ATF6, IRE1, Apoptosis
Introduction
Protein folding is strategically important to cellular function. Secreted, membrane-bound and organelle-targeted proteins are typically processed and folded in the endoplasmic reticulum (ER) in eukaryotes (Kaufman et al., 2002; Naidoo, 2009; Ron, 2002). Intracellular perturbations caused by a variety of stressors disturb the specialized environment of the ER leading to the accumulation of unfolded proteins (Ellgaard and Helenius, 2003; Fonseca et al., 2009). Glucose deprivation, aberrant calcium regulation, viral infection and hypoxia can all alter protein folding and induce ER stress (Kaufman et al., 2002; Ron, 2002). Physiological processes such as aging can also influence protein folding (Naidoo, 2009). Normally, cells ensure that proteins are correctly folded using a combination of molecular chaperones, foldases and lectins (Naidoo, 2009). However, when proper folding can not be restored, incorrectly folded proteins are targeted to ER Associated Degradation (ERAD) pathways for processing (Kaufman et al., 2002). If unfolded or misfolded proteins continue to accumulate, eukaryotes induce the unfolded protein response (UPR).
In mammalian cells, UPR is a complex signaling program mediated by three ER trans-membrane receptors: activating transcription factor 6 (ATF6), inositol requiring kinase 1 (IRE1) and double-stranded RNA-activated protein kinase (PKR)-like endoplasmic reticulum kinase (PERK). UPR performs three functions, adaptation, alarm and apoptosis. During adaptation, the UPR tries to reestablish folding homeostasis by inducing the expression of chaperones that enhance protein folding. Simultaneously, translation is globally attenuated to reduce the ER folding load while the degradation of unfolded proteins is increased. If these steps fail, the UPR induces a cellular alarm and apoptosis program. The alarm phase involves several signal transduction events, ultimately leading to the removal of the translational block and the down-regulation of the expression and activity of pro-survival factors such as the B-cell lymphoma 2 (Bcl2) protein. After the alarm phase, cells can undergo apoptosis, although ER stress can also initiate autophagy (Bernales et al., 2006; Fujita et al., 2007; Høyer-Hansen et al., 2007; Kamimoto et al., 2006; Kouroku et al., 2007; Ogata et al., 2006; Yorimitsu et al., 2006). Thus, ER folding homeostasis strongly influences physiology (Fonseca et al., 2009). Aberrant protein folding and UPR have been implicated in a number of pathologies. For example, the onset of diabetes (Schnell, 2009) as well as myocardial ischaemia, cardiac hypertrophy, atherosclerosis and heart failure (Glembotski, 2007) have all been linked with aberrant folding or UPR signaling.
UPR reprogramming could also be a means to increase recombinant monoclonal antibody (MAb) production in mammalian cells. Therapeutic protein production using recombinant mammalian cell lines has been a critical component of the biologics industry since the mid 1980s with drugs such as tissue plasminogen activator (tPA) (Becker et al., 2008, Mohan et al., 2008). Several other current biopharmaceutical products are also secreted proteins, for example interferon-γ (IFNγ) and erythropoeitin (EPO) (Ku et al., 2008). Typical mammalian production hosts for this class of product include immortalized Chinese Hamster Ovary (CHO) cells (Wurm, 2004) or other mammalian cell types, such as murine lymphoid cells (NS0, SP2/0) (Khoo et al., 2007). These cell lines are often engineered or selected for high specific protein production rates (Seth et al., 2007, Barnes and Dickson, 2006), high growth rates (Khoo et al., 2007) and reduced apoptosis (Connor et al., 2009). However, beyond the robustness of the production host, protein production may also be sensitive to intracellular processing bottlenecks affecting transcription, translation or post-translational modifications. Thus, engineering the folding subsystems of mammalian cells might also be a promising route to increase the production of secreted protein biopharmaceuticals.
The folding cycle, quality control and ER associated degradation (ERAD)
Newly synthesized polypeptide chains enter the ER through a peptide translocon in the ER membrane composed of four proteins, Sec61α,β,γ and TRAM (Matlack et al., 1998). Upon entering the ER, these nascent chains begin to fold, often as they are being co-translationally modified (Fedorov and Baldwin, 1997). In the cytosol, protein folding is largely driven by the collapse of hydrophobic side chains which ultimately form the core of the folded protein. Another key factor, the burial of electrostatic interactions, e.g., salt bridges or hydrogen bonds in the hydrophobic core, constrains possible folding choices in a very complex free energy landscape. The forces that govern cytosolic folding are also active in the ER. However, protein folding in the ER is more complex because of post-translational modifications such as disulfide bond formation or N-linked glycosylation. Interestingly, the folding landscape in the ER can sometimes be traversed quickly; small proteins like the Semliki Forest virus capsid protein can fold in approximately 50ms (Sanchez et al., 2004). However, for larger and more complex proteins, e.g., the human coagulation factors V and VIII, folding can take several minutes to hours to complete (Pittman et al., 1994).
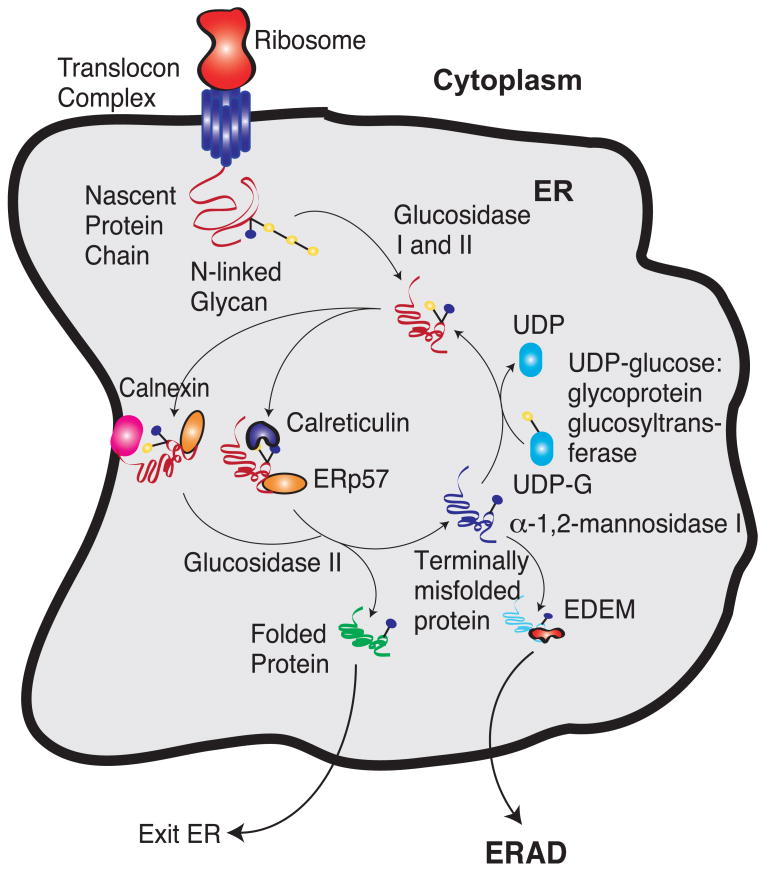
The folding quality of proteins in the ER is maintained by an in-built quality control (QC) system (Fig. 1) which ensures proteins are in their functional folded state before exiting the ER (Ellgaard and Helenius, 2003; Ellgaard et al., 1999). A protein is correctly folded if it has attained its native conformation after required co-or post-translational modifications. On the other hand, exposed hydrophobic regions, unpaired cysteine residues, or aggregation are all markers of an unfolded or misfolded conformation (Ellgaard et al., 1999). The best characterized QC system in the ER is the so-called glycan-code (Hebert et al., 2005). Most polypeptides entering the ER are modified by adding preassembled oligosaccharides to asparagine side chains appearing in ASN-X-SER/THR motifs (Helenius and Aebi, 2004). Once attached, these oligosaccharide groups can be sequentially modified by glucosidases I and II to form mono-glucosylated intermediates that are recognized by the ER lectins calnexin and calreticulin (Hammond et al., 1994) and the associated oxidoreductase ERp57 (Hebert et al., 1995; Liu et al., 1999; Oliver et al., 1999, 1997; Ware et al., 1995). Calnexin is a type I membrane protein with a β-sandwich carbohydrate binding domain and a hairpin, called the P-domain, extending away from the carbohydrate binding domain (Schrag et al., 2001). Calreticulin is the soluble paralog of calnexin with only minor structural differences in the P-domain (Ellgaard et al., 2001a,b; Schrag et al., 2001). Calnexin and calreticulin binding increases the efficiency of glycoprotein folding by protecting against aggregation (Hebert et al., 1996) and ensures that misfolded proteins are retained in the ER (Rajagopalan et al., 1994). These ER glycoprotein chaperones also promote disulfide bond formation through their interaction with ERp57; ERp57 binds the P-domain of both calnexin and calreticulin thereby promoting disulfide bond formation at specific glycoprotein locations (Frickel et al., 2002; Oliver et al., 1999, 1997). Glycoproteins are released from calnexin and calreticulin by further glucose residue cleavage by glucosidase II. Once released, these proteins can fold into their native conformation, they can be glycosylated and re-processed by the calnexin/calreticulin cycle, or they can be targeted for ER-associated degradation (ERAD) (recently reviewed in Maattanen et al., 2010). Interestingly, while calnexin and calreticulin ultimately promote folding, they do not recognize misfolded proteins. Misfolded proteins can be targeted (or re-targeted) to the calnexin/calreticulin cycle by glucosylation with UDP-glucose; the C-terminal domain of glycoprotein glucosyltransferase (GT) glucosylates near-native conformations by using its N-terminal protein sensor to locate exposed hydrophobic residues (Caramelo et al., 2004; D'Alessio et al., 2010; Sousa and Parodi, 1995; Trombetta and Parodi, 2003). In this way, GT acts as an adapter allowing attachment of calnexin/calreticulin and re-processing in the calnexin/calreticulin cycle (Parodi, 2000). Repeated glycosylation and deglycosylation cycles ensure misfolded glycoproteins spend sufficient time in the ER to correctly fold. Exit from the calnexin/calreticulin cycle can also lead to ERAD. In this case, terminal mannose residues are removed from the attached oligosaccharides by ER α-mannosidase I, which leads to interaction with membrane localized ER degradation enhancing α-mannosidase like proteins (EDEM and EDEM2) (Hosokawa et al., 2001; Olivari et al., 2005) and subsequent retro-translocation to the cytosol. The retro-translocon responsible for transporting unfolded or misfolded proteins out of the ER is uncertain (Tsai et al., 2002); several ER membrane proteins have been proposed including Sec61 components, Derlin family members and E3-ubiquitin ligases (reviewed in (Hebert et al., 2010)). Mannose removal decreases the likelihood that a unfolded protein will be processed in the calnexin/calreticulin cycle (Ellgaard et al., 1999), thus, increasing the probability of terminal mannose cleavage and retro-translocation. Once in the cytosol, these unfolded or misfolded proteins are degraded by the ubiquitin proteasome system (Hershko et al., 2000).
Fig. 1.
The calnexin/calreticulin protein folding cycle. Yellow circles denote glucose groups and while blue circles denote mannose groups. After entering the ER lumen, glucosidase I and II remove two glucose groups. The monoglucosylated glycoprotein then interacts with calnexin/calreticulin. These chaperones interact with the thiol-disulphide oxidoreductase ERp57. Cleavage of the last glucose residue by glucosidase II leads to the release of the chaperones. At this time, the protein could have either folded and left the ER or it could have attained an incorrect state. The incorrectly folded proteins are then the substrates of UDP glucose:glycoprotein glucosyltransferase, which puts a glucose residue back to the incorrectly folded protein. This enables the protein to spend some more time in folding in the ER. If the protein fails to fold in a repeated number of cycles, the mannose residue is removed by α-1,2-mannosidase I. This enables the protein to be recognized by ER-degradation-enhancing 1,2-mannosidase-like protein (EDEM). This targets the unfolded proteins for ER-associated degradation (ERAD).
Hydrophobic unfolded or misfolded proteins are recognized in the ER by molecular chaperones which bind these proteins and increase the probability of correct folding (Fra et al., 1993; Helenius et al., 1997; Hellman et al., 1999). Similar to calnexin and calreticulin, hydrophobic chaperone binding increases the residence time of unfolded proteins in the ER lumen, giving these proteins a chance to fold (Fig. 2A). For example, the HSP70 family of chaperones recognize, in an ATP-dependent manner, exposed hydrophobic patches on a broad spectrum of unfolded or misfolded proteins (Kaufman et al., 2002). Repeated binding and release of HSP70 chaperones ensures that incorrectly folded proteins do not exit the ER (Kaufman et al., 2002). One critical member of the HSP70 family is BiP or GRP78. BiP consists of an N-terminal ATPase domain and a C-terminal peptide binding domain (Gething, 1999). When bound to ATP, BiP binds unfolded hydrophobic stretches with low-affinity. However, unfolded or misfolded protein binding stimulates the N-terminal ATPase activity of BiP resulting in an ADP-bound form with much higher affinity for hydrophobic motifs (Gething, 1999). Interestingly, the affinity of BiP (and other HSP70 family members) for ADP is approximately six-fold greater than for ATP. Thus, nucleotide exchange factors (NEFs) such as BiP associated protein (BAP), Sil1/S1s1p or GrpE-like proteins are required to catalyze the ADP/ATP exchange needed for the dissociation of BiP from unfolded proteins (Tyson and Stirling, 2000). In addition to its role as a folding chaperone, BiP also functions as an ER stress regulator by buffering Ca2+ levels (Reddy et al., 2003). BiP interacts with ER localized caspase-7 (Reddy et al., 2003) and prevents the activation of pro-apoptotic Bcl2 family members such as Bax (Fu et al., 2007; Ranganathan et al., 2006). Beyond these activities, BiP also regulates the activation of the three transmembrane ER stress transducers: PERK, ATF6 and IRE1. Normally, BiP is bound to these ER receptors, blocking their activation. However, in the presence of exposed hydrophobic residues BiP disassociates, allowing PERK, ATF6 and IRE1 activation (Fig. 2B). Overexpression of BiP leads to reduced activation of IRE1 and PERK (Bertolotti et al., 2000; Kohno et al., 1993). The PERK and ATF6 branches are thought to be activated before IRE1 (Szegezdi et al., 2006); this ordering is consistent with the signals that each branch transduces. The PERK and ATF6 pathways largely promote ER adaptation to misfolding, while IRE1 has a dual role, transmitting both survival and pro-apoptotic signals.
Fig. 2.
An array of cellular stressors can perturb the folding environment in the endoplasmic reticulum (ER) leading to unfolded or misfolded protein. In response to the folding imbalance, cells initiate the cytoprotective unfolded protein response (UPR). A: The problem of unfolded or misfolded proteins in the ER is addressed by increasing the folding capacity through the up-regulation of the expression of chaperone proteins, attenuating translation by regulating eIF2α, and promoting the degradation of misfolded proteins through ER-associated degradation (ERAD). If UPR is unable to restore the folding balance, ER stress will eventually lead to apoptotic cell-death. B: The three signal transduction pathways mediating the unfolded protein response in higher eukaryotes. First, the PRKR-like ER kinase (PERK) pathway is initiated after BiP dissociation from PERK. While PERK transduces both pro-and anti-apoptotic signals, its main function is translation attenuation through the phosphorylation of eIF2α. Next, the activating transcription factor 6 (ATF6) pathway is activated following BiP dissociation. ATF6 induces the expression of chaperones e.g., BiP as well as apoptosis effectors such as CHOP. Lastly, the inositol-requiring kinase 1 (IRE1) pathway is activated following BiP dissociation from IRE1. Activated IRE1 has both an endoribonuclease and a serine-threonine kinase activity that drive can pro-apoptotic signals.
Double-stranded RNA-activated protein kinase (PKR)-like endoplasmic reticulum kinase (PERK) pathway
The PERK branch of UPR transduces both pro-survival as well as pro-apoptotic signals following the accumulation of unfolded or misfolded protein in the ER. However, its main function is to modulate translation. PERK is a type I trans-membrane protein, composed of a ER luminal stress sensor and a cytosolic protein kinase domain. Dissociation of BiP from the N-terminus of PERK initiates dimerization and autophosphorylation of the kinase domain at T981 (Kebache et al., 2004). The C-terminal kinase domain shares homology with the eukaryotic translation initiation factor 2α (eIF2α) and the kinases, protein kinase R (PKR), heme-regulated inhibitor (HRI) kinase, and the gene control non-derepressible-2 (GCN2) kinase (Harding et al., 1999; Shi et al., 1999, 1998). The eIF2α protein, which is composed of three subunits, is critical to translation initiation in eukaryotes, including GTP-dependent start-site recognition (Merrick, 2004). Phosphorylation of the α subunit of eIF2α blocks the exchange of GDP bound to eIF2α; hence, eIF2α remains bound to partner initiation factors (eIF2B) and translation initiation is blocked (Gebauer and Hentze, 2004). Activated PERK can phosphorylate eIF2α at S51 (Harding et al., 1999; Raven et al., 2008), which leads to three downstream effects. First, phosphorylated eIF2α globally attenuates translation initiation. Decreased translation reduces the influx of protein into the ER, hence diminishing the folding load. Translation attenuation is followed by increased clearance of the accumulated proteins from the ER by ERAD and expression of pro-survival genes. For example, PERK activation induces the expression of cellular inhibitor of apoptosis (cIAP) (Hamanaka et al., 2009). Interestingly, decreased protein translation is not universal; genes with internal ribosome entry site (IRES) sequences in the 5′ untranslated regions bypass the eIF2α translational block (Schroder and Kaufman, 2005). One of the most well-studied of these, ATF4, encodes a cAMP response element-binding transcription factor (C/EBP) (Lu et al., 2004) ATF4 that drives the expression of pro-survival functions such as amino acid transport and synthesis, redox reactions and protein secretion (Harding et al., 2003). Taken together, these effects seem to be largely pro-survival. However, ATF4 can also induce the expression of pro-apoptotic factors. For example, ATF4 induces the expression of the transcription factor C/EBP homologous protein (CHOP), which is associated with apoptotic cell-death. CHOP (also known as GADD153) is a 29 kDa protein composed of an N-terminal transcriptional activation domain and a C-terminal basic-leucine zipper (bZIP) domain that is normally present at low levels in mammalian cells (Ron and Habener, 1992). The transcriptional activator domain is positively regulated by phosphorylation at S78 and S81 by p38 MAPK family members (Maytin et al., 2001; Wang and Ron, 1996) while the bZIP domain plays a key role in the homodimerization of the protein (Matsumoto et al., 1996; Maytin et al., 2001). CHOP activity promotes apoptosis primarily by repression of Bcl2 expression and the sensitization of cells to ER-stress inducing agents (Gotoh et al., 2001; McCullough et al., 2001). For example, Matsumoto et al. showed that ectopic expression of CHOP in M1 myeloblastic leukemia cells reduced Bcl2 protein concentrations, while Bax levels remained constant (Matsumoto et al., 1996). They further established a link between CHOP expression and apoptosis in these cells. However, while CHOP expression is negatively correlated with Bcl2 levels, there is no CHOP binding site in the bcl2 promoter (McCullough et al., 2001). McCullough et al. have suggested that the bZIP domain of CHOP could act with other bZIP transcription factors to regulate bcl2 expression (McCullough et al., 2001). Thus, it's likely that the connection between CHOP expression and apoptosis is more complex than simple down-regulation of Bcl2 expression. Given its central role in translation attenuation, cells have evolved multiple axes to regulate PERK activity. First, the cytosolic kinase domain of PERK can be inhibited by the action of the DNAJ family member P58IPK, which was initially discovered as an inhibitor of the eIF2α protein kinase PKR (Lee et al., 1990). P58IPK, whose expression is induced following ATF6 activation, binds to the cytosolic kinase domain of PERK, inhibiting its activity (van Huizen et al., 2003; Yan et al., 2002). Inhibition of PERK kinase activity relieves eIF2α phosphorylation, thereby removing the translational block. Interestingly, P58IPK expression occurs several hours after PERK activation and eIF2α phosphorylation. Thus, P58IPK induction may mark the end of UPR adaptation, and the beginning of the alarm/apoptosis phase of the response (Szegezdi et al., 2006). In addition to its direct interaction with PERK, P58IPK is also involved in cotranslational protein degradation (Oyadomari et al., 2006). In this role, P58IPK is thought to recruit Hsp70 to the cytosolic opening of the ER translocon in an effort to extract stalled nascent proteins. Interestingly, P58IPK has also been identified as an ER-luminal co-chaperone acting in conjunction with BiP, although BiP-independent association between P58IPK and a mutant vesicular stomatitis virus envelope glycoprotein (VSV-Gts045) was also observed (Petrova et al., 2008). The activity of P58IPK appears to be regulated; recently, Ni et al. reported a novel UPR-inducible cytosolic BiP isoform (GRP78va), generated by alternative splicing and IRES mediated translation, that antagonizes cytosolic P58IPK in several human and mouse cell-lines (Ni et al., 2009). Second, PERK induces a negative feedback loop, through its downstream effector CHOP, involving the direct de-phosphorylation of eIF2α. CHOP induces the expression of GADD34 which, in conjunction with protein phosphatase 1 (PP1), assembles into a phosphatase which dephosphorylates the S51 residue of eIF2α (Novoa et al., 2001). GADD34 is a member of the GADD family of genes which are induced by DNA damage and a variety of other cellular stresses (Zhan et al., 1994). The GADD34 binding partner in this complex appears to be responsible for PP1α recognition and targeting of the phosphatase complex to the ER. Association between GADD34 and PP1 is encoded by a C-terminal canonical PP1 binding motif, KVRF, while approximately 180 residues, near the N-terminus of GADD34, appear to be responsible for ER localization (Brush et al., 2003).
Activating transcription factor 6 (ATF6) pathway
ATF6 activation involves a complex series of translocation and irreversible proteolytic processing steps, ultimately leading to the up-regulation of a pro-survival transcriptional program, in the presence of unfolded or misfolded proteins. ATF6 is a 90 kDa ER transmembrane protein with two homologs: ATF6α (Hai et al., 1989; Haze et al., 1999) and ATF6β (Haze et al., 2001; Khanna and Campbell, 1996; Min et al., 1995). The ATF6α homolog is thought to be primarily responsible for transcriptional regulation of pro-survival genes following ER stress, however, ATF6β may also play a role (Haze et al., 2001; Thuerauf et al., 2007; Yamamoto et al., 2007). Similar to IRE1 and PERK, ER stress leads to the dissociation of BIP from the N-terminus of ATF6, followed by translocation and activation. However, the mechanism of BiP interaction with ATF6 and the factors controlling ATF6 translocation to the golgi remain uncertain. N-terminal golgi localization sequences (GLS1 and GLS2) seem to be involved with BiP regulation of ATF6. BiP binding to the N-terminal GLS1 promotes the retention of ATF6 in the ER (Shen et al., 2002). On the other hand, the GLS2 domain was required to target ATF6 to the golgi body following BiP dissociation from GLS1 (Shen et al., 2002). The lectin CRT might also have a role in keeping the ATF6 in the ER (Hong et al., 2004). Unlike the previous two kinase pathways, ATF6 activation does not involve phosphorylation of a C-terminal kinase domain. Rather, after translocated to the golgi, ATF6 undergoes regulated intramembrane proteolysis (RIP); the luminal domain is first cleaved by serine protease site-1 protease (S1P) followed by metalloprotease site-2 protease (S2P) cleavage (Chen et al., 2002; Haze et al., 1999; Shen and Prywes, 2004; Ye et al., 2000). Cleavage at the juxtamembrane site allows the 50 kDa transcriptional domain of ATF6 to be translocated to the nucleus where it regulates the expression of genes with ATF/cAMP response elements (CREs) (Wang et al., 2000) and ER stress response elements (ERSE) in their promoters (Kokame et al., 2001; Yoshida et al., 1998).
Cleaved ATF6 induces a gene expression program, in conjunction with other bZIP transcription factors and required co-regulators, such as nuclear factor Y (NF-Y) (Kokame et al., 2001; Yoshida et al., 2000), that increases chaperone activity as well as the degradation of unfolded proteins (Wu et al., 2007; Yamamoto et al., 2007). For example, ATF6 upregulates BiP, protein disulfide isomerase (PDI) and ER degradation-enhancing alpha-mannosidase-like protein 1 (EDEM1) expression. Additionally, ATF6 induces the expression of the X box-binding protein 1 (XBP1) which, after processing by activated IRE1α, induces the expression of chaperones as well as control elements such as P58INK (Yoshida et al., 2001). The ATF6-induced gene expression program is also cytoprotective. For example, ATF6 induces regulator of calcineurin 1 (RCAN1) expression (Belmont et al., 2008). RCAN1 sequesters calcineurin (Belmont et al., 2008), a calcium activated protein-phosphotase B, that dephosphorylates Bcl2-antagonist of cell death (BAD) at S75 or S99 (Wang et al., 1999). This leads to sequestering of Bcl2 by Bad, which inhibits its downstream anti-apoptotic activity (Wang et al., 1999). Recently, a number of ATF6 homologs have been identified, e.g., OASIS, CREBH, LUMAN/CREB3, CREB4 and BBF2H7 that are processed in a similar way as ATF6, yet their function remains unknown (Ron and Walter, 2007). Thus, ER-stress induced ATF6 signaling may be responsible for additional undiscovered functionality.
Currently, little is known about deactivation of ATF6. Recently, XBP1u, the unspliced form of XBP1, has been implicated as a negative regulator for ATF6 (Yoshida et al., 2009). Following, the induction of ER stress, two versions of XBP1 exist: XBP1u and sXBP1 (Yoshida et al., 2009). In the recovery phase following ER stress, high levels of XBP1u may play a dual role. First, XBP1u binds sXBP1, promoting complex degradation (Tirosh et al., 2006; Yoshida et al., 2006). Second, XBP1u can bind ATF6α rendering it more prone to proteasomal degradation (Yoshida et al., 2009). Taken together, these two steps may slow the transcription of ER chaperones and ERAD components during the recovery phase following ER stress.
Inositol-requiring kinase 1 (IRE1) pathway
IRE1 is the most evolutionarily conserved branch of UPR and its interactome has recently been reviewed by Hetz and Glimcher (Hetz and Glimcher, 2009). IRE1 initiates a program with both pro-survival and pro-apoptotic components in the presence of misfolded or unfolded proteins. IRE1 is a 100 kDa type I ER transmembrane protein with both an endoribonuclease and a serine-threonine kinase domain (Kaufman et al., 2002). IRE1 has two homologs, IRE1α and IRE1β; IRE1α is expressed in a variety of tissues (Tirasophon et al., 1998) while IRE1β is found only in the intestinal epithelia (Tirasophon et al., 1998; Wang et al., 1998). The N-terminus of IRE1, located in the ER lumen, senses unfolded or misfolded proteins through its interaction with BiP (Cox et al., 1993; Shamu and Walter, 1996; Sidrauski and Walter, 1997). There has been some controversy surrounding the mechanism of unfolding-induced BiP dissociation and the role of possible direct interaction of unfolded proteins with IRE1. Normally BiP is bound to the N-terminus of IRE1 (Bertolotti et al., 2000; Liu et al., 2003; Okamura et al., 2000). However, in the presence of unfolding cues BiP dissociates and is sequestered by the unfolded or misfolded proteins (Kimata et al., 2003). IRE1 may also sense unfolded cues in a BiP independent manner through a N-terminal peptide binding domain (Liu et al., 2002, 2003). Interaction of this domain with unfolded or misfolded proteins has been suggested as a precursor to BiP dissociation. Current studies in yeast have suggested that BiP first dissociates, allowing IRE1 dimerization. This step is then followed by direct sensing of unfolded motifs which work to orient IRE1 into an active signaling cluster (Kimata et al., 2007). In either case, BiP dissociation allows IRE1 activation. IRE1 is activated by homo-oligomerization followed by autophosphorylation of the C-terminal kinase domain at S724 (Papa et al., 2003; Shamu and Walter, 1996; Weiss and Schlessinger, 1998; Welihinda and Kaufman, 1996). IRE1 activation enables both its kinase and endoribonuclease activities to transduce signals simultaneously through two distinct signaling axes. The endoribonuclease activity cleaves a 26-nucleotide intron from the XBP1-mRNA (Lee et al., 2002; Shen et al., 2001; Yoshida et al., 2001) which generates a 41 kDa frameshift variant (sXBP1) that acts as a potent transcription factor. sXBP1 homodimers, along with co-regulators such as nuclear factor Y (NF-Y), regulate the expression of a variety of ER chaperones and protein degradation related genes (Malhotra and Kaufman, 2007; Rao and Bredesen, 2004). sXBP1 also upregulates the expression of P58IPK; as discussed previously, P58IPK is a member of the DNAJ protein family that negatively regulates PERK activity, forming one of the many modes of crosstalk between the UPR branches (Yan et al., 2002).
The cytosolic domains of activated IRE1α transduce late-phase UPR signals. Cytosolic IRE1α dimers interact with adaptors such as tumor necrosis factor receptor-associated factor 2 (TRAF2) to drive signal-regulating kinase (ASK1) activation and then subsequently cJUN NH2-terminal kinase (JNK) and p38MAPK activation (Urano et al., 2000). IRE1α also modulates the activation of other kinases such as extracellular signal-regulated kinases (ERKs) as well as nuclear factor κB (NF-κB) pathways (Hu et al., 2006; Nguyen et al., 2004). However, the role of these additional effectors in UPR is not well understood. ASK1 activity is regulated by phosphorylation/de-phosphorylation at several sites as well as by physical interaction with other proteins. The current model for ASK1 activation, at least for TNF-mediated activation, involves release of ASK1 from inhibitory proteins such as 14-3-3 (Nishitoh et al., 1998), TRAF-dependent homodimerization (Gotoh and Cooper, 1998) and autophosphorylation at T845 (Tobiume et al., 2002). Phosphorylation of ASK1 at S83 by Akt/PKB (protein kinase B) (Kim et al., 2001) and dephosphorylation at S845 by protein phosphatase 5 (PP5) (Morita et al., 2001) both decrease ASK1 activity. More recently, the kinase PIM1 has also been shown to phosphorylate ASK1 at S83 (Gu et al., 2009). Interaction with CDC25A and 14-3-3 proteins have also been shown to decrease ASK1 activity (Zhang et al., 1999; Zou et al., 2001). ASK1 phosphorylates and activates two downstream kinases, MMK4 and MMK3 which in turn activate JNK and p38 MAP kinase, respectively. JNK is activated by dual phosphorylation at T183 and Y185 by MMK4 (Derijard et al., 1995). Once activated, JNK performs a number of functions including activation of the pro-apoptotic Bim protein (Lei and Davis, 2003; Putcha et al., 2003) and the inhibition of Bcl2 (Yamamoto et al., 1999). Activated JNK activates the proapoptotic Bcl-2 family member Bim by phosphorylation at S65 (Lei and Davis, 2003; Putcha et al., 2003). Bim is normally sequestered by motor complexes interacting with the cytoskeleton (Puthalakath et al., 1999). Following JNK-mediated phosphorylation, Bim translocates to the mitochondrial outer membrane, where it promotes cytochrome c release and caspase activation (Chen and Zhou, 2004; Puthalakath et al., 1999). Interestingly, a positive feedback loop exists between Bim and caspase 3 activation; phosphorylated Bim is a caspase 3 target, that once cleaved, is a more potent inducer of cytochrome c release (Chen and Zhou, 2004; Corazza et al., 2006). Recent ER stress studies in MCF-7 breast carcinoma-derived cells using thapsigargin (Tg) suggested that Bim expression was regulated by CHOP; following Tg treatment, a two-fold increase in Bim mRNA and a five-fold increase in Bim proteins were observed (Puthalakath et al., 2007). JNK activation also regulates the activity of anti-apoptotic protein Bcl2 (Wei et al., 2008; Yamamoto et al., 1999). Active JNK1 inhibits Bcl2 via phosphorylation at sites T69, S70 and S87 (Wei et al., 2008). In contrast, other stress induced proteins like p38 family members phosphorylate Bcl2 at S87 and T56 only (De Chiara et al., 2006). Ultimately, inhibition of Bcl2 and the activation of Bim leads to BAX/BAK dependent apoptosis. Thus, signals initiated from the cytosolic kinase domain of IRE1α are largely pro-apoptotic.
IRE1α activity is regulated by several proteins, including tyrosine phosphatase 1B (PTP-1B), ASK1-interactive protein 1 (AIP1) and members of the Bcl2 protein family. PTP-1B has been implicated in a number of IRE1α signaling events. The absence of PTP-1B reduced IRE1α dependent JNK activation, XBP1 splicing and EDEM transcription in immortalized and primary mouse embryonic fibroblasts (Gu et al., 2004). However, no physical interaction between IRE1α and PTP-1B was established. On the other hand, AIP1 physically interacts with both TRAF2 and IRE1α, suggesting a model in which AIP1 facilitates IRE1α dimerization and activation (Luo et al., 2008). The C-terminal period-like domain (PER) of AIF1 binds the N-terminal RING finger domain of TRAF2, followed by ASK1-JNK signaling (Zhang et al., 2004). Thus, based on these findings, Luo et al. postulated that AIF1 may be directly involved in the IRE1α-TRAF2 complex and its activation of the ASK1-JNK signaling axis (Luo et al., 2008). This hypothesis was validated in AIP1KO mouse studies; AIP1-knockout mouse embryonic fibroblasts and vascular endothelial cells showed significant reductions in ER-stress induced ASK1-JNK activation that was rescued in AIP1 knock-in cells (Luo et al., 2008). IRE1α has also been shown to directly interact with Bcl-2 family members Bax and Bak. Hetz et al. showed that Bax and Bak complex with the cytosolic domain of IRE1α and modulate IRE1α signaling (Hetz et al., 2006). Bax and Bak double knockout mice failed to signal through the IRE1α UPR branch following tunicamycin-induced ER stress; however, PERK signaling markers, e.g., eIF2α phosphorylation, responded normally (Hetz et al., 2006). This pro-activation role of Bak and Bax may be modulated by one of the few negative regulators of IRE1α activity, Bax inhibitor 1 (BI-1). BI-1 is an anti-apoptotic protein that enhances cell survival following several intrinsic death stimuli (Xu and Reed, 1998). Bailly-Maitre et al. were the first to suggest that BI-1 may downregulate IRE1α and possibly ATF6 activity (Bailly-Maitre et al., 2006). BI-1 deficient mice displayed increased XBP1s and enhanced JNK activity in the liver and kidney, while eIF2α phosphorylation remained normal under ER-stress conditions (Bailly-Maitre et al., 2006). Lisbona et al. later showed that BI-1 directly interacts with the cytosolic domain of IRE1α, inhibiting its endoribonuclease activity (Lisbona et al., 2009). Interestingly, BI-1 interacts with several members of the Bcl2 protein family e.g., Bcl2 and Bcl-XL, even though it has no homology (Xu and Reed, 1998). Members of the HSP family of proteins have also been shown to regulate IRE1α. For example, HSP90 interacts with the cytosolic domain of IRE1α, potentially protecting it from degradation by the proteasome (Marcu et al., 2002). HSP72 interaction with the cytosolic IRE1α domain has also recently been shown to enhance IRE1α endoribonuclease activity (Gupta et al., 2010). Taken together, these modes of IRE1α regulation with the exception of B1-1, largely promote or enhance IRE1α signaling.
ER stress-induced autophagy and apoptosis
Ultimately, if UPR fails to restore ER homeostasis, cells initiate terminal programs such as autophagy or apoptosis. Several recent studies indicate that ER stress can trigger autophagy (Bernales et al., 2006; Fujita et al., 2007; Høyer-Hansen et al., 2007; Kamimoto et al., 2006; Kouroku et al., 2007; Ogata et al., 2006; Wei et al., 2008; Yorimitsu et al., 2006). Autophagy is an evolutionarily conserved cellular pathway, in which a cell recycles its macromolecules and organelles (Levine and Klionsky, 2004). Autophagy is initiated by the formation of an autophagosome, composed of part of the cytosol or cellular organelles, encased in a double membrane (Codogno and Meijer, 2005; Kroemer and Jaattela, 2005; Levine and Klionsky, 2004). Autophagosomes then bind endolysosomal vesicles, leading to the creation of the autolysosome. The autolysosome is then degraded, completing the autophagy cycle. The key regulatory proteins involved in the nucleation and formation of the autophagosomal membrane are class III phospha-tidylinositol 3-kinase (PI3K), p150 myristylated protein kinase and Beclin-1 (Levine and Klionsky, 2004). On the other hand, the ER-stress sensors PERK, IRE1 and cytosolic Ca2+ all act as effectors initiating autophagy in ER stressed cells (Fujita et al., 2007; Høyer-Hansen et al., 2007; Kouroku et al., 2007; Ogata et al., 2006). JNK1 mediated phosphorylation of Bcl2 at T69, S70 and S87 may also be important; phosphorylation at these sites leads to the dissociation of Bcl2 from Beclin-1, and the activation of autophagy (Wei et al., 2008). Association of Beclin-1 with PI3K and other proteins promotes the localization of other autophagy related proteins to the preautophagosomal membrane (Levine and Kroemer, 2008). Autophagy can be inhibited in eukaryotic cells by several factors, including the mammalian target of rapamycin complex 1 (mTORC1) protein complex (Meijer and Codogno, 2006; Noda and Ohsumi, 1998).
The autophagy program is a cellular reboot mechanism while apoptosis is a terminal death program. Apoptosis is characterized by nuclear and cytoplasmic condensation, blebbing of the plasma membrane and DNA fragmentation (Kerr et al., 1972). The dying cell eventually disintegrates into membrane-enclosed apoptotic bodies which are quickly destroyed by phagocytes or neighboring cells. A common biomarker of apoptosis is the activation of aspartate-specific proteases, collectively known as caspases (Alnemri et al., 1996). Caspases rapidly dismantle cell cycle, cytoskeletal and organelle proteins by proteolytic cleavage. There are two pathways that result in caspase activation in response to apoptotic signals; the death-receptor and the stress mediated pathways. The death-receptor pathway is marked by ligand-mediated activation of death receptors on the plasma membrane. The alternative pathway for caspase activation is mediated by cellular stress e.g., ER stress. Caspases are activated from their zymogens (procaspases), in response to various death cues. First, the initiator caspases, caspase-8 and caspase-9, are activated in response to death cues (Muzio et al., 1998). This is followed by the activation of executioner caspases, such as caspase-3, caspase-6 and caspase-7. Activated executioner caspases proteolytically process several substrates, facilitating cell death. They also activate initiator caspases, forming a positive feedback loop.
Activation of both the PERK and IRE1 pathways modulate stress-induced apoptosis through their regulation of Bcl2 expression and activity. Overall, stress induced apoptosis can occur through both mitochondrial-dependent and independent pathways. Stress signals cause oligomerization of pro-apoptotic proteins, such as Bax and Bak. These proteins are normally sequestered at the mitochondrial outer membrane by the survival protein Bcl2, under non-apoptotic conditions (Wei et al., 2001). Once Bax and Bak oligomerize, they insert into the mitochondrial membrane and breach membrane integrity (Nechushtan et al., 1999). This results in a net efflux of cytochrome-c from the mitochondria to the cytosol and the initiation of the well-studied Apaf-1 mediated caspase-9 activation pathway. Recently, crosstalk between the ER and mitochondria during apoptosis, which might initiate mitochondrial apoptotic events, has also been explored (Demaurex and Distelhorst, 2003). This crosstalk may be facilitated via the release of Ca2+ ions from the ER into the cytosol, in response to ER stress. Mitochondrial uptake of Ca2+ initiates membrane fission and caspase activation via an uncertain mechanism. However, a few key drivers of ER Ca2+ release in response to stress signals have been identified (Nicotera and Orrenius, 1998). Fas-mediated caspase-8 activation leads to cleavage of the membrane-bound ER protein, Bap31. Although Bap31 is membrane-bound, its caspase-recognition sequence is cytosolic, thus it is accessible by caspase-8. The truncated product, Bap20, is believed to promote the release of ER Ca2+ (Demaurex and Distelhorst, 2003; Distelhorst and Shore, 2004; Thomenius and Distelhorst, 2003). Interestingly, Bcl2 is also localized to the ER-membrane, but its role is uncertain (Adams, 2003). Once in the mitochondria, Ca2+ modulates the permeability transition (PT) pore (PTP) thereby increasing the efflux of cytochrome-c (Bernardi, 1999; Decaudin et al., 1997). Stress induced mitochondrial-independent apoptotic pathways are not well understood. Currently, caspase 12 has been suggested as a possible ER-stress apoptotic mediator in mice (Nakagawa et al., 2000; Szegezdi et al., 2006; Yoneda et al., 2001). However, caspase 12 is not expressed in human cells. Moreover, there is considerable debate about its role in stress-induced apoptotic cell-death (Saleh et al., 2006).
The pathophysiology of ER-stress and aberrant UPR
ER stress plays an important role in a spectrum of diseases ranging from neurodegeneration, cardiac diseases, cancer, diabetes to muscle degeneration (Table 1). Understanding ER-stress mechanisms in the context of these diseases presents a unique opportunity for drug discovery. Current therapeutic efforts have largely focused on amplifying the adaptive, pro-survival components of the UPR signal, for example, by inducing chaperone expression, which works to restore ER homeostasis. However, in the context of cancer, the opposite outcome is sought; in this case a number of strategies have focused on inhibiting proteasome function (Table 2).
Table 1.
Relevance of ER stress in human disease states
| Disease state | Role of ER-Stress and UPR | Key proteins | Reference |
|---|---|---|---|
| Alzheimer's disease (AD) | AD-induced Presenilin 1 regulates BiP. | Presenilin 1, PERK, eIF2α, BiP | Katayama et al. (1999); Milhavet et al. (2002); Niwa et al. (1999); Terro et al. (2002); Unterberger et al. (2006) |
| Parkinson's disease (PD) | Suppressed ER-stress- induced apoptosis and aggregation of α-synuclein. | Parkin,α-synuclein | Dawson and Dawson (2003); Imai et al. (2000); Petrucelli et al. (2002); Ryu et al. (2002); Takahashi et al. (2003) |
| Amyotrophic lateral sclerosis (ALS) | Altered ERAD machinery and activation of ASK1. | Cu/Zn-superoxide dismutase (SOD), ASK1 and Derlin-1 | Nishitoh et al. (2008) |
| Bipolar Disorder | Current medications target induction of UPR. | XBP1 | Cichon et al. (2004); Kakiuchi et al. (2005, 2003) |
| Stroke | CHOP and UPR mediates neuronal response in Ischemia. | PERK-eIF2α, ASK1, CHOP | Kumar et al. (2001); Paschen et al. (1998); Tajiri et al. (2004); Tanaka et al. (2000) |
| Heart Disease | Degeneration of cardiac myocytes, transaortic constriction. Myocardial infarction induces UPR. | IRE1, PERK- eIF2α, ASK1 | Glembotski (2008); Okada et al. (2004); Pan et al. (2004); Shintani-Ishida et al. (2006); Thuerauf et al. (2006); Vitadello et al. (2003) |
| Atherosclerosis | Regulates inflammatory genes in vascular cells. | IRE1, JNK, TRAF2, XBP1 | Gargalovic et al. (2006); Harding et al. (2001); Lee et al. (2008); Zhang et al. (2001) |
| Type 1 diabetes | Regulators in normal conditions and triggers β cell dysfunction and apoptosis in ER Stress. | PERK-eIF2α, JNK and Calcium | Araki et al. (2003); Fonseca et al. (2005); Ishihara et al. (2004); Oyadomari et al. (2002, 2001); Scheuner et al. (2001); Yamada et al. (2006) |
| Type 2 diabetes | ER stress is a key aspect of type 2 diabetes. | XBP1, JNK | Ozcan et al. (2004) |
| Cancer | Cytoprotective branches of UPR vital to survival and progression of tumors. | BiP, XBP1, ATF6, PERK | Bi et al. (2005); Fujimoto et al. (2003); Jamora et al. (1996); Romero-Ramirez et al. (2004); Shuda et al. (2003) |
| Autoimmune disease | Development of plasma cells and dendritic cells. | XBP1, HLA-B27 | Blass et al. (2001); Corrigall et al. (2001); Todd et al. (2008) |
| Glomerulonephritis, acute kidney injury | Upregulation of BiP, CHOP and down- regulation of Bcl-2 in primary glomerular diseases. | BiP, CHOP, Bcl2 | Blass et al. (2001); Corrigall et al. (2001); Todd et al. (2008) Bando et al. (2004); Markan et al. (2009) |
Table 2.
Therapeutic interventions targeting ER stress.
| Compound | Pathway | Function | Reference |
|---|---|---|---|
| Valporate (VPA), lithium | ER chaperones (BiP, GRP94, Calreticulin) | Remove ER Stress; ER chaperone induction in Neuro-protection in Bipolar Disorders. | Bown et al. (2002); Hiroi et al. (2005); Shao et al. (2006) |
| BiP inducer X (BIX) | ER chaperones (BiP) | Remove ER Stress; Small-molecule inducer of BiP in Celebral ischemia; stroke resulting in reduced cell death. | Kudo et al. (2008) |
| Salubrinal | eIF2α, Bcl2 | Phosphatase inhibitor; PERK- eIF2α pathway agonist in Parkinson's disease; anti-stroke; regulator of Bcl2. | Araki et al. (2003); Boyce et al. (2005); Harding et al. (2001); Kessel (2006); Kumar et al. (2001); Reijonen et al. (2008); Scheuner et al. (2001) |
| p38 MAPK antagonists | p38 MAPK (CHOP activity) | Apoptosis up; p38 dependent CHOP phosphorylation regulator in stroke, diabetes. | Milhavet et al. (2002); Paschen et al. (1998); Tajiri et al. (2004); Wang and Ron (1996); Yoshida (2007) |
| Versipelostatin (VST) | BiP | Apoptosis up; Alters BiP production in glucose-deprived solid tumors. | Park et al. (2004) |
| Bortezomib, Nelfavir, Atazanavir | Proteasome, XBP1 | Apoptosis up; Inhibition of HIV-1 proteasome activity. CHOP mediated cell death. | Gills et al. (2007); Lee et al. (2003); Neubert et al. (2008); Pyrko et al. (2007) |
| Brefeldin A | CHOP, BiP | Apoptosis up; Inhibition of protein trafficking, ER swelling. | Carew et al. (2006) |
Diabetes
In the context of diseases such as diabetes, pancreatic β-cells depend on efficient UPR signaling to meet the demands for constantly varying levels of insulin synthesis. Type 1 diabetes is marked by excessive loss of pancreatic β-cells, while type 2 diabetes is marked by pancreatic β-cell dysfunction. The large biosynthetic load placed on the ER because of insulin production in response to food uptake (glucose) can overwhelm the folding capacity of the ER, resulting in ER stress. This leads to subsequent PERK activation and reduction of protein synthesis. In PERK -/- cells, protein synthesis is unresponsive to the stress and leads to accumulation of unfolded proteins (e.g. proinsulin) and ultimately cell death. It has been shown that PERK deficient mice are more prone to diabetes and progressive hyperglycemia (Harding et al., 2001). In type II diabetes, ER stress leads to JNK-mediated phosphorylation of insulin receptor substrate 1 (IRS1) at S307 (Ozcan et al., 2004). IRS1 is a substrate of Insulin receptor. Phosphorylation of IRS1 therefore inhibits insulin action. Nitric oxide (NO), is also a key player in β-cell death in type-1 diabetes and vascular complications in type-2 diabetes. NO depletes ER Ca2+ leading to ER stress and ultimately apoptosis. Pancreatic β-cells have shown that NO induced apoptosis is CHOP dependent (Oyadomari et al., 2001). Thus, ER stress is a critical feature of both type-I and type-II diabetes at the molecular, cellular and organismal level.
Role of UPR in hypoxia and cancers
The ER not only acts as the center for maturation of proteins, but also as a critical node for oxygen sensing and signaling. In rapidly growing tumors, cells face stressors like hypoxia and nutrient deprivation both of which can lead to ER stress and ultimately UPR. For example, oxygen deprivation has recently been shown to be an initiator of UPR (Koumenis and Wouters, 2006). Interestingly, the connection between hypoxia and UPR is through hypoxia inducible factor (HIF) independent pathways. Hypoxia leads to PERK activation and the transient phosphorylation of eIF2α (Koritzinsky et al., 2006; Koumenis et al., 2002; Ma and Hendershot, 2003). This effect occurs on a time scale of minutes in the case of anoxia, and is slower for moderate (pO2 ≤ 1%) hypoxic conditions (Koritzinsky et al., 2006). Because of their ability to transduce pro-apoptotic signals, the modulation of PERK and IRE1 activity has been explored as a potential anti-cancer strategy. Versipelostatin, a repressor of BiP expression, has been shown to produce anti-tumor activity in MKN-74 xenograft mouse models (Park et al., 2004). Enhanced apoptosis has also been observed in BiP-deficient fibrosarcoma cells, and XBP1-and PERK-deficient mouse fibroblasts (Bi et al., 2005; Jamora et al., 1996; Romero-Ramirez et al., 2004).
Another related anti-cancer strategy is the induction of ER-stress using proteasome or protein trafficking inhibitors. Bortezomib, a selective inhibitor of the 26S proteasome, is used for the treatment of relapsed multiple myeloma, a plasma cell neoplasia (Neubert et al., 2008). Anti-tumor activity was also observed with Bortezomib in preclinical pancreatic cancer studies using the L3.6pl cell-line (Nawrocki et al., 2005). Addition of Bortezomib to splenic and bone marrow plasma cells led to a 40-fold increase in CHOP expression, and a subsequent decrease in Bcl2 levels (Neubert et al., 2008). HIV type 1 (HIV-1) protease inhibitors (PI) like Nelfavir and Atazanavir are also currently being studied for the treatment of malignant gliomas (Pyrko et al., 2007). Studies in glioblastoma cells suggested that Nelfavir or Atazanavir led to inhibition of proteasome activity and subsequently CHOP-induced cell death (Pyrko et al., 2007). Thus, although no mechanistic link between proteasome inhibitors and UPR has been established, anecdotal evidence, such as CHOP induction, suggests these inhibitors are at least partially inducing ER-stress and subsequent UPR-mediated cell death. Recently, studies targeting intracellular trafficking of proteins have also been explored as an anti-cancer strategy. Brefeldin A treated chronic lymphocytic leukemia (CLL) cells showed induction of ER stress, caused by inhibition of protein trafficking, ultimately resulting in Golgi collapse and cell death (Carew et al., 2006). Inhibition of protein trafficking from the ER can also lead to ER swelling and p53-independent apoptosis (Carew et al., 2006).
Neurodegenerative disorders
Several neurodegenerative diseases like Parkinson's disease, Alzheimer's disease; have been implicated to have accumulation of misfolded proteins leading to altered neuronal connectivity, neuronal death and and dysfunctional ERAD machinery (Soto, 2003; Bence et al., 2001; Katayama et al,. 1999) (Table 1). The implications of UPR in neurodegenerative diseases is yet to be determined (Lindhom et al., 2006). Alzheimer's disease (AD) is characterized by progressive decline of cognitive functions, marked by loss of neurons from different brain regions and extracellular senile plaques formed by aggregates of amyloid b-peptide (Ab) and neurofibrillary tangles (NFT) of protein tau (Selkoe, 2001). Mutations in Ab precursor protein (APP) and Presenillins (PS1 and PS2) are associated with early onset of AD (Selkoe, 2001; Katayama et al,. 1999). PS1 and PS2 mediate the cleavage of APP and Notch (Wilquet and Strooper, 2004). Mutant PS1 studies have been implicated in increased Ab production and ER Stress mediated apoptosis (Verkhratsky, 2005; Mattson, 2000; LaFarla, 2002). Mutant PS1 was shown to bind to IRE1, thereby suppressing the activation of a key branch of UPR and reduced transcriptional regulation of BiP (Katayama et al., 2004). On the contrary, increased levels of BiP and PERK was seen in AD human brain specimens (Hoozemans et al., 2005). Whether UPR is initiated for neuro-protection or for neuronal death, is still to be determined.
Immune and Inflammatory disorders
Unfolded or misfolded protein accumulation and UPR-induction is observed in autoimmune diseases like rheumatoid arthritis, inflammatory bowel diseases and multiple sclerosis. UPR and the inflammatory response are connected through several modalities including increased reactive oxygen species (ROS), increased calcium release from the ER, activation of JNK and the activation of NF-κB (Zhang and Kaufman, 2008). NF-κB plays a central role in inflammation (Rius et al., 2008). In unstressed cells, NF-κB is inactive, sequestered by the inhibitor of NF-κB (IκB). UPR induction leads to phosphorylation and subsequent degradation of IκB and the expression of inflammatory genes (Rius et al., 2008; Tak and Firestein, 2001). UPR also plays a key role in autoimmune diseases. CREBH, an ATF6 homolog, translocates to the golgi and undergoes cleavage by S1p and S2p proteases in the presence of ER stress. Proteolytic processing releases an N-terminal fragment that migrates to the nucleus and induces C-reactive protein and serum amyloid P-component expression (Zhang et al., 2006). Both of these genes are associated with the activation of the acute inflammatory response (Zhang et al., 2006). CREBH can also induce pro-inflammatory cytokines such as IL-6 and IL-1β (Zhang et al., 2006).
Summary and Conclusions
Protein folding is strategically important to cellular function. Perturbations to the unique folding environment of the endoplasmic reticulum (ER) lead to the accumulation of unfolded or misfolded proteins. Higher eukaryotes have evolved a complex three-pronged response to correct aberrant folding, known as the unfolded protein response. The unfolded protein response acts to restore ER homeostasis. However, following prolonged or severe ER stress, cells initiate an alarm phase ultimately leading to apoptotic cell death. Malfunctions in the folding state of critical proteins have been linked with cancer, diabetes and other diseases. Thus, the ability of UPR to regulate cell fate, has made it a primary area of study for pathophysiology and a potential therapeutic axis for the treatment of a diverse array of diseases.
Acknowledgments
The project described was supported by Award Number #U54CA143876 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Finally, the authors thank Ryan Tasseff and Joshua Lequieu for their helpful suggestions, and the development of the pathway schematics.
References
- Adams JM. Ways of dying: multiple pathways to apoptosis. Genes Dev. 2003;17:2481–95. doi: 10.1101/gad.1126903. [DOI] [PubMed] [Google Scholar]
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Human ice/ced-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- Araki E, Oyadomari S, Mori M. Endoplasmic reticulum stress and diabetes mellitus. Intern Med. 2003;42:7–14. doi: 10.2169/internalmedicine.42.7. [DOI] [PubMed] [Google Scholar]
- Bailly-Maitre B, Fondevila C, Kaldas F, Droin N, Luciano F, Ricci JE, Croxton R, Krajewska M, Zapata JM, Kupiec-Weglinski JW, Farmer D, Reed JC. Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:2809–14. doi: 10.1073/pnas.0506854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando Y, Tsukamoto Y, Katayama T, Ozawa K, Kitao Y, Hori O, Stern DM, Yamauchi A, Ogawa S. Orp150/hsp12a protects renal tubular epithelium from ischemia-induced cell death. FASEB J. 2004;18:1401–3. doi: 10.1096/fj.03-1161fje. [DOI] [PubMed] [Google Scholar]
- Barnes LM, Dickson AJ. Mammalian cell factories for efficient and stable protein expression. Curr Opin Biotechnol. 2006;4:381–86. doi: 10.1016/j.copbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Becker E, Florin L, Pfizenmaier K, Kaufmann H. An XBP-1 dependent bottle-neck in production of IgG subtype antibodies in chemically defined serum-free Chinese hamster ovary (CHO) fed-batch processes. J Biotechnol. 2008;2:217–33. doi: 10.1016/j.jbiotec.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Belmont PJ, Tadimalla A, Chen WJ, Martindale JJ, Thuerauf DJ, Marcinko M, Gude N, Sussman MA, Glembotski CC. Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (rcan1), a novel atf6-inducible gene. J Biol Chem. 2008;283:14012–21. doi: 10.1074/jbc.M709776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin- proteasome system by protein aggregation. Science. 2001;292:1552–55. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi P. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev. 1999;79:1127–55. doi: 10.1152/physrev.1999.79.4.1127. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of bip and er stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–32. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. Er stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–81. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blass S, Union A, Raymackers J, Schumann F, Ungethum U, Muller-Steinbach S, De Keyser F, Engel JM, Burmester GR. The stress protein bip is overexpressed and is a major b and t cell target in rheumatoid arthritis. Arthritis Rheum. 2001;44:761–71. doi: 10.1002/1529-0131(200104)44:4<761::AID-ANR132>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Bown CD, Wang JF, Chen B, Young LT. Regulation of er stress proteins by valproate: therapeutic implications. Bipolar Disord. 2002;4:145–51. doi: 10.1034/j.1399-5618.2002.t01-1-40201.x. [DOI] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eif2alpha dephosphorylation protects cells from er stress. Science. 2005;307:935–9. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. Growth arrest and dna damage-inducible protein gadd34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo JJ, Castro OA, de Prat-Gay G, Parodi AJ. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J Biol Chem. 2004;279:46280–5. doi: 10.1074/jbc.M408404200. [DOI] [PubMed] [Google Scholar]
- Carew JS, Nawrocki ST, Krupnik YV, Dunner K, Jr, McConkey DJ, Keating MJ, Huang P. Targeting endoplasmic reticulum protein transport: a novel strategy to kill malignant b cells and overcome fludarabine resistance in cll. Blood. 2006;107:222–31. doi: 10.1182/blood-2005-05-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhou Q. Caspase cleavage of bimel triggers a positive feedback amplification of apoptotic signaling. Proc Natl Acad Sci U S A. 2004;101:1235–40. doi: 10.1073/pnas.0308050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen J, Prywes R. The luminal domain of atf6 senses endoplasmic reticulum (er) stress and causes translocation of atf6 from the er to the golgi. J Biol Chem. 2002;277:13045–52. doi: 10.1074/jbc.M110636200. [DOI] [PubMed] [Google Scholar]
- Cichon S, Buervenich S, Kirov G, Akula N, Dimitrova A, Green E, Schumacher J, Klopp N, Becker T, Ohlraun S, Schulze TG, Tullius M, Gross MM, Jones L, Krastev S, Nikolov I, Hamshere M, Jones I, Czerski PM, Leszczynska-Rodziewicz A, Kapelski P, Bogaert AVD, Illig T, Hauser J, Maier W, Berrettini W, Byerley W, Coryell W, Gershon ES, Kelsoe JR, McInnis MG, Murphy DL, Nurnberger JI, Reich T, Scheftner W, O'Donovan MC, Propping P, Owen MJ, Rietschel M, Nothen MM, McMahon FJ, Craddock N. Lack of support for a genetic association of the xbp1 promoter polymorphism with bipolar disorder in probands of european origin. Nat Genet. 2004;36:783–4. doi: 10.1038/ng0804-783. [DOI] [PubMed] [Google Scholar]
- Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;2:1509–18. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- Corazza N, Jakob S, Schaer C, Frese S, Keogh A, Stroka D, Kassahn D, Torgler R, Mueller C, Schneider P, Brunner T. Trail receptor-mediated jnk activation and bim phosphorylation critically regulate fas-mediated liver damage and lethality. J Clin Invest. 2006;116:2493–9. doi: 10.1172/JCI27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigall VM, Bodman-Smith MD, Fife MS, Canas B, Myers LK, Wooley P, Soh C, Staines NA, Pappin DJ, Berlo SE, van Eden W, van Der Zee R, Lanchbury JS, Panayi GS. The human endoplasmic reticulum molecular chaperone bip is an autoantigen for rheumatoid arthritis and prevents the induction of experimental arthritis. J Immunol. 2001;166:1492–8. doi: 10.4049/jimmunol.166.3.1492. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- D'Alessio C, Caramelo JJ, Parodi AJ. Udp-glc:glycoprotein glucosyltransferaseglucosidase ii, the ying-yang of the er quality control. Semin Cell Dev Biol. 2010;21:491–9. doi: 10.1016/j.semcdb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Rare genetic mutations shed light on the pathogenesis of parkinson disease. J Clin Invest. 2003;111:145–51. doi: 10.1172/JCI17575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chiara G, Marcocci ME, Torcia M, Lucibello M, Rosini P, Bonini P, Higashimoto Y, Damonte G, Armirotti A, Amodei S, Palamara AT, Russo T, Garaci E, Cozzolino F. Bcl-2 phosphorylation by p38 mapk: identification of target sites and biologic consequences. J Biol Chem. 2006;281:21353–61. doi: 10.1074/jbc.M511052200. [DOI] [PubMed] [Google Scholar]
- Decaudin D, Geley S, Hirsch T, Castedo M, Marchetti P, Macho A, Kofler R, Kroemer G. Bcl-2 and bcl-xl antagonize the mitochondrial dysfunction preceding nuclear apoptosis induced by chemotherapeutic agents. Cancer Res. 1997;57:62–7. [PubMed] [Google Scholar]
- Demaurex N, Distelhorst C. Cell biology. apoptosis–the calcium connection. Science. 2003;300:65–7. doi: 10.1126/science.1083628. [DOI] [PubMed] [Google Scholar]
- Derijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, Davis RJ. Independent human map-kinase signal transduction pathways defined by mek and mkk isoforms. Science. 1995;267:682–5. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- Distelhorst CW, Shore GC. Bcl-2 and calcium: controversy beneath the surface. Oncogene. 2004;23:2875–80. doi: 10.1038/sj.onc.1207519. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–91. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–8. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Riek R, Braun D, Herrmann T, Helenius A, Wuthrich K. Three-dimensional structure topology of the calreticulin p-domain based on nmr assignment. FEBS Lett. 2001a;488:69–73. doi: 10.1016/s0014-5793(00)02382-6. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Riek R, Herrmann T, Guntert P, Braun D, Helenius A, Wuthrich K. Nmr structure of the calreticulin p-domain. Proc Natl Acad Sci U S A. 2001b;98:3133–8. doi: 10.1073/pnas.051630098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov AN, Baldwin TO. Cotranslational protein folding. J Biol Chem. 1997;272:32715–8. doi: 10.1074/jbc.272.52.32715. [DOI] [PubMed] [Google Scholar]
- Fonseca SG, Burcin M, Gromada J, Urano F. Endoplasmic reticulum stress in beta-cells and development of diabetes. Curr Opin Pharmacol. 2009;9:763–70. doi: 10.1016/j.coph.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, Oka Y, Urano F. Wfs1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. J Biol Chem. 2005;280:39609–15. doi: 10.1074/jbc.M507426200. [DOI] [PubMed] [Google Scholar]
- Fra AM, Fagioli C, Finazzi D, Sitia R, Alberini CM. Quality control of er synthesized proteins: an exposed thiol group as a three-way switch mediating assembly, retention and degradation. EMBO J. 1993;12:4755–61. doi: 10.1002/j.1460-2075.1993.tb06164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. Trosy-nmr reveals interaction between erp57 and the tip of the calreticulin p-domain. Proc Natl Acad Sci U S A. 2002;99:1954–9. doi: 10.1073/pnas.042699099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li J, Lee AS. Grp78/bip inhibits endoplasmic reticulum bik and protects human breast cancer cells against estrogen starvation-induced apoptosis. Cancer Res. 2007;67:3734–40. doi: 10.1158/0008-5472.CAN-06-4594. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Onda M, Nagai H, Nagahata T, Ogawa K, Emi M. Upregulation and overexpression of human x-box binding protein 1 (hxbp-1) gene in primary breast cancers. Breast Cancer. 2003;10:301–6. doi: 10.1007/BF02967649. [DOI] [PubMed] [Google Scholar]
- Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. Two endoplasmic reticulum-associated degradation (erad) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome erad(i) and autophagy/lysosome erad(ii) Hum Mol Genet. 2007;16:618–29. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- Gargalovic PS, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Baruch-Oren T, Berliner JA, Kirchgessner TG, Lusis AJ. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:2490–6. doi: 10.1161/01.ATV.0000242903.41158.a1. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- Gills JJ, Lopiccolo J, Tsurutani J, Shoemaker RH, Best CJM, Abu-Asab MS, Borojerdi J, Warfel NA, Gardner ER, Danish M, Hollander MC, Kawabata S, Tsokos M, Figg WD, Steeg PS, Dennis PA. Nelfinavir, a lead hiv protease inhibitor, is a broad-spectrum, anticancer agent that induces endoplasmic reticulum stress, autophagy, and apoptosis in vitro and in vivo. Clin Cancer Res. 2007;13:5183–94. doi: 10.1158/1078-0432.CCR-07-0161. [DOI] [PubMed] [Google Scholar]
- Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101:975–84. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- Glembotski CC. The role of the unfolded protein response in the heart. J Mol Cell Cardiol. 2008;44:453–9. doi: 10.1016/j.yjmcc.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T, Terada K, Mori M. hsp70-dnaj chaperone pairs prevent nitric oxide-mediated apoptosis in raw 264.7 macrophages. Cell Death Differ. 2001;8:357–66. doi: 10.1038/sj.cdd.4400829. [DOI] [PubMed] [Google Scholar]
- Gotoh Y, Cooper JA. Reactive oxygen species-and dimerization-induced activation of apoptosis signal-regulating kinase 1 in tumor necrosis factor-alpha signal transduction. J Biol Chem. 1998;273:17477–82. doi: 10.1074/jbc.273.28.17477. [DOI] [PubMed] [Google Scholar]
- Gu F, Nguyen DT, Stuible M, Dube N, Tremblay ML, Chevet E. Protein-tyrosine phosphatase 1b potentiates ire1 signaling during endoplasmic reticulum stress. J Biol Chem. 2004;279:49689–93. doi: 10.1074/jbc.C400261200. [DOI] [PubMed] [Google Scholar]
- Gu JJ, Wang Z, Reeves R, Magnuson NS. Pim1 phosphorylates and negatively regulates ask1-mediated apoptosis. Oncogene. 2009;28:4261–71. doi: 10.1038/onc.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A. Hsp72 protects cells from er stress-induced apoptosis via enhancement of ire1alpha-xbp1 signaling through a physical interaction. PLoS Biol. 2010;8:e1000410. doi: 10.1371/journal.pbio.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor atf cdna clones: an extensive family of leucine zipper proteins able to selectively form dna-binding heterodimers. Genes Dev. 1989;3:2083–90. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Bobrovnikova-Marjon E, Ji X, Liebhaber SA, Diehl JA. Perk-dependent regulation of iap translation during er stress. Oncogene. 2009;28:910–20. doi: 10.1038/onc.2008.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C, Braakman I, Helenius A. Role of n-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci U S A. 1994;91:913–7. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–63. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, Mori K. Identification of the g13 (camp-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor atf6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Bernasconi R, Molinari M. Erad substrates: which way out? Semin Cell Dev Biol. 2010;21:526–32. doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–33. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 1996;15:2961–8. [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–70. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of n-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–49. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Helenius A, Trombetta E, Hebert D, Simons J. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7:193–200. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- Hellman R, Vanhove M, Lejeune A, Stevens FJ, Hendershot LM. The in vivo association of bip with newly synthesized proteins is dependent on the rate and stability of folding and not simply on the presence of sequences that can bind to bip. J Cell Biol. 1999;144:21–30. doi: 10.1083/jcb.144.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A. Basic medical research award. the ubiquitin system. Nat Med. 2000;6:1073–81. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Proapoptotic bax and bak modulate the unfolded protein response by a direct interaction with ire1alpha. Science. 2006;312:572–6. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the ire1alpha interactome. Mol Cell. 2009;35:551–61. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi T, Wei H, Hough C, Leeds P, Chuang DM. Protracted lithium treatment protects against the er stress elicited by thapsigargin in rat pc12 cells: roles of intracellular calcium, grp78 and bcl-2. Pharmacogenomics J. 2005;5:102–11. doi: 10.1038/sj.tpj.6500296. [DOI] [PubMed] [Google Scholar]
- Hong M, Luo S, Baumeister P, Huang JM, Gogia RK, Li M, Lee AS. Underglycosylation of atf6 as a novel sensing mechanism for activation of the unfolded protein response. J Biol Chem. 2004;279:11354–63. doi: 10.1074/jbc.M309804200. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, Veerhuis R, Van Haastert ES, Rozemuller JM, Baas F, Eikelenboom P, Scheper W. The unfolded protein response is activated in Alzheimer's disease. Acta Neuropathol. 2005;110:165–72. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. A novel er alpha-mannosidase-like protein accelerates er-associated degradation. EMBO Rep. 2001;2:415–22. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jaattela M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hu P, Han Z, Couvillon A, Kaufman R, Exton J. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1α-mediated NF-κB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006;26:3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its e3 ubiquitin-protein ligase activity. J Biol Chem. 2000;275:35661–4. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- Ishihara H, Takeda S, Tamura A, Takahashi R, Yamaguchi S, Takei D, Yamada T, Inoue H, Soga H, Katagiri H, Tanizawa Y, Oka Y. Disruption of the wfs1 gene in mice causes progressive beta-cell loss and impaired stimulus-secretion coupling in insulin secretion. Hum Mol Genet. 2004;13:1159–70. doi: 10.1093/hmg/ddh125. [DOI] [PubMed] [Google Scholar]
- Jamora C, Dennert G, Lee AS. Inhibition of tumor progression by suppression of stress protein grp78/bip induction in fibrosarcoma b/c10me. Proc Natl Acad Sci U S A. 1996;93:7690–4. doi: 10.1073/pnas.93.15.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi C, Ishiwata M, Nanko S, Kunugi H, Minabe Y, Nakamura K, Mori N, Fujii K, Umekage T, Tochigi M, Kohda K, Sasaki T, Yamada K, Yoshikawa T, Kato T. Functional polymorphisms of hspa5: possible association with bipolar disorder. Biochem Biophys Res Commun. 2005;336:1136–43. doi: 10.1016/j.bbrc.2005.08.248. [DOI] [PubMed] [Google Scholar]
- Kakiuchi C, Iwamoto K, Ishiwata M, Bundo M, Kasahara T, Kusumi I, Tsujita T, Okazaki Y, Nanko S, Kunugi H, Sasaki T, Kato T. Impaired feedback regulation of xbp1 as a genetic risk factor for bipolar disorder. Nat Genet. 2003;35:171–5. doi: 10.1038/ng1235. [DOI] [PubMed] [Google Scholar]
- Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–76. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Sato N, Miyoshi K, Kudo T, Hitomi J, Morihara T, Yoneda T, Gomi F, Mori Y, Nakano Y, Takeda J, Tsuda T, Itoyama Y, Murayama O, Takashima A, St George-Hyslop P, Takeda M, Tohyama M. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat Cell Biol. 1999;1:479–85. doi: 10.1038/70265. [DOI] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Manabe T, Hitomi J, Kudo T, Tohyama M. Induction of neuronal death by ER stress in Alzheimer's disease. J Chem Neuroanat. 2004;28:67–78. doi: 10.1016/j.jchemneu.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, Arnold SM. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol. 2002;3:411–21. doi: 10.1038/nrm829. [DOI] [PubMed] [Google Scholar]
- Kebache S, Cardin E, Nguyen DT, Chevet E, Larose L. Nck-1 antagonizes the endoplasmic reticulum stress-induced inhibition of translation. J Biol Chem. 2004;279:9662–71. doi: 10.1074/jbc.M310535200. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel D. Protection of bcl-2 by salubrinal. Biochem Biophys Res Commun. 2006;346:1320–3. doi: 10.1016/j.bbrc.2006.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A, Campbell RD. The gene g13 in the class iii region of the human mhc encodes a potential dna-binding protein. Biochem J. 1996;319:81–9. doi: 10.1042/bj3190081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo SHG, Falciani F, Al-Rubeai M. A genome-wide transcriptional analysis of producer and non-producer NS0 myeloma cell lines. Biotechnol Appl Biochem. 2007;47:85–95. doi: 10.1042/BA20060185. [DOI] [PubMed] [Google Scholar]
- Kim AH, Khursigara G, Sun X, Franke TF, Chao MV. Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol Cell Biol. 2001;21:893–901. doi: 10.1128/MCB.21.3.893-901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. Two regulatory steps of er-stress sensor ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. Genetic evidence for a role of bip/kar2 that regulates ire1 in response to accumulation of unfolded proteins. Mol Biol Cell. 2003;14:2559–69. doi: 10.1091/mbc.E02-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast kar2 (bip) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–90. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokame K, Kato H, Miyata T. Identification of erse-ii, a new cis-acting element responsible for the atf6-dependent mammalian unfolded protein response. J Biol Chem. 2001;276:9199–205. doi: 10.1074/jbc.M010486200. [DOI] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–25. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase perk and phosphorylation of the translation initiation factor eif2alpha. Mol Cell Biol. 2002;22:7405–16. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C, Wouters BG. “translating” tumor hypoxia: unfolded protein response (upr)-dependent and upr-independent pathways. Mol Cancer Res. 2006;4:423–36. doi: 10.1158/1541-7786.MCR-06-0150. [DOI] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. Er stress (perk/eif2alpha phosphorylation) mediates the polyglutamine-induced lc3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–9. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–97. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- Kudo T, Kanemoto S, Hara H, Morimoto N, Morihara T, Kimura R, Tabira T, Imaizumi K, Takeda M. A molecular chaperone inducer protects neurons from er stress. Cell Death Differ. 2008;15:364–75. doi: 10.1038/sj.cdd.4402276. [DOI] [PubMed] [Google Scholar]
- Kumar R, Azam S, Sullivan JM, Owen C, Cavener DR, Zhang P, Ron D, Harding HP, Chen JJ, Han A, White BC, Krause GS, DeGracia DJ. Brain ischemia and reperfusion activates the eukaryotic initiation factor 2alpha kinase, perk. J Neurochem. 2001;77:1418–21. doi: 10.1046/j.1471-4159.2001.00387.x. [DOI] [PubMed] [Google Scholar]
- LaFerla F. Calcium dyshomeostasis and intracellular signalling in Alzheimer's disease. Nat Rev Neurosci. 2002;3:862–72. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100:9946–51. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor xbp1. Science. 2008;320:1492–6. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. Ire1-mediated unconventional mrna splicing and s2p-mediated atf6 cleavage merge to regulate xbp1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–66. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TG, Tomita J, Hovanessian AG, Katze MG. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of mr 68,000 from influenza virus-infected cells. Proc Natl Acad Sci U S A. 1990;87:6208–12. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Davis RJ. Jnk phosphorylation of bim-related members of the bcl2 family induces bax-dependent apoptosis. Proc Natl Acad Sci U S A. 2003;100:2432–7. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Wootz H, Korhonen L. ER stress and neurogenerative diseases. Cell Death Differ. 2006;13:385–92. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, Reed JC, Glimcher LH, Hetz C. Bax inhibitor-1 is a negative regulator of the er stress sensor ire1alpha. Mol Cell. 2009;33:679–91. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Wong HN, Schauerte JA, Kaufman RJ. The protein kinase/endoribonuclease ire1alpha that signals the unfolded protein response has a luminal n-terminal ligand-independent dimerization domain. J Biol Chem. 2002;277:18346–56. doi: 10.1074/jbc.M112454200. [DOI] [PubMed] [Google Scholar]
- Liu CY, Xu Z, Kaufman RJ. Structure and intermolecular interactions of the luminal dimerization domain of human ire1alpha. J Biol Chem. 2003;278:17680–7. doi: 10.1074/jbc.M300418200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem. 1999;274:5861–7. doi: 10.1074/jbc.274.9.5861. [DOI] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, He Y, Zhang H, Yu L, Chen H, Xu Z, Tang S, Urano F, Min W. Aip1 is critical in transducing ire1-mediated endoplasmic reticulum stress response. J Biol Chem. 2008;283:11905–12. doi: 10.1074/jbc.M710557200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278:34864–73. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- Maattanen P, Gehring K, Bergeron JJM, Thomas DY. Protein quality control in the ER: The recognition of misfolded proteins. Semin Cell Dev Biol. 2010;21:500–11. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–31. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu MG, Doyle M, Bertolotti A, Ron D, Hendershot L, Neckers L. Heat shock protein 90 modulates the unfolded protein response by stabilizing ire1alpha. Mol Cell Biol. 2002;22:8506–13. doi: 10.1128/MCB.22.24.8506-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markan S, Kohli HS, Joshi K, Minz RW, Sud K, Ahuja M, Anand S, Khullar M. Up regulation of the grp-78 and gadd-153 and down regulation of bcl-2 proteins in primary glomerular diseases: a possible involvement of the er stress pathway in glomerulonephritis. Mol Cell Biochem. 2009;324:131–8. doi: 10.1007/s11010-008-9991-2. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Mothes W, Rapoport TA. Protein translocation: tunnel vision. Cell. 1998;92:381–90. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Minami M, Takeda K, Sakao Y, Akira S. Ectopic expression of chop (gadd153) induces apoptosis in m1 myeloblastic leukemia cells. FEBS Lett. 1996;395:143–7. doi: 10.1016/0014-5793(96)01016-2. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Apoptosis in neurodegenerative disorders. Nat Rev Mol Cell Biol. 2000;1:120–29. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- Maytin EV, Ubeda M, Lin JC, Habener JF. Stress-inducible transcription factor chop/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms. Exp Cell Res. 2001;267:193–204. doi: 10.1006/excr.2001.5248. [DOI] [PubMed] [Google Scholar]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–59. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Signalling and autophagy regulation in health Aging and disease. Mol Aspects Med. 2006;27:411–25. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Milhavet O, Martindale JL, Camandola S, Chan SL, Gary DS, Cheng A, Holbrook NJ, Mattson MP. Involvement of gadd153 in the pathogenic action of presenilin-1 mutations. J Neurochem. 2002;83:673–81. doi: 10.1046/j.1471-4159.2002.01165.x. [DOI] [PubMed] [Google Scholar]
- Min J, Shukla H, Kozono H, Bronson SK, Weissman SM, Chaplin DD. A novel creb family gene telomeric of hla-dra in the hla complex. Genomics. 1995;30:149–56. doi: 10.1006/geno.1995.9891. [DOI] [PubMed] [Google Scholar]
- Mohan C, Kim YG, Koo J, Lee GM. Assessment of cell engineering strategies for improved therapeutic protein production in CHO cells. Biotechnol J. 2008;5:624–30. doi: 10.1002/biot.200700249. [DOI] [PubMed] [Google Scholar]
- Morita K, Saitoh M, Tobiume K, Matsuura H, Enomoto S, Nishitoh H, Ichijo H. Negative feedback regulation of ask1 by protein phosphatase 5 (pp5) in response to oxidative stress. EMBO J. 2001;20:6028–36. doi: 10.1093/emboj/20.21.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem. 1998;273:2926–30. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- Naidoo N. Er and aging-protein folding and the er stress response. Ageing Res Rev. 2009;8:150–9. doi: 10.1016/j.arr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Caspase12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- Nawrocki ST, Carew JS, Dunner K, Jr, Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ. Bortezomib inhibits pkr-like endoplasmic reticulum (er) kinase and induces apoptosis via er stress in human pancreatic cancer cells. Cancer Res. 2005;65:11510–9. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- Nechushtan A, Smith CL, Hsu YT, Youle RJ. Conformation of the bax c-terminus regulates subcellular location and cell death. EMBO J. 1999;18:2330–41. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert K, Meister S, Moser K, Weisel F, Maseda D, Amann K, Wiethe C, Winkler TH, Kalden JR, Manz RA, Voll RE. The proteasome inhibitor bortezomib depletes plasma cells and protects mice with lupus-like disease from nephritis. Nat Med. 2008;14:748–55. doi: 10.1038/nm1763. [DOI] [PubMed] [Google Scholar]
- Nguyen DT, Kebache S, Fazel A, Wong HN, Jenna S, EMadali A, Lee EH, Bergeron JJM, Kaufman RJ, Larose L, Chevet E. Nck-dependent activation of extracellular signal-regulated kinase-1 and regulation of cell survival during endoplasmic reticulum stress. Mol Biol Cell. 2004;15:4248–60. doi: 10.1091/mbc.E03-11-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Zhou H, Wey S, Baumeister P, Lee AS. Regulation of perk signaling and leukemic cell survival by a novel cytosolic isoform of the upr regulator grp78/bip. PLoS One. 2009;4:e6868. doi: 10.1371/journal.pone.0006868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotera P, Orrenius S. The role of calcium in apoptosis. Cell Calcium. 1998;23:173–80. doi: 10.1016/s0143-4160(98)90116-6. [DOI] [PubMed] [Google Scholar]
- Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, Fukutomi H, Noguchi T, Matsuzawa A, Takeda K, Ichijo H. Als-linked mutant sod1 induces er stress-and ask1-dependent motor neuron death by targeting derlin-1. Genes Dev. 2008;22:1451–64. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, Miyazono K, Ichijo H. Ask1 is essential for jnk/sapk activation by traf2. Mol Cell. 1998;2:389–95. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- Niwa M, Sidrauski C, Kaufman RJ, Walter P. A role for presenilin-1 in nuclear accumulation of ire1 fragments and induction of the mammalian unfolded protein response. Cell. 1999;99:691–702. doi: 10.1016/s0092-8674(00)81667-0. [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by gadd34-mediated dephosphorylation of eif2alpha. J Cell Biol. 2001;153:1011–22. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor S, Li E, Majors BS, He L, Placone J, Baycin D, Betenbaugh MJ, Hristova K. Increased expression of the integral membrane protein ErbB2 in Chinese hamster ovary cells expressing the anti-apoptotic gene Bcl-Xl. Protein Expres Pur. 2009;67:41–47. doi: 10.1016/j.pep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino Si, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Ki, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, Yutani C, Ozawa K, Ogawa S, Tomoike H, Hori M, Kitakaze M. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–12. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K. Dissociation of kar2p/bip from an er sensory molecule, ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun. 2000;279:445–50. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- Olivari S, Galli C, Alanen H, Ruddock L, Molinari M. A novel stress-induced edem variant regulating endoplasmic reticulum-associated glycoprotein degradation. J Biol Chem. 2005;280:2424–8. doi: 10.1074/jbc.C400534200. [DOI] [PubMed] [Google Scholar]
- Oliver JD, Roderick HL, Llewellyn DH, High S. Erp57 functions as a subunit of specific complexes formed with the er lectins calreticulin and calnexin. Mol Biol Cell. 1999;10:2573–82. doi: 10.1091/mbc.10.8.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, van der Wal FJ, Bulleid NJ, High S. Interaction of the thiol-dependent reductase erp57 with nascent glycoproteins. Science. 1997;275:86–8. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Targeted disruption of the chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109:525–32. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, Wada I, Akira S, Araki E, Mori M. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci U S A. 2001;98:10845–50. doi: 10.1073/pnas.191207498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, Harding HP, Goodman AG, Harant H, Garrison JL, Taunton J, Katze MG, Ron D. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–39. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–61. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Pan YX, Lin L, Ren AJ, Pan XJ, Chen H, Tang CS, Yuan WJ. Hsp70 and grp78 induced by endothelin-1 pretreatment enhance tolerance to hypoxia in cultured neonatal rat cardiomyocytes. J Cardiovasc Pharmacol. 2004;44:S117–20. doi: 10.1097/01.fjc.0000166234.11336.a9. [DOI] [PubMed] [Google Scholar]
- Papa FR, Zhang C, Shokat K, Walter P. Bypassing a kinase activity with an atp-competitive drug. Science. 2003;302:1533–7. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- Park HR, Tomida A, Sato S, Tsukumo Y, Yun J, Yamori T, Hayakawa Y, Tsuruo T, Shin-ya K. Effect on tumor cells of blocking survival response to glucose deprivation. J Natl Cancer Inst. 2004;96:1300–10. doi: 10.1093/jnci/djh243. [DOI] [PubMed] [Google Scholar]
- Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- Paschen W, Gissel C, Linden T, Althausen S, Doutheil J. Activation of gadd153 expression through transient cerebral ischemia: evidence that ischemia causes endoplasmic reticulum dysfunction. Brain Res Mol Brain Res. 1998;60:115–22. doi: 10.1016/s0169-328x(98)00180-6. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with p58/dnajc3. EMBO J. 2008;27:2862–72. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli L, O'Farrell C, Lockhart PJ, Baptista M, Kehoe K, Vink L, Choi P, Wolozin B, Farrer M, Hardy J, Cookson MR. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36:1007–19. doi: 10.1016/s0896-6273(02)01125-x. [DOI] [PubMed] [Google Scholar]
- Pittman DD, Tomkinson KN, Kaufman RJ. Post-translational requirements for functional factor v and factor viii secretion in mammalian cells. J Biol Chem. 1994;269:17329–37. [PubMed] [Google Scholar]
- Putcha GV, Le S, Frank S, Besirli CG, Clark K, Chu B, Alix S, Youle RJ, LaMarche A, Maroney AC, Johnson EM., Jr Jnk-mediated bim phosphorylation potentiates bax-dependent apoptosis. Neuron. 2003;38:899–914. doi: 10.1016/s0896-6273(03)00355-6. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. The proapoptotic activity of the bcl-2 family member bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–96. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A. Er stress triggers apoptosis by activating bh3-only protein bim. Cell. 2007;129:1337–49. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Pyrko P, Kardosh A, Wang W, Xiong W, Schonthal AH, Chen TC. Hiv-1 protease inhibitors nelfinavir and atazanavir induce malignant glioma death by triggering endoplasmic reticulum stress. Cancer Res. 2007;67:10920–8. doi: 10.1158/0008-5472.CAN-07-0796. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Xu Y, Brenner MB. Retention of unassembled components of integral membrane proteins by calnexin. Science. 1994;263:387–90. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of bip and activation of rna-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 2006;66:1702–11. doi: 10.1158/0008-5472.CAN-05-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RV, Bredesen DE. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol. 2004;16:653–62. doi: 10.1016/j.ceb.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JF, Baltzis D, Wang S, Mounir Z, Papadakis AI, Gao HQ, Koromilas AE. Pkr and pkr-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin d1 via a mechanism requiring eukaryotic initiation factor 2alpha phosphorylation. J Biol Chem. 2008;283:3097–108. doi: 10.1074/jbc.M709677200. [DOI] [PubMed] [Google Scholar]
- Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein grp78 protects cells from apoptosis induced by topoisomerase inhibitors: role of atp binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–24. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- Reijonen S, Putkonen N, Nørremølle A, Lindholm D, Korhonen L. Inhibition of endoplasmic reticulum stress counteracts neuronal cell death and protein aggregation caused by n-terminal mutant huntingtin proteins. Exp Cell Res. 2008;314:950–60. doi: 10.1016/j.yexcr.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. Nf-kappab links innate immunity to the hypoxic response through transcriptional regulation of hif-1alpha. Nature. 2008;453:807–11. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, Mori K, Glimcher LH, Denko NC, Giaccia AJ, Le QT, Koong AC. Xbp1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–7. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- Ron D. Translational control in the endoplasmic reticulum stress response. J Clin Invest. 2002;110:1383–8. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Habener JF. Chop, a novel developmentally regulated nuclear protein that dimerizes with transcription factors c/ebp and lap and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–53. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ryu EJ, Harding HP, Angelastro JM, Vitolo OV, Ron D, Greene LA. Endoplasmic reticulum stress and the unfolded protein response in cellular models of parkinson's disease. J Neurosci. 2002;22:10690–8. doi: 10.1523/JNEUROSCI.22-24-10690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M, Mathison JC, Wolinski MK, Bensinger SJ, Fitzgerald P, Droin N, Ulevitch RJ, Green DR, Nicholson DW. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature. 2006;440:1064–8. doi: 10.1038/nature04656. [DOI] [PubMed] [Google Scholar]
- Sanchez IE, Morillas M, Zobeley E, Kiefhaber T, Glockshuber R. Fast folding of the two-domain semliki forest virus capsid protein explains co-translational proteolytic activity. J Mol Biol. 2004;338:159–67. doi: 10.1016/j.jmb.2004.02.037. [DOI] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–76. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Schnell S. A model of the unfolded protein response: pancreatic beta-cell as a case study. Cell Physiol Biochem. 2009;23:233–44. doi: 10.1159/000218170. [DOI] [PubMed] [Google Scholar]
- Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. The structure of calnexin, an er chaperone involved in quality control of protein folding. Mol Cell. 2001;8:633–44. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–66. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Seth G, Charaniya S, Wlaschin KF, Hu WS. In pursuit of a super producer--alternative paths to high producing recombinant mammalian cells. Curr Opin Biotechnol. 2007;6:557–64. doi: 10.1016/j.copbio.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–39. [PMC free article] [PubMed] [Google Scholar]
- Shao L, Sun X, Xu L, Young LT, Wang JF. Mood stabilizing drug lithium increases expression of endoplasmic reticulum stress proteins in primary cultured rat cerebral cortical cells. Life Sci. 2006;78:1317–23. doi: 10.1016/j.lfs.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. Er stress regulation of atf6 localization by dissociation of bip/grp78 binding and unmasking of golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shen J, Prywes R. Dependence of site-2 protease cleavage of atf6 on prior site-1 protease digestion is determined by the size of the luminal domain of atf6. J Biol Chem. 2004;279:43046–51. doi: 10.1074/jbc.M408466200. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for c. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shi Y, An J, Liang J, Hayes SE, Sandusky GE, Stramm LE, Yang NN. Characterization of a mutant pancreatic eif-2alpha kinase, pek, and co-localization with somatostatin in islet delta cells. J Biol Chem. 1999;274:5723–30. doi: 10.1074/jbc.274.9.5723. [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, pek, involved in translational control. Mol Cell Biol. 1998;18:7499–509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani-Ishida K, Nakajima M, Uemura K, Yoshida Ki. Ischemic preconditioning protects cardiomyocytes against ischemic injury by inducing grp78. Biochem Biophys Res Commun. 2006;345:1600–5. doi: 10.1016/j.bbrc.2006.05.077. [DOI] [PubMed] [Google Scholar]
- Shuda M, Kondoh N, Imazeki N, Tanaka K, Okada T, Mori K, Hada A, Arai M, Wakatsuki T, Matsubara O, Yamamoto N, Yamamoto M. Activation of the atf6, xbp1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the er stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–14. doi: 10.1016/s0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase ire1p is a site-specific endonuclease that initiates mrna splicing in the unfolded protein response. Cell. 1997;90:1031–9. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- Sousa M, Parodi AJ. The molecular basis for the recognition of misfolded glycoproteins by the udp-glc:glycoprotein glucosyltransferase. EMBO J. 1995;14:4196–203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–5. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajiri S, Oyadomari S, Yano S, Morioka M, Gotoh T, Hamada JI, Ushio Y, Mori M. Ischemia-induced neuronal cell death is mediated by the endoplasmic reticulum stress pathway involving chop. Cell Death Differ. 2004;11:403–15. doi: 10.1038/sj.cdd.4401365. [DOI] [PubMed] [Google Scholar]
- Tak PP, Firestein GS. Nf-kappab: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Imai Y, Hattori N, Mizuno Y. Parkin and endoplasmic reticulum stress. Ann N Y Acad Sci. 2003;991:101–6. doi: 10.1111/j.1749-6632.2003.tb07467.x. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Uehara T, Nomura Y. Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J Biol Chem. 2000;275:10388–93. doi: 10.1074/jbc.275.14.10388. [DOI] [PubMed] [Google Scholar]
- Terro F, Czech C, Esclaire F, Elyaman W, Yardin C, Baclet MC, Touchet N, Tremp G, Pradier L, Hugon J. Neurons overexpressing mutant presenilin-1 are more sensitive to apoptosis induced by endoplasmic reticulum-golgi stress. J Neurosci Res. 2002;69:530–9. doi: 10.1002/jnr.10312. [DOI] [PubMed] [Google Scholar]
- Thomenius MJ, Distelhorst CW. Bcl-2 on the endoplasmic reticulum: protecting the mitochondria from a distance. J Cell Sci. 2003;116:4493–9. doi: 10.1242/jcs.00829. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Marcinko M, Belmont PJ, Glembotski CC. Effects of the isoformspecific characteristics of atf6 alpha and atf6 beta on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem. 2007;282:22865–78. doi: 10.1074/jbc.M701213200. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC. Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res. 2006;99:275–82. doi: 10.1161/01.RES.0000233317.70421.03. [DOI] [PubMed] [Google Scholar]
- Tigges M, Fussenegger M. Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Metab Eng. 2006;3:264–72. doi: 10.1016/j.ymben.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (ire1p) in mammalian cells. Genes Dev. 1998;12:1812–24. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. Rapid turnover of unspliced xbp-1 as a factor that modulates the unfolded protein response. J Biol Chem. 2006;281:5852–60. doi: 10.1074/jbc.M509061200. [DOI] [PubMed] [Google Scholar]
- Tobiume K, Saitoh M, Ichijo H. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J Cell Physiol. 2002;191:95–104. doi: 10.1002/jcp.10080. [DOI] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–74. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–76. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–55. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Tyson JR, Stirling CJ. Lhs1 and sil1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–52. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterberger U, Hoftberger R, Gelpi E, Flicker H, Budka H, Voigtlander T. Endoplasmic reticulum stress features are prominent in alzheimer disease but not in prion diseases in vivo. J Neuropathol Exp Neurol. 2006;65:348–57. doi: 10.1097/01.jnen.0000218445.30535.6f. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the er to activation of jnk protein kinases by transmembrane protein kinase ire1. Science. 2000;287:664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- van Huizen R, Martindale JL, Gorospe M, Holbrook NJ. P58ipk, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eif2alpha signaling. J Biol Chem. 2003;278:15558–64. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A. Physiology and pathophysiology of the calcium store in the endoplasmic reticulum of neurons. Physiol Rev. 2005;85:201–79. doi: 10.1152/physrev.00004.2004. [DOI] [PubMed] [Google Scholar]
- Vitadello M, Penzo D, Petronilli V, Michieli G, Gomirato S, Menabo R, Di Lisa F, Gorza L. Overexpression of the stress protein grp94 reduces cardiomyocyte necrosis due to calcium overload and simulated ischemia. FASEB J. 2003;17:923–5. doi: 10.1096/fj.02-0644fje. [DOI] [PubMed] [Google Scholar]
- Wang HG, Pathan N, Ethell IM, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke TF, Reed JC. Ca2+-induced apoptosis through calcineurin dephosphorylation of bad. Science. 1999;284:339–43. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian ire1 reveals diversity in the er stress responses. EMBO J. 1998;17:5708–17. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Ron D. Stress-induced phosphorylation and activation of the transcription factor chop (gadd153) by p38 map kinase. Science. 1996;272:1347–9. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of atf6 and an atf6 dna binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–20. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Ware FE, Vassilakos A, Peterson PA, Jackson MR, Lehrman MA, Williams DB. The molecular chaperone calnexin binds glc1man9glcnac2 oligosaccharide as an initial step in recognizing unfolded glycoproteins. J Biol Chem. 1995;270:4697–704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, Mac-Gregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic bax and bak: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. Jnk1-mediated phosphorylation of bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94:277–80. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ. The unfolded protein response pathway in saccharomyces cerevisiae. oligomerization and trans-phosphorylation of ire1p (ern1p) are required for kinase activation. J Biol Chem. 1996;271:18181–7. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- Wilquet V, De Strooper B. Amyloid-beta precursor protein processing in neurodegeneration. Curr Opin Neurobiol. 2004;14:582–84. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GDY, Kaufman RJ. Atf6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Wurn FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;11:1393–98. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–46. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- Yamada T, Ishihara H, Tamura A, Takahashi R, Yamaguchi S, Takei D, Tokita A, Satake C, Tashiro F, Katagiri H, Aburatani H, Miyazaki Ji, Oka Y. Wfs1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic beta-cells. Hum Mol Genet. 2006;15:1600–9. doi: 10.1093/hmg/ddl081. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Ichijo H, Korsmeyer SJ. Bcl-2 is phosphorylated and inactivated by an ask1/jun n-terminal protein kinase pathway normally activated at g/m. Mol Cell Biol. 1999;19:8469–78. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian er quality control proteins is mediated by single or combined action of atf6alpha and xbp1. Dev Cell. 2007;13:365–76. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of perk eif2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone p58ipk. Proc Natl Acad Sci U S A. 2002;99:15920–5. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. Er stress induces cleavage of membrane-bound atf6 by the same proteases that process srebps. Mol Cell. 2000;6:1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. Activation of caspase-12, an endoplastic reticulum (er) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the er stress. J Biol Chem. 2001;276:13935–40. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. Er stress and diseases. FEBS J. 2007;274:630–58. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–9. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. Xbp1 mrna is induced by atf6 and spliced by ire1 in response to er stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. Atf6 activated by proteolysis binds in the presence of nf-y (cbf) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–67. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Oku M, Suzuki M, Mori K. pxbp1(u) encoded in xbp1 pre-mrna negatively regulates unfolded protein response activator pxbp1(s) in mammalian er stress response. J Cell Biol. 2006;172:565–75. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Uemura A, Mori K. pxbp1(u), a negative regulator of the unfolded protein response activator pxbp1(s), targets atf6 but not atf4 in proteasome-mediated degradation. Cell Struct Funct. 2009;34:1–10. doi: 10.1247/csf.06028. [DOI] [PubMed] [Google Scholar]
- Zhan Q, Lord KA, Alamo I, Jr, Hollander MC, Carrier F, Ron D, Kohn KW, Hoffman B, Liebermann DA, Fornace AJ., Jr The gadd and myd genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol Cell Biol. 1994;14:2361–71. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kawauchi J, Adachi MT, Hashimoto Y, Oshiro S, Aso T, Kitajima S. Activation of jnk and transcriptional repressor atf3/lrf1 through the ire1/traf2 pathway is implicated in human vascular endothelial cell death by homocysteine. Biochem Biophys Res Commun. 2001;289:718–24. doi: 10.1006/bbrc.2001.6044. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang R, Luo Y, D'Alessio A, Pober JS, Min W. Aip1/dab2ip, a novel member of the ras-gap family, transduces traf2-induced ask1-jnk activation. J Biol Chem. 2004;279:44955–65. doi: 10.1074/jbc.M407617200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–62. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of crebh to induce a systemic inflammatory response. Cell. 2006;124:587–99. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc Natl Acad Sci U S A. 1999;96:8511–8515. doi: 10.1073/pnas.96.15.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Tsutsui T, Ray D, Blomquist JF, Ichijo H, Ucker DS, Kiyokawa H. The cell cycle-regulatory cdc25a phosphatase inhibits apoptosis signal-regulating kinase 1. Mol Cell Biol. 2001;21:4818–28. doi: 10.1128/MCB.21.14.4818-4828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]