Abstract
The bacterial flagellum is a complex molecular machine that is made up of over forty proteins and is rotated to propel cells either through liquids or over solid surfaces. Flagellar gene expression is extensively regulated to coordinate flagellar assembly in both space and time. In Bacillus subtilis, the proteins of unknown function, SwrA and SwrB, and the alternative sigma factor σD are required to activate expression of the flagellar filament protein, flagellin. Here we determine that in the absence of SwrA and SwrB, the phosphorylated form of the response regulator DegU inhibits σD-dependent gene expression indirectly by binding to the PflgM promoter region and activating expression of the anti-sigma factor FlgM. We further demonstrate that DegU-P-dependent activation of FlgM is essential to inhibit flagellin expression when flagellar basal body assembly is disrupted. Regulation of FlgM is poorly understood outside of Salmonella, and differential control of FlgM expression may be a common means of coupling flagellin expression to flagellar assembly.
Keywords: SigD, bistability, motility, flagella, SwrA
INTRODUCTION
Bacteria rotate flagella to swim through liquid or swarm over solid surfaces. The flagellum is a complex structure composed of more than forty proteins and is assembled in three primary domains: the basal body, the hook, and the filament (Macnab, 2003). The cell first assembles the basal body in the membrane to serve as an anchor for the rest of the flagellum and act as the secretion conduit for the more distal flagellum components. Hook subunits are secreted through the basal body and assembled on the outside of the cell to form a flexible universal joint. Once the hook is assembled, thousands of subunits of flagellin are secreted through the basal body and the hook to be polymerized in a propeller-like helical filament. Bacteria use a complex hierarchy of gene regulation to control the synthesis and secretion of flagellar structural components and coordinate flagellum assembly (Chevance and Hughes, 2008).
The coupling of flagellar gene expression to the structure of the flagellum is well-studied in the Gram-negative bacterium Salmonella enterica serovar typhimurium. Flagellar genes are expressed as a short cascade of sigma factors in which RNA polymerase and the vegetative sigma factor σ70 direct expression of the basal body components and the alternative sigma factor σ28 (Arnosti and Chamberlin, 1989; Ohnishi et al., 1990). Once made, σ28 is held inactive by direct interaction with the anti-sigma factor FlgM until basal body assembly is completed, FlgM is secreted, and inhibition is relieved (Hughes et al., 1993; Katsukake, 1994; Karlinsey et al., 2000). Once activated, σ28 directs RNA polymerase to express the gene that encodes the flagellar filament subunit, flagellin (Ohnishi et al., 1990; Chen and Helmann, 1992). Thus, FlgM couples late flagellar gene expression directly to the completion status of flagellum synthesis such that flagellin is not expressed until it is ready to be secreted and assembled. The paradigm of FlgM control by secretion is assumed to be widespread but has not been proven to occur outside of the γ-proteobacteria (Correa et al., 2004; Rust et al., 2009).
The regulation of motility gene expression is poorly understood in the Gram-positive bacterium Bacillus subtilis. Many of the genes required for flagellar basal body assembly are co-expressed in the long 27 kb, 31-gene fla/che operon expressed primarily by RNA polymerase and the vegetative σ70 homolog, σA (Márquez-Magaña and Chamberlin, 1994; Estacio et al., 1998; West et al., 2000). The penultimate gene in the fla/che operon is sigD that encodes the σ28 homolog, σD (Helmann et al., 1988; Chen and Helmann, 1992). Once activated, σD directs the expression of the flagellar hook components, the flagellar motor components, the flagellar filament protein, and a suite of autolysins that promote cell separation after binary fission (Mirel and Chamberlin, 1989; Márquez et al., 1990; Serizawa et al., 2004). σD also directs the expression of the anti-sigma factor FlgM that binds to σD and inhibits σD-dependent gene expression (Caramori et al., 1996; Bertero et al., 1999; Kearns and Losick, 2005; Cozy and Kearns, 2010). FlgM activity is antagonized by the flagellar basal body but FlgM has not yet been demonstrated to be secreted by the flagellum in B. subtilis (Mirel et al., 1994; Caramori et al., 1996; Cozy and Kearns, 2010). The mechanism(s) that controls FlgM activity is unknown.
In addition to being antagonized by FlgM, σD is also activated by two proteins of unknown function, SwrA and SwrB. SwrA is a small cytoplasmic protein that activates the transcription of the fla/che operon that contains both the sigD and swrB genes (Kearns and Losick, 2005; Calvio et al., 2005). SwrB is a single-pass transmembrane protein that is required for maximal σD-dependent gene expression (Werhane et al., 2004; Kearns and Losick, 2005). Motility is heterogeneous in undomesticated wild-type cells such that a majority of cells express σD-dependent genes and a minority does not (Kearns and Losick, 2005; Chen et al., 2009). Mutation of either swrA or swrB increases the relative number of cells that fail to express σD-dependent genes. In cells simultaneously mutated for both swrA and swrB, however, σD-dependent genes are uniformly inactivated, neither flagellin nor autolysins are expressed, and the population grows exclusively as long sessile chains (Kearns and Losick, 2005). Thus SwrA and SwrB activate σD-dependent gene expression in individual cells and synergize at the population level.
Here we investigate the question of why σD-dependent gene expression is inactivated in a subpopulation of cells. To address this question, we use a strain doubly mutated for SwrA and SwrB to produce a uniform population of non-motile chaining cells that represent the subpopulation of non-motile cells normally found as a small minority in the wild type. We determine that mutation of the two-component system response regulator transcription factor DegU restores flagellin expression to a swrA swrB double mutant. The effect of DegU on flagellin is indirect, and we demonstrate that phosphorylated DegU directly activates transcription of the gene encoding the anti-sigma factor FlgM. Finally, we find that DegU-P dependent activation of FlgM is essential to inhibit flagellin expression when flagellar assembly is disrupted. Our results suggest that FlgM is regulated differently than other members of the σD regulon, that DegU becomes phosphorylated in the non-motile, chaining subpopulation, and that DegU may be part of a novel mechanism for coordinating flagellar assembly with flagellar gene expression.
RESULTS
Mutation of degU increases flagellin expression in a swrA swrB double mutant
The gene encoding flagellin (hag), is expressed from the Phag promoter by RNA polymerase and the alternative sigma factor σD (Mirel and Chamberlin, 1989). Colonies of wild-type cells that expressed a reporter (Phag-lacZ) in which the Phag promoter was transcriptionally fused to the gene encoding β-galactosidase (lacZ) at the ectopic amyE site, turned dark blue when grown on media containing the chromogenic β-galactosidase substrate X-gal (Fig 1A). When B. subtilis cells were mutated for both the swrA and swrB genes, Phag-lacZ gene expression was reduced 10-fold and colonies containing Phag- lacZ were pale blue on media containing X-gal (Fig. 1A). The reason why high level flagellin expression requires SwrA and SwrB is unknown.
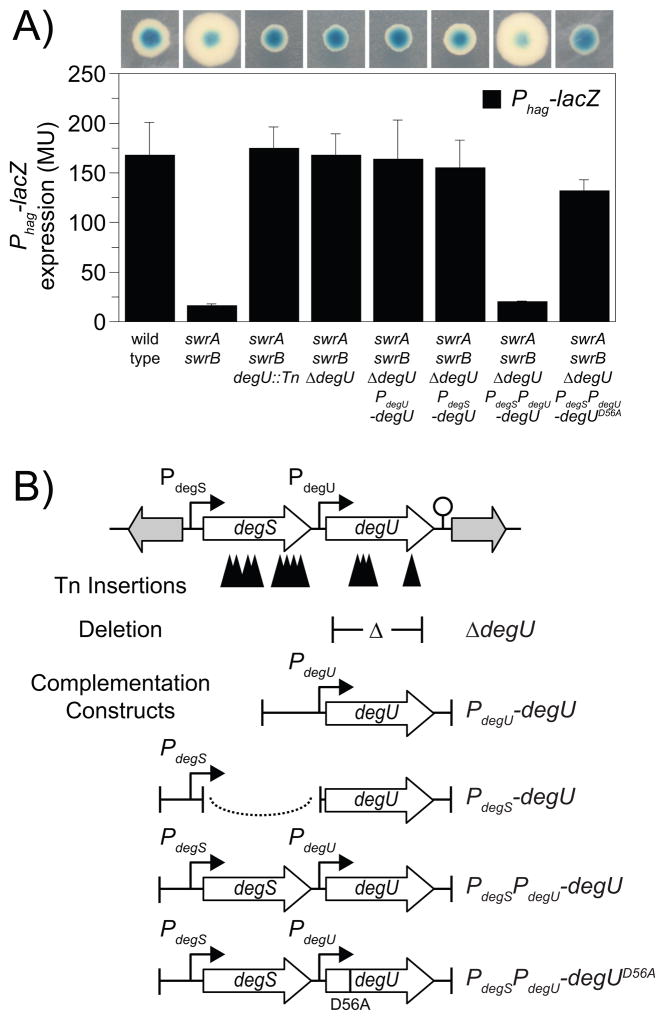
Figure 1. Mutations in degU increase expression from the Phag promoter in a swrA swrB double mutant background.
A) Phag-lacZ activity expressed either as blue/white colony color on media containing X-gal (top) or in Miller units (bottom). The indicated genotypes on the X-axis correspond both to the bars and to the colonies directly above. The following strains were used to generate this figure: wild type (DS793), swrA swrB (DS3789), swrA swrB degU::Tn (DS3442), swrA swrB ΔdegU (DS3794), swrA swrB ΔdegU (PdegU-degU) (DS5643), swrA swrB ΔdegU (PdegS-degU) (DS5502), swrA swrB ΔdegU (PdegSPdegU-degU) (DS5503), and swrA swrB ΔdegU (PdegSPdegU-degUD56A) (DS6563). Each bar is the average of 6 replicas and error bars are standard deviations. Raw data is found in Table S1. The basis for the increased colony size of swrA swrB mutants is unknown and was not studied here. B) Genetic map of the degS-degU operon. Open arrows represent open reading frames. Bent arrows represent promoters. Lollipop represents putative rho-independent terminator. Solid triangles represent the sites of transposon insertions that restored blue colony color to a swrA swrB Phag-lacZ genetic construct on media containing X-gal. Δ symbol within T-bars represent the boundaries of an in-frame markerless deletion mutation in degU. Dotted line represents the fusion of two genetic fragments.
We hypothesized that, in the absence of SwrA and SwrB, one or more proteins might inhibit flagellin expression. To screen for genes that inhibit flagellin expression, we generated a transposon insertion library in a swrA swrB null mutant containing the Phag-lacZ reporter, and we screened for colonies with enhanced blue color on media containing X-gal. Thirty thousand transposon mutant colonies were generated and seventeen transposon mutants with enhanced blue color were identified. To confirm that the enhanced blue color in each strain was due to a single transposon insertion, each transposon was backcrossed into the parental swrA swrB background by SPP1 phage-mediated generalized transduction. In each case, the transposon insertion was found to be genetically inseparable from the mutant phenotype.
Fourteen transposon insertions that enhanced blue colony color in the swrA swrB double mutant were located within a two gene operon (degS-degU), encoding the histidine kinase DegS and the cognate response regulator DegU (Msadek et al., 1990; Dahl et al., 1991) (Fig. 1; Table 1). We focused on mutations within degU because i) the insertions in degS were likely polar on downstream degU gene expression and ii) the DegU protein is phosphorylated by, and therefore biochemically downstream of, the DegS kinase. When degU was mutated in the swrA swrB double mutant parent, either by transposon insertion or by an in-frame markerless deletion, Phag-lacZ expression was increased 10-fold (Fig. 1). To determine whether mutation of degU was solely responsible for the increase in Phag expression we attempted to complement the degU gene using the native promoters from which it is expressed (Yasumura et al., 2008) (Fig 1B). Complementation constructs were generated in which the degU gene was cloned downstream of PdegS, PdegU, or PdegSPdegU together and introduced at the ectopic thrC locus. Only the PdegSPdegU-degU construct complemented the degU phenotype, and reduced the activity of Phag-lacZ in the swrA swrB degU triple mutant (Fig. 1A). We conclude that one reason that flagellin expression is reduced in the SwrA and SwrB double mutant background is that DegU either directly or indirectly inhibits the Phag promoter.
Table 1.
Transposon insertions that enhanced Phag-lacZ expression in a swrA swrB double mutant background.
| Gene | Encoded Function | Transposon insertion | Transposon insertion site |
|---|---|---|---|
| degS | histidine kinase | TnΩ3438 | TATTGTCAT |
| TnΩ3439 | TAATAAATG | ||
| TnΩ3440 | TAACATTCG | ||
| TnΩ3443 | TAATCCTTC | ||
| TnΩ3445 | TAATGATTC | ||
| TnΩ3448 | TATTTTCGA | ||
| TnΩ3449 | TAAATGTCC | ||
| TnΩ3450 | TATAATCCT | ||
| TnΩ3451 | TATATAATC | ||
| TnΩ3456 | TATTTTCTT | ||
| degU | response regulator | TnΩ3442 | TAATGGTCT |
| TnΩ3453 | TAATTACTT | ||
| TnΩ3454 | TAATGTATC | ||
| TnΩ3458 | TATCCATGA | ||
| yvyF | unknown function (transposon falls between PflgM and the DegU-P binding sites DegUsite1 and DegUsite2) | TnΩ2723 | TACTATGCC |
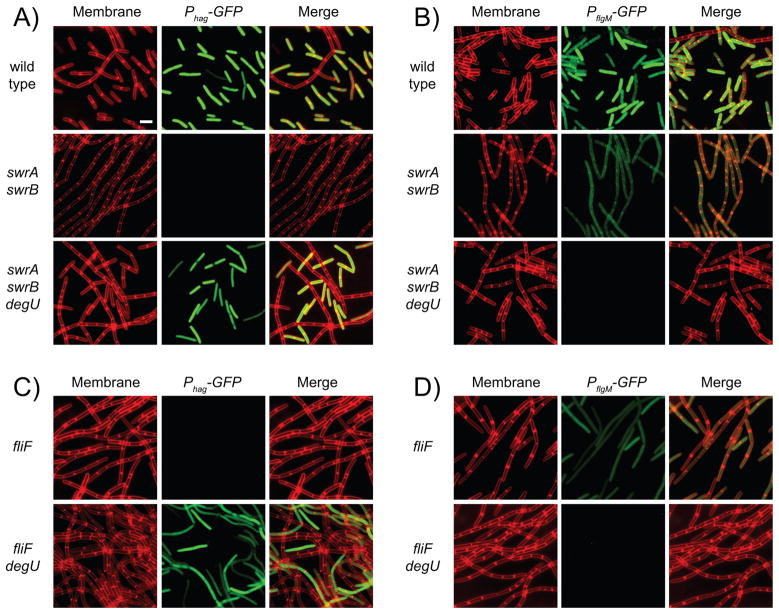
Expression from the Phag promoter is not homogenous and occurs in a subpopulation of wild-type cells (Kearns and Losick, 2005). To measure the effects of swrA, swrB, and degU on subpopulation level gene expression, expression from the Phag promoter was evaluated cytologically by fusing the promoter to the gene encoding green fluorescent protein (Phag-GFP). The majority of cells in the wild-type expressed the Phag- GFP reporter, whereas simultaneous mutation of SwrA and SwrB resulted in a population that was uniformly diminished for Phag-GFP expression (Kearns and Losick, 2005) (Fig. 2A). Mutation of degU restored Phag-GFP expression to a subpopulation of swrA swrB mutant cells (Fig. 2A). We conclude that DegU inhibits Phag-GFP in a subpopulation of cells mutated for SwrA and SwrB.
Figure 2. DegU inhibits Phag-GFP expression in a subpopulation of swrA swrB cells.
Fluorescent micrographs of cells expressing either Phag-GFP (A and C) or PflgM-GFP (B and D) in backgrounds of the indicated genotype. Membranes false colored red. GFP reporter expression false colored green. The following strains were used to generate this figure: wild type (DS908, DS7014), swrA swrB (DS4882, DS7015), swrA swrB ΔdegU (DS5533, DS7015), fliF (DS7223, DS7224) and fliF degU (DS7230, DS7231). Scale bar is 2 um.
DegU indirectly inhibits σD-dependent gene expression by activating flgM
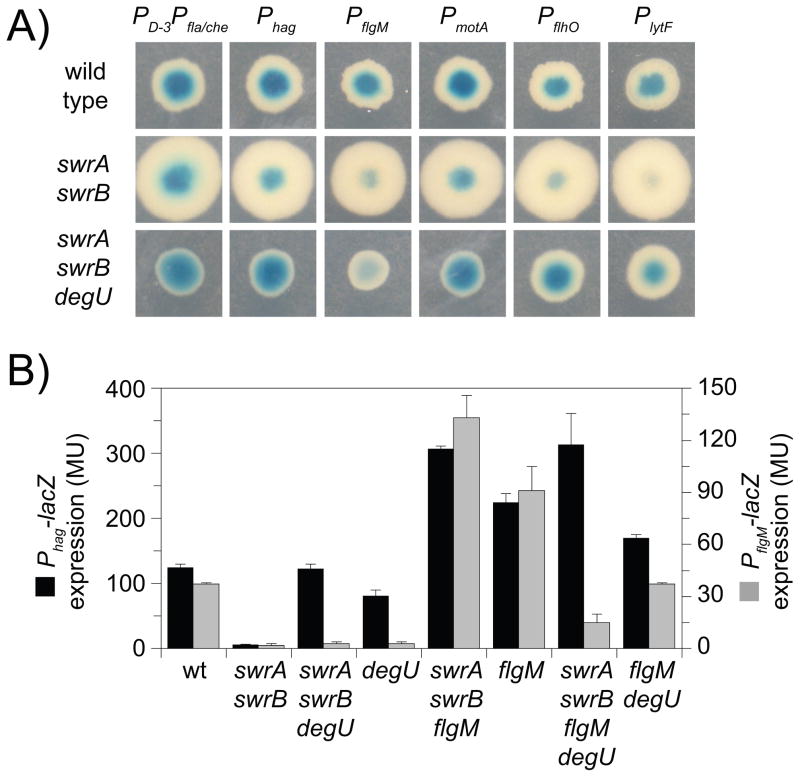
To determine whether DegU inhibited other σD-dependent genes besides flagellin, a series of σD-dependent reporters were generated in which lacZ was fused to the promoter regions of the genes encoding the anti-σD anti-sigma factor FlgM (PflgM), the flagellar motor components MotA and MotB (PmotA), the genes encoding the putative flagellar hook protein homologs FlhO and FlhP (PflhO), and the gene encoding the primary cell separating enzyme LytF (PlytF) (Mirel et al., 1992; Margot et al., 1999; Kearns and Losick, 2005; Chen et al., 2009). In addition, the lacZ gene was fused to the dual promoters of the fla/che operon (PD-3Pfla./che); the former PD-3 is a weak promoter transcribed by σD and the latter Pfla/che is a strong promoter transcribed by the vegetative σA (Estacio et al., 1998; West et al., 2000). Wild-type colonies containing the σD-dependent promoters were blue on media containing X-gal, and the blue color was reduced in colonies mutated for swrA and swrB (Fig. 3A). Mutation of degU enhanced blue colony color for all σD-dependent reporters except for PflgM in the swrA swrB background (Fig. 3A). We conclude that DegU is an inhibitor of σD-dependent gene expression, and we infer that flgM is regulated differently than other σD-dependent genes.
Figure 3. DegU activates expression of PflgM.
A) A grid of colonies of the genotype indicated vertically containing the promoters indicated horizontally fused to lacZ and grown on media containing X-Gal. Strains containing the following reporters were used to generate this panel: PD-3Pfla/che (DS791, DS3790, DS3795); Phag (DS793, DS3789, DS3794); PflgM (DS811, DS3792, DS3797); PmotA (DS1849, DS3793, DS3798); PflhO (DS5776, DS5778, DS5780); and PlytF (DS5775, DS5777, DS5779). B) Expression of Phag-lacZ (black bars) and PflgM-lacZ (gray bars) expressed as Miller units. The following strains were used to generate this figure: wild type (DS793, DS811), swrA swrB (DS3789, DS3792), swrA swrB degU (DS3794, DS3797), degU (DS4654, DS3658), swrA swrB flgM (DS6385, DS6386), flgM (DS4752, DS4754), and swrA swrB flgM degU (DS6408, DS6409). Columns are the average of six replicas and error bars are standard deviations. Raw data is found in Table S2.
To explore why the transcriptional level of flgM was not restored in the swrA swrB degU triple mutant background, we assayed and compared Phag-lacZ and PflgM-lacZ reporter activities. Mutation of swrA and swrB reduced activity of the Phag-lacZ and PflgM-lacZ reporters approximately 10-fold (Fig 3B). When degU was mutated in a swrA swrB null, the activity of the Phag-lacZ reporter was restored to wild-type levels but PflgM- lacZ expression remained low (Fig 3B). To further investigate the epistatic relationship of degU, we measured expression of Phag and PflgM in a degU single mutant background. Expression from Phag remained at near wild type levels, but expression from PflgM was dramatically reduced (Fig 3B). Thus, mutation of degU was epistatic to mutations in swrA and swrB for the Phag reporter and abolished expression of PflgM in otherwise wild type cells. We conclude that PflgM differs from other σD-dependent promoters because DegU activates PflgM expression.
To determine the consequence of DegU on gene expression at the single cell level, a strain was built that contained the PflgM promoter fused to the gene encoding green fluorescent protein (PflgM-GFP). Most cells of the wild-type exhibited strong expression of PflgM, but expression from PflgM was weak when SwrA and SwrB were mutated (Fig. 2B). Whereas the swrA swrB degU triple mutant had restored Phag expression, expression of PflgM was abolished in the same genetic background (Fig. 2B). We conclude that DegU activates the expression of PflgM and that the loss of PflgM expression was correlated with the rescue of Phag expression in the swrA swrB degU triple mutant.
Phosphorylated DegU binds two sites upstream of the PflgM promoter
One way in which DegU might activate PflgM expression is if DegU bound directly to the region upstream of the PflgM promoter. To explore the regulation of PflgM by DegU, we first carried out primer extension analysis to map the transcriptional start site of the flgM gene. The transcriptional start site, designated as position +1, was located 40 bp upstream of the flgM open reading frame (Fig. S1). Using the transcriptional start site we were able to identify sequences at positions −35 (CTAAA) and −10 (GTCGATAA) of PflgM similar to the consensus −35 box (CTAAA) and −10 box (GCCGATAT) of the σD-dependent promoter sequence (Gilman et al., 1981; Serizawa et al., 2004; Kearns and Losick, 2005) (Fig. 4). We conclude that we have identified the sequence of PflgM, a new σD-dependent promoter that drives expression of the flgM gene.
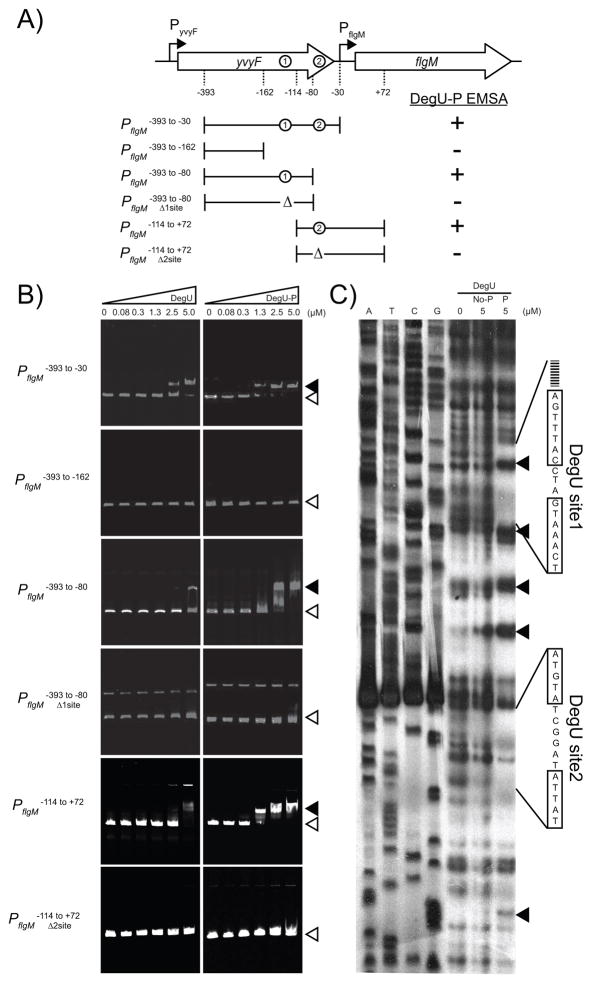
Figure 4. The PflgM promoter region.
The sequence is of the 3′ end of the yvyF gene and the 5′ end of the flgM gene from B. subtilis strain 3610 (gray arrows behind text). Underlined sequences indicate the predicted “−35” and “−10” promoter elements of the σD consensus sequence and the +1 transcriptional start predicted for the flgM gene in Fig S2. Boxes indicate inverted repeat sequences protected by DegU-P in the DNase footprint assay in Fig. 5B. Inverted triangle and bolded, capitalized sequence indicates the location of the TnΩ2723 transposon insertion that phenocopies a degU mutation and restores flagellin expression to cells mutated for SwrA and SwrB. Dashed lines indicate important positions relative to the electrophoretic mobility shift experiments in Fig. 5A.
To determine whether DegU bound to the region upstream of flgM, an electrophoretic mobility shift assay (EMSA) was conducted. When a DNA fragment from base pair −393 to −30 upstream of the flgM transcriptional start site was used as a target, DegU binding caused a mobility shift (Fig. 5A,B). Phosphorylation of DegU by DegS and ATP (Fig. S2) enhanced the mobility shift such that lower protein concentrations were required (Fig. 5B). DegU did not cause a mobility shift when a DNA fragment from base pair −393 to −162 upstream of the flgM transcriptional start site was used as the target (Fig. 5A,B). We conclude that DegU bound to a region upstream of flgM, and that DNA binding was enhanced when DegU was phosphorylated. We infer that there is one or more DegU binding sites in the region between bases −162 and −30 upstream of the flgM transcriptional start site.
Figure 5. DegU-P binds to the promoter region of PflgM.
A) A map of the PflgM promoter region. Large open arrows indicate open reading frames. Bent arrows indicate promoters. Dashed lines indicate important positions with respect to the EMSA experiments in panel B. Circles indicate the positions of DegUsite1 and DegUsite2. Brackets indicate the boundaries of the fragments used in the EMSA experiments in panel B. Δ indicates deletion of a DegU binding site. Results of the EMSA experiments in panel B are summarized as either + (indicating a the presence of a mobility shift) or − (indicating the absence of a mobility shift. B) Electrophoretic mobility shift experiments. The target DNA fragments are indicated vertically. The left hand series of panels includes an increasing concentration of DegU protein. The right hand series of panels includes an increasing concentration of DegU-P protein that was phosphorylated by DegS and ATP. Concentrations of DegU are listed across the top in μM. Open triangles indicate the position of of the unbound fragment; closed triangles indicate the position of the shifted fragment. C) DNase footprint protection experiment of PflgM promoter region. Left hand lanes are DNA sequencing lanes of the indicated base. Right hand lanes include increasing amounts of either 0 μM DegU, 5 μM DegU (No-P), or 5 μM DegU-P (P) phosphorylated by incubation with ATP and DegS. Lines indicate the boundaries of protection and the boxes indicate the sequences of the protected inverted repeats. Closed triangles indicate sites that became hypersensitive to DNase digestion. Corresponding regions of protection from DNase digestion were detected on the other DNA strand (data not shown).
To determine the site at which DegU bound to the PflgM promoter region, we conducted footprinting experiments in which PflgM was radiolabeled and mixed with various concentrations of either DegU or DegU that was phosphorylated by incubation with ATP and DegS. The protein-DNA complexes were then treated with DNase and the digestion products were resolved by electrophoresis and detected by autoradiography. Unphosphorylated DegU did not protect the PflgM promoter fragment from DNase (Fig. 5C). Phosphorylated DegU, however, protected two regions upstream of PflgM; one region corresponded to an inverted repeat “DegUsite1” that was similar to a DegU binding sequence predicted by Tsukuhara and Ogura, 2008, and the other region corresponded to an inverted repeat “DegUsite2” that was similar to a DegU binding sequence predicted by Hamoen et al., 2000. Furthermore, binding of DegU-P caused the appearance of digestion sites with enhanced intensity perhaps indicative of a conformational change in the DNA fragment (Fig 5C). We conclude that DegU-P protects two different regions upstream of PflgM from DNase digestion and that these regions correspond to two different inverted repeat sequences.
To determine the biochemical relevance of the two different putative DegU binding sites, two new EMSA DNA fragments were generated to separate the two sites. Phosphorlyated DegU bound to a fragment containing only DegUsite1 (base pairs −393 to −80) and bound to a fragment containing only DegUsite2 (base pairs −114 to +72) (Fig 5A, 5B). When DNA fragments were used in which each DegU binding site was deleted respectively, DegU-P no longer caused a mobility shift of the corresponding fragment. Thus, DegU-P binds to both DegUsite1 and DegUsite2 and either site was sufficient for DegU-P to bind to DNA and cause a mobility shift.
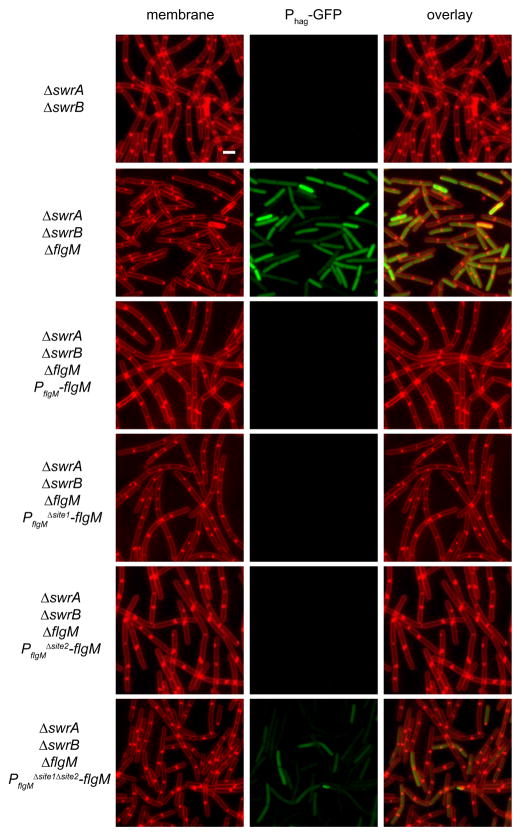
To determine the biological relevance of the two different DegU binding sites, we first generated a construct which could complement a flgM mutation in trans by inserting the flgM gene and 300 base pairs of upstream DNA at the ectopic amyE site (amyE::PflgM- flgM). A Phag-GFP reporter was used to detect flgM function (Fig. 6). In the absence of swrA and swrB cells failed to express the Phag-GFP, because FlgM antagonized σD-dependent gene expression. A swrA swrB flgM triple mutant was restored for Phag-GFP expression. Introduction of the PflgM-flgM complementation to the swrA swrB flgM triple mutant rescued FlgM activity as indicated by the inactivation of Phag-GFP. Whereas mutation of either DegUsite1 or DegUsite2 alone had no effect, simultaneous mutation of both DegUsite1 and DegUsite2 rendered flgM complementation incomplete as indicated by partial restoration of Phag-GFP expression (Fig. 6). We conclude that either DegUsite1 or DegUsite2 is sufficient to activate flgM expression and we infer that DegU-P binds to both sites to activate transcription in vivo.
Figure 6. Either of the DegU binding sites upstream of PflgM are sufficient to activate flgM expression.
Fluorescent micrographs of cells expressing Phag-GFP in backgrounds of the indicated genotype. Membranes were false-colored red. GFP reporter expression was false-colored green. The following strains were used to generate this figure: swrA swrB (DS8014), swrA swrB ΔflgM (DS7740), swrA swrB ΔflgM PflgM-flgM (DS7762), swrA swrB ΔflgM PflgMΔsite1-flgM (DS7763), swrA swrB ΔflgM PflgMΔsite2-flgM (DS8430), and swrA swrB ΔflgM PflgMΔsite1Δsite2-flgM (DS8662). Scale bar is 2 um.
Phosphorylated DegU activates flgM expression in response to flagellum completion status
DegU appears to inhibit σD-dependent gene expression, at least in part, by directly activating the expression of the FlgM anti-σD anti-sigma factor. If true, FlgM should be genetically downstream of DegU for regulating σD. FlgM was downstream of SwrA and SwrB as mutation of flgM increased the expression of the σD- dependent promoters Phag and PflgM in both the wild-type and the swrA swrB double mutant backgrounds (Fig 3B). Likewise, FlgM was downstream of DegU as the swrA swrB degU flgM quadruple mutant resembled mutation of flgM with respect to Phag expression (Fig. 3B). Conversely, DegU appeared to be downstream of FlgM with respect to PflgM expression as the swrA swrB degU flgM quadruple mutant resembled mutation of degU alone (Fig. 3B). Thus, despite the fact that PflgM is a σD-dependent promoter and that σD is uninhibited in the absence of FlgM, PflgM was unable to be fully activated in the absence of DegU in the swrA swrB degU flgM quadruple mutant background. Together, these results support the conclusion that in the absence of SwrA and SwrB, phosphorylated DegU activates FlgM expression, which leads to inhibition of σD-dependent genes.
FlgM inhibits σD-dependent gene expression when assembly of the flagellum is incomplete (Mirel et al., 1994; Barilla et al., 1994; Caramori et al., 1996; Bertero et al., 1999; Cozy and Kearns, 2010). Regulation of FlgM by DegU-P suggests that the signal that results in phosphorylation of DegU may be related to flagellar assembly. When flagellar basal body components, such as the membrane ring protein FliF, were mutated, the expression of Phag was abolished but expression from PflgM was not (Fig 2C,D). When degU was mutated in the fliF background, however, Phag expression was restored and PflgM expression was abolished (Fig 2C, D). We conclude that activation of PflgM by DegU-P is not specific to the swrA swrB double mutant background, and is also relevant when flagellar basal body assembly is abrogated. We infer that DegS and DegU may play an important role in sensing basal body completion and coordinating flagellar gene expression with flagellar assembly.
DISCUSSION
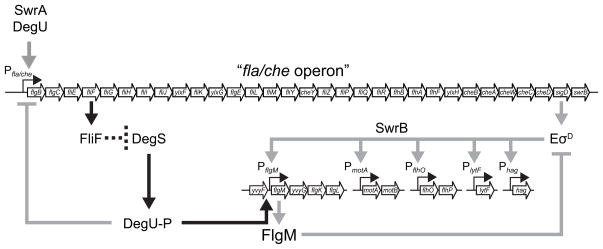
The expression of flagellin is heterogeneous in B. subtilis and requires two proteins of unknown function, SwrA and SwrB, and the assembly of the flagellar basal body. Here we identify an inhibitor of σD-dependent gene expression, the response regulator DegU, which when mutated, restores flagellin expression to cells lacking either SwrA and SwrB or lacking the basal body protein FliF. We discovered that the phosphorylated form of DegU inhibits σD indirectly by activating a newly characterized promoter, PflgM, that drives expression of the FlgM anti-sigma factor (Fig. 7). Our work not only adds a new target and a new role for DegU regulation, it also provides a new mechanism for controlling the activity of FlgM, an important protein in many bacteria that couples gene expression to the structural completion state of the flagellum.
Figure 7. DegU-P inhibits σD indirectly by activating the expression of the anti-sigma factor FlgM.
A model of motility gene regulation in B. subtilis. Open arrows indicate open reading frames. Bent thin arrows indicate promoters. Thick arrows indicate activation. Thick T-bars indicate repression. Heavy gray lines indicate previously published data; heavy black lines indicate the newly proposed model. The dashed line indicates the genetic inference that basal body completion, or the FliF basal body protein, antagonizes DegS by an unknown mechanism.
DegU activates the expression of flgM by binding DNA upstream of the PflgM promoter. Several different consensus sequences for DegU binding have been proposed and here we show that DegU binds to two different sites each supported by separate reports in the literature (Tsukuhara and Ogura, 2008; Hamoen et al., 2000). The relevance of the DegU binding sites is supported by the observation that one transposon insertion that restored flagellin expression to cells mutated for SwrA and SwrB, fell within the promoter proximal DegUsite2 inverted repeat (Table 1, Fig. 4). The insertion phenocopies disruption of degU itself, and we infer that the insertion activates flagellin expression indirectly by separating PflgM from both sites of DegU-dependent activation. The DegU binding site is over 80 base pairs away from PflgM and DegU-P may activate PflgM expression directly by DNA bending and interaction with RNA polymerase or indirectly by displacing an as yet unidentified negative regulator. Finally, although sequence analysis predicts only one helix-turn-helix binding domain, we wonder whether DegU has a second cryptic DNA binding domain to account for the two consensus binding sites. If so, two binding domains may account for the difficulty identifying a DegU binding consensus and resolve apparent disagreements in the literature.
DegU has been shown to regulate gene expression in either the phosphorylated or the unphosphorylated form. Phosphorylation of DegU seems to be important for activating the expression of flgM because: i) phosphorylation of DegU increased the affinity of DegU for the PflgM promoter region in EMSA and footprint analysis (Fig. 5), ii) mutation of the aspartate56 phosphorylation site to an alanine that cannot not be phosphorylated resulted in a DegU loss-of-function phenotype (Fig. 1, swrA swrB ΔdegU PdegSPdegU-degUD56A, Dahl et al., 1991), and iii) an in-frame deletion of degS, which encodes the cognate DegS histidine kinase of DegU, phenocopied an in-frame deletion of degU for restoring flagellin expression to cells mutated for SwrA and SwrB (data not shown). Phosphorylated DegU has been shown to activate a variety of gene products including the expression of extracellular proteases and gene products required for complex colony architecture (Mukai et al., 1990, Verhamme et al., 2007; Kobayashi, 2007). The signal that is detected by the cytoplasmic DegS kinase to trigger phosphorylation of DegU is unknown, but regulation of flgM by DegU-P suggests that the DegS-DegU two-component system may either directly or indirectly sense the completion state of the flagellum.
FlgM couples incomplete flagellar assembly to the inhibition of flagellin expression through binding to the alternative sigma factors σ28 and σD, in S. typhimurium and B. subtilis respectively (Ohnishi et al., 1992, Caramori et al., 1996; Bertero et al., 1999). FlgM is antagonized in S. typhimurium by completion of the flagellar basal body, through which FlgM is secreted leaving behind free sigma factor (Hughes et al., 1993). In B. subtilis, FlgM is also antagonized by flagellar basal body components, but FlgM has never been shown to be secreted (Mirel et al., 1994; Barilla et al., 1994; Caramori et al., 1996; Bertero et al., 1999; Cozy and Kearns, 2010). The FlgM proteins of B. subtilis and S. typhimurium differ in both primary and tertiary structure (Fig S3). Whereas the N-terminus of FlgM from S. typhimurium is disordered to promote secretion, the N-terminus of FlgM from B. subtilis is structured, perhaps suggesting an alternative mechanism of control (Daughdrill et al., 1997; Bertero et al., 1999). Alternatives to secretion-mediated control of FlgM may be common as FlgM has been shown to be regulated independently of secretion in the ε-proteobacterium, Helicobacter pylori (Josenhans et al., 2002; Rust et al., 2009). While the mechanism by which flagellar completion state is coupled to FlgM activity in B. subtilis remains poorly understood, here we demonstrate that at least part of FlgM control is transcriptional and regulated by a two-component signal transduction system.
DegU-P appears to act at multiple levels in the regulation of motility in B. subtilis. In addition to binding to the PflgM promoter region, DegU also binds to the promoter region of the fla/che operon. Our results that DegU-P activates an inhibitor of σD activity, flgM, synergize with the observation that DegU-P also binds to Pfla/che and inhibits sigD gene expression (Mader et al., 2002, Amati et al., 2004; Verhamme et al., 2007). Conversely, DegU in the unphosphorylated form has been shown to activate Pfla/che expression (Tokunaga, et al., 1994; Ogura et al., 2001; Tsukuhara and Ogura, 2008). We found that DegU binds to the fla/che promoter region in both the phosphorylated and unphosphorylated forms, but that mutation of DegU had very modest effects on PD-3Pfla/che expression (Fig. S4). We infer that the positive and negative regulation of the fla/che operon by DegU and DegU-P, respectively, neutralizes in the degU null mutant background. Finally, mutation of degU abolishes swarming motility, a social, flagella-mediated form of surface migration, for unknown reasons (Kearns and Losick, 2003; Verhamme et al., 2007; Kobayashi, 2007; Patrick and Kearns, 2009). Together, these observations suggest that the regulation of PflgM by DegU-P is one aspect of the complex role of DegU in controlling motility.
Studying the inactivation of σD is complicated in the undomesticated wild-type because σD-dependent gene expression is OFF in only a minority of cells. Recent work demonstrated that a subpopulation was OFF for flagellin expression because sigD expression and σD protein fell below a threshold level (Cozy and Kearns, 2010). In the absence of SwrA and SwrB, the expression of PflgM persisted despite low levels of σD (Fig 3; Cozy and Kearns, 2010). We therefore infer that DegU-P functions to lower the minimal threshold of σD required to activate the PflgM promoter specifically. The special requirement of phosphorylated DegU to activate flgM may permit sensory input on the system and help resolve the seemingly contradictory fact that σD is required for the expression of its own antagonist. The sensory input that controls DegU may be related to flagellar synthesis as DegU-P is also essential to activate FlgM expression when basal body assembly is abrogated. In sum, DegU phosphorylation promotes low level expression of flgM, inhibits residual σD activity, and stabilizes the OFF state of a subpopulation, perhaps in response to the completion state of the flagellum structure. Population heterogeneity in bacteria is becoming more commonly recognized (Smits et al., 2006), and our work suggests that other two-component systems may act similarly to reduce thresholds and buffer heterogeneous gene expression at specific promoters.
EXPERIMENTAL PROCEDURES
Strains and growth conditions
B. subtilis strains were grown in Luria-Bertani (LB) (10 g tryptone, 5 g yeast extract, 5 g NaCl per L) broth or on LB plates containing 1.5% Bacto agar at 37 °C. When appropriate, antibiotics were included at the following concentrations: 10 μg/ml tetracycline, 100 μg/ml spectinomycin, 5 μg/ml chloramphenicol, 5 μg/ml kanamycin, and 1 μg/ml erythromycin plus 25 μg/ml lincomycin (mls). Isopropyl β-D-thiogalactopyranoside (IPTG, Sigma) was added to the medium at the indicated concentration when appropriate.
Microscopy
Fluorescence microscopywas performed with a Nikon 80i microscope with a phase contrast objective Nikon Plan Apo 100X and an Excite 120 metal halide lamp. Dyes or fluorescent proteins were visualized using the following filter cubes: FM4-64 (C-FL HYQ Texas Red Filter Cube; excitation filter 532–587 nm, barrierfilter >590 nm) and GFP (C-FL HYQ FITC Filter Cube (FITC, excitation filter 460–500 nm, barrier filter 515–550 nm). Images were captured with a Photometrics Coolsnap HQ2 camera in black and white, false colored and superimposed using Metamorph image software.
For GFP microscopy, cells were grown overnight at 22 °C on LB agar plates and a single colony was selected and grown at 37 °C to OD600 0.6–1.0 LB broth. One ml of the culture was collected by centrifugation, and washed once in T-Base buffer (15 mM (NH4)2SO4, 80 mM K2HPO4, 44 mM KH2PO4, 3.4 mM sodium citrate, and 3.0 mM MgSO4·6H20). Following centrifugation, the pellet was resuspended in 50μl T-Base buffer containing 5 μg/ml FM 4–64 and incubated for 10 min at room temperature. Samples were observed by spotting 3 μl of suspension on a slideand immobilized with a poly-L-lysine-treated glass coverslip. Images were captured with Metamorph software.
Strain construction
All constructs were first introduced into the domesticated strain PY79 by natural competence and then transferred to the 3610 background using SPP1-mediated generalized phage transduction (Yasbin and Young, 1974). All strains used in this study are listed in Table 2. All plasmids used in this study are listed in Supplemental Table S3. All primers used in this study are listed in Supplemental Table S4.
Table 2.
Strainsa
| Strain | Genotype |
|---|---|
| 168 | sfp swrA trpC2 |
| 3610 | Wild type |
| PY79 | sfp swrA |
| DS791 | amyE::PD-3Pfla/che-lacZ cat (Kearns and Losick, 2005) |
| DS793 | amyE::Phag-lacZ cat (Kearns and Losick, 2005) |
| DS811 | amyE::PflgM-lacZ cat (Kearns and Losick, 2005) |
| DS908 | amyE::Phag-GFP cat (Kearns and Losick, 2005) |
| DS1849 | amyE::PmotA-lacZ cat |
| DS2655 | ΔswrA ΔswrB amyE::Phag-lacZ cat lacA::tet |
| DS2723 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat yvyF::TnYLB kan |
| DS3438 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3439 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3440 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3442 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degU::TnYLB kan |
| DS3443 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3445 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3448 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3449 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3450 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3453 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degU::TnYLB kan |
| DS3454 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degU::TnYLB kan |
| DS3451 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3456 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degS::TnYLB kan |
| DS3458 | ΔswrA ΔswrB lacA::tet amyE::Phag-lacZ cat degU::TnYLB kan |
| DS3654 | ΔdegU amyE::Phag-lacZ cat |
| DS3658 | ΔdegU amyE::PflgM-lacZ cat |
| DS3713 | ΔdegU amyE::PD-3Pfla/che-lacZ cat |
| DS3789 | ΔswrA ΔswrB amyE::Phag-lacZ cat |
| DS3790 | ΔswrA ΔswrB amyE::PD-3Pfla/che-lacZ cat |
| DS3792 | ΔswrA ΔswrB amyE::PflgM-lacZ cat |
| DS3793 | ΔswrA ΔswrB amyE::PmotA-lacZ cat |
| DS3794 | ΔswrA ΔswrB ΔdegU amyE::Phag-lacZ cat |
| DS3795 | ΔswrA ΔswrB ΔdegU amyE::PD-3Pfla/che-lacZ cat |
| DS3797 | ΔswrA ΔswrB ΔdegU amyE::PflgM-lacZ cat |
| DS3798 | ΔswrA ΔswrB ΔdegU amyE::PmotA-lacZ cat |
| DS4754 | ΔflgM amyE::PflgM-lacZ cat |
| DS4882 | ΔswrA ΔswrB amyE::Phag-GFP cat |
| DS5502 | ΔswrA ΔswrB ΔdegU amyE::Phag-lacZ cat thrC::PdegS-degU mls |
| DS5503 | ΔswrA ΔswrB ΔdegU amyE::Phag-lacZ cat thrC::PdegSPdegU-degU mls |
| DS5533 | ΔswrA ΔswrB ΔdegU amyE::Phag-GFP cat |
| DS5643 | ΔswrA ΔswrB ΔdegU amyE::Phag-lacZ cat thrC::PdegU-degU mls |
| DS5775 | amyE::PlytF-lacZ cat |
| DS5776 | amyE::PflhO-lacZ cat |
| DS5777 | ΔswrA ΔswrB amyE::PlytF-lacZ cat |
| DS5778 | ΔswrA ΔswrB amyE::PflhO-lacZ cat |
| DS5779 | ΔswrA ΔswrB ΔdegU amyE::PlytF-lacZ cat |
| DS5780 | ΔswrA ΔswrB ΔdegU amyE::PflhO-lacZ cat |
| DS6385 | ΔflgM ΔswrA ΔswrB amyE::Phag-lacZ cat |
| DS6386 | ΔflgM ΔswrA ΔswrB amyE::PflgM-lacZ cat |
| DS6408 | ΔflgM ΔswrA ΔswrB ΔdegU amyE::Phag-lacZ cat |
| DS6409 | ΔflgM ΔswrA ΔswrB ΔdegU amyE::PflgM-lacZ cat |
| DS6563 | ΔswrA ΔswrB ΔdegU amyE::Phag-lacZ cat thrC::PdegSPdegU-degUD56A mls |
| DS6859 | ΔdegU ΔflgM amyE::Phag-lacZ cat |
| DS6860 | ΔdegU ΔflgM amyE::PflgM-lacZ cat |
| DS7014 | amyE::PflgM-GFP spec |
| DS7015 | ΔswrA ΔswrB amyE::PflgM-GFP spec |
| DS7017 | ΔswrA ΔswrB ΔdegU amyE::PflgM-GFP spec |
| DS7198 | [168] thrC::PswrA-swrA mls amyE::PD-3P fla/che-lacZ cat |
| DS7202 | [168] degU::TnYLB kan thrC::PswrA-swrA mls amyE::PD-3Pfla/che-lacZ cat |
| DS7223 | ΔfliF amyE::Phag-GFP cat |
| DS7224 | ΔfliF amyE::PflgM-GFP spec |
| DS7230 | ΔfliF degU::TnYLB kan amyE::Phag-GFP cat |
| DS7231 | ΔfliF degU::TnYLB kan amyE::PflgM-GFP spec |
| DS7740 | ΔswrA ΔswrB thrC::Phag-GFP mls ΔflgM |
| DS7762 | ΔswrA ΔswrB thrC::Phag-GFP mls ΔflgM amyE::PflgM-flgM cat |
| DS7763 | ΔswrA ΔswrB thrC::Phag-GFP mls ΔflgM amyE::PflgMΔsite1-flgM cat |
| DS8014 | ΔswrA ΔswrB thrC::Phag-GFP mls |
| DS8430 | ΔswrA ΔswrB thrC::Phag-GFP mls ΔflgM amyE::PflgMΔsite2-flgM cat |
| DS8662 | ΔswrA ΔswrB thrC::Phag-GFP mls ΔflgM amyE::PflgMΔsite1Δsite2-flgM cat |
All strains are in the 3610 genetic background unless an alternative parent is indicated in brackets.
In-frame deletions
To generate the ΔdegU in frame marker-less deletion construct, the region upstream of degU was amplified using the primer pair 1109/1110, and digested with KpnI and BamHI. The region downstream of degU was PCR amplified respectively using the primer pairs 1111/1112 and digested with BamHI and SalI. The two fragments were then simultaneously ligated respectively into the KpnI/SalI sites of pMiniMAD which carries a temperature sensitive origin of replication and an erythromycin resistance cassette to generate pJH8 (Patrick and Kearns, 2008). The plasmid was introduced to PY79 by single cross-over integration by transformation at the restrictive temperature for plasmid replication (37°C) using mls resistance as a selection. The integrated plasmid was then transduced into DS3610. To evict the plasmid, the strain was incubated in 3ml LB broth at a permissive temperature for plasmid replication (22 °C) for 14 hours, diluted 30-fold in fresh LB broth, and incubated at 22 C for another 8 hours. Dilution and outgrowth was repeated 2 more times. Cells were then serially diluted and plated on LB agar at 37 °C. 100 individual colonies were patched on LB plates and LB plates containing mls to identify mls sensitive colonies that had evicted the plasmid. Chromosomal DNA from colonies that had excised the plasmid was purified and screened by PCR using primers 1109/1112 to determine which isolate had retained the ΔdegU allele.
To generate the ΔswrB in frame marker-less deletion construct, the region upstream of swrB was amplified using the primer pair 740/741, and digested with EcoRI and SalI. The region downstream of swrB was PCR amplified respectively using the primer pairs 839/840 and digested with SalI and BamHI. The two fragments were then simultaneously ligated respectively into the EcoRI and BamHI sites of pMiniMAD to generate pDP242. The plasmid was introduced to PY79 by single cross-over integration by transformation at the restrictive temperature for plasmid replication (37°C) using mls resistance as a selection. The integrated plasmid was then transduced into various strains prior to eviction as described above. Chromosomal DNA from colonies that had excised the plasmid was purified and screened by PCR using primers 740/840 to determine which isolate had retained the ΔswrB allele.
Complementation constructs
To generate the PdegU-degU complementation construct pLC1, a PCR product containing the degU gene plus approximately 500 base pairs upstream was amplified from B. subtilis 3610 chromosomal DNA using primer pairs 596/597, digested with HinDIII and BamHI and cloned into the HinDIII and BamHI sites of pDG1664 containing a polylinker and erythromycin resistance cassette between two arms of the thrC (Guerout-Fleury et al., 1996).
To generate the PdegS-degU complementation construct pLC2, a PCR product containing the degU gene was amplified from B. subtilis 3610 chromosomal DNA using primer pairs 598/597 and digested with NcoI and BamHI. Another PCR product containing the PdegS promoter region was amplified from B. subtilis 3610 chromosomal DNA using primer pairs 599/600 and digested with EcoRI and NcoI. The two digested PCR fragments were and cloned simultaneously into the EcoRI/BamHI sites of pDG1664 (Guerout-Fleury et al., 1996).
To generate the PdegSPdegU-degU complementation construct pYH5, a PCR product containing the degS and degU genes plus approximately 500 base pairs upstream was amplified from B. subtilis 3610 chromosomal DNA using primer pairs 599/597, digested with EcoRI and BamHI and cloned into the EcoRI and BamHI sites of pDG1664 (Guerout-Fleury et al., 1996). To generate the PdegS-degSdegUD56A mutant, degU was PCR amplified using pYH5 as template and the primer pair 1664/1665 to generate pYH6 by using the Quick change II XL site-directed mutagenesis kit (Stratagene). In a second step, degU was PCR amplified from pYH6 as template and the primer pair 1666/1667 to generate pYH7 by using the Quick change II XL site-directed mutagenesis kit. The D56A mutagenesis site was confirmed by DNA sequencing.
To generate the PflgM-flgM complementation construct, a PCR product containing flgM was PCR amplified using 3610 chromosomal DNA as a template and primer pair 2365/2366, digested with EcoRI and BamHI and cloned into the EcoRI and BamHI sites of pDG364 to create plasmid pRC2. To generate the PflgMΔdegUsite1-flgM complementation construct, a PCR product containing flgM was PCR amplified using 3610 chromosomal DNA as a template and primer pair 2365/2475, digested with EcoRI and XhoI and a PCR product was similarly amplified using primer pair 2476/2366 and digested with XhoI and BamHI. Both digested fragments were simultaneously ligated into the EcoRI and BamHI sites of pDG364 to generate pDP365. To generate the PflgMΔdegUsite2-flgM complementation construct, a PCR product containing flgM was PCR amplified using 3610 chromosomal DNA as a template and primer pair 2365/2644, digested with EcoRI and XhoI and a PCR product was similarly amplified using primer pair 2645/2366 and digested with XhoI and BamHI. Both digested fragments were simultaneously ligated into the EcoRI and BamHI sites of pDG364 to generate pRC7. To generate the PflgMΔdegUsite1ΔdegUsite2-flgM complementation construct, a PCR product containing flgM was PCR amplified using pDP365 plasmid DNA as a template and primer pair 2365/2786, digested with EcoRI and SalI and a PCR product was similarly amplified using primer pair 2787/2366 and digested with SalI and BamHI. Both digested fragments were simultaneously ligated into the EcoRI and BamHI sites of pDG364 to generate pRC9.
LacZ reporter constructs
To generate the β-galactosidase (lacZ) reporter constructs pCC1, pKB17, and pLC126, PCR products containing the following promoters were amplified from B. subtilis 3610 chromosomal DNA using the primers indicated in parentheses: PflhO (1251/1252), PmotA (798/799), and PlytF (917/1771). Each PCR product was digested with EcoRI and BamHI and cloned independently into the EcoRI and BamHI sites of plasmid pDG268, which carries a chloramphenicol-resistance marker and a polylinker upstream of the lacZ gene between two arms of the amyE gene (Antoniewski et al., 1990).
PflgM-GFP transcriptional fusion
To generate the transcriptional fusion of PflgM to GFP, a 444 base pair fragment containing the promoter region of flgM gene was amplified using 3610 as a template and primer pair 2258/2259. A 767 base pair fragment containing the full gfp gene was amplified using the plasmid, pMF35, as template and primer pair 2256/2257. PflgM and gfp gene PCR fragments were digested with SphI and XhoI, and XhoI and BamHI respectively, and simultaneously ligated into the SphI and BamHI sites of pAH25 containing a spectinomycin resistance cassette to generate plasmids, pYH9. pYH9 plasmid was integrated into PY79 at the ectopic site of amyE and transduced to other recipients.
DegS-His6 translation fusion construct
To generate the translational fusion of degS with His6, a 1173 base pair fragment containing the full coding region of degS gene was amplified using 3610 as a template and primer pair 1227/1228 and was digested with NcoI and HindIII and ligated into the their compatible cohesive sites of pET28a containing an His6 tag and a kanamycin resistance cassette to generate plasmids, pYH8.
SPP1 phage transduction
To 0.2 ml of dense culture grown in TY broth (LB broth supplemented after autoclaving with 10 mM MgSO4 and 100 μM MnSO4), serial dilutions of SPP1 phage stock were added and statically incubated for 15 minutes at 37 C. To each mixture, 3 ml TYSA (molten TY supplemented with 0.5% agar) was added, poured atop fresh TY plates, and incubated at 37 C overnight. Top agar from the plate containing near confluent plaques was harvested by scraping into a 50 ml conical tube, vortexed, and centrifuged at 5,000 × g for 10 minutes. The supernatant was treated with 25 μg/ml DNase final concentration before being passed through a 0.45 μm syringe filter and stored at 4 °C.
Recipient cells were grown to stationary phase in 2 ml TY broth at 37°C. 0.9 ml cells were mixed with 5 μl of SPP1 donor phage stock. 9 ml of TY broth was added to the mixture and allowed to stand at 37°C for 30 minutes. The transduction mixture was then centrifuged at 5,000 × g for 10 minutes, the supernatant was discarded and the pellet was resuspended in the remaining volume. 100 μl of cell suspension was then plated on TY fortified with 1.5% agar, the appropriate antibiotic, and 10 mM sodium citrate.
Transposon mutagenesis
To generate bypass mutants of ΔswrAΔswrB, the pMarA plasmid was introduced into strain DS2655 by SPP1 phage transduction (Le Breton et al., 2006). Mutagenesis was performed on each isolate by growing cells in 2ml LB broth supplemented with kanamycin at 22°C for 24 hours. Cells were diluted serially to 10−1, 10−2, and 10−3, and 100μl of each dilution was plated on prewarmed LB plates containing 1.5% agar and supplemented with kanamycin and X-Gal, and grown at the non permissive temperature (42°C) overnight. Blue colonies were selected and confirm that the transposon was linked to the suppressor mutation, a lysate was generated on the suppressor mutant and the transposon was transduced to the parent strain. Transposon insertion sites were identified by partially degenerate touchdown PCR using primer 766 and hybrid degenerate primer 749, 50 ng of purified chromosomal DNA, and Phusion polymerase (New England Biolabs) (Levano-Garcia et al., 2005).
β-Galactosidase assay
Cells were grown until 0.5 to 1.0 OD 600 at 37°C in LB broth and collected in 1 ml aliquots and suspended in an equal volume of Z buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 10 mM KCl and 38 mM 2-mercaptoethanol). Lysozyme was added to each sample to a final concentration of 0.2 mg/ml and incubated at 30°C for 30 min. Each sample was diluted in Z-buffer to a final volume of 1 ml and the reaction was started with 200 μl of 4 mg/ml 2-nitrophenyl β-D-galactopyranoside in Z buffer and stopped with 500 μl of 1M Na2CO3. The OD420 of the reaction mixture was measured. The β-galactosidase-specific activity was calculated according to the equation: [OD420/(time × OD600)] × dilution factor × 1000].
Primer extension mapping of the flgM transcription start site
Total RNA was prepared from exponentially growing strain 3610 using the method described in (Ramos-Montanez et al., 2008). The 5′-end of the flgM transcript was determined using the Primer Extension System-AMV reverse transcriptase (Promega) with modifications in (Ramos-Montanez et al., 2008). Primer extension reactions contained 0.33 pmole of 32P-end-labeled primer 1409 and 6.6 μg of total 3610 RNA. A sequencing ladder of the PCR amplicon synthesized with primers 1375/1411 from 3610 genomic DNA was generated by using the Sequenase PCR Product Sequencing Kit (USB Corp.) and primer 1409 according to the manufacturer’s instructions.
Overexpression and purification of DegS-His6 and DegU-His6 proteins
To purify the DegU-His6 protein, an overnight culture of E. coli strain BL21 (DE3) carrying plasmid pNW43 was diluted into 500 ml LB broth supplemented with 100 μg/ml ampicillin and 0.2% (wt/vol) glucose grown to an OD600 of 0.9. The culture was induced with 1 mM IPTG and grown for an additional hour at 37°C. Cells were centrifuged, collected and washed with buffer A (20 mM Tris-HCl (pH 8), 200 mM NaCl). Cell pellets were frozen and stored at −70°C. Pellets were resuspended in buffer B (20 mM Tris-HCl (pH 8), 200 mM NaCl and 0.25% (vol/vol) Tween-20). Cells were broken with a French press at 20,000 psi (138 MPa) and centrifuged at 15,000 rpm for 30 min (Beckman T 17 rotor). The supernatant was mixed with 2 ml of Ni-nitrilotriacetate (NTA) resin (Qiagen) equilibrated with buffer B by continuous rotation for 1 hour at 4°C. The mixture was loaded into a column and washed with buffer B containing 30 mM imidazole and subsequently with buffer C (20 mM Tris-HCl (pH 8), 300 mM NaCl). The DegU-His6 fusion protein was eluted with a 30–500 mM imidazole gradient in buffer C.
To purify the DegS-His6 protein, an overnight culture of E. coli strain BL21 (DE3) harboring plasmid pYH8 was diluted in 500 ml LB broth supplemented with 30 μg/ml of kanamycin and grown at 30 °C to an OD 600 of 0.5. The culture was induced with 1 mM IPTG and grown for an additional 3 hours at 30°C. Cells were centrifuged, collected and washed with 10 mM Tris-HCl (pH 7.6) buffer and pellets were frozen and stored at −70°C. Pellets were resuspended in Tris-HCl (pH 7.6) buffer, cells were broken with a French press at 20,000 psi (138 MPa) and centrifuged at 15,000 rpm for 30 min (Beckman T 17 rotor). Insoluble pellets were resuspended in 3 ml of buffer D (6 M guanidine-HCl, 50 mM Tris-HCl (pH 7.6), 150 mM NaCl), stored on ice for 30 min and centrifuged at 30,000 g for 15 min. The supernatant was mixed with 2 ml of Ni-nitrilotriacetate (NTA) resin (Qiagen) and equilibrated by continuous rotation in buffer A (6 M guanidine-HCl, 50 mM Tris-HCl (pH 7.6) and 150 mM NaCl) for 20 min at room temperature. The mixture was loaded into a column and washed with 10 ml of buffer R1 (8 M urea, 50 mM Tris-HCl (pH 7.6) 150 mM NaCl). To renature DegS-His6, the column was washed serially with 10 ml of buffer containing 6M, 4M, 2M urea and subsequently with 10 ml of wash buffer (50 mM Tris-HCl (pH 7.6), 150 mM NaCl). DegS-His6 fusion protein was eluted with wash buffer containing 500 mM imidazole. Purified DegS-His6 was dialysed against storage buffer (50 mM Tris-HCl (pH 7.6), 200 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 50% (vol/vol) glycerol) and stored at −70°C.
Phosphorylation of DegS and DegU
Autophosphorylation of DegS-His6 was carried our as described in (Gutu, et. al., 2010) with the following modifications. DegS-His6 was diluted to a final concentration of 0.5 μM with kinase buffer (50 mM Tris-HCl (pH 7.6), 0.1 mM EDTA, 10 mM MgCl2, 1 mM DTT, 50 mM KCl, 0.1 mg/ml BSA, 10% (vol/vol) glycerol) and autophosphorylation reactions were initiated by [γ-32P] ATP (specific activity = 2.5 Ci/mmol; Perkin-Elmer BLU502Z) to a final concentration of 6.0 μM. Reactions were carried out at room temperature for 30 sec to 20 min. At designated times (Fig. S2), 15 μL samples were removed and reactions were stopped by adding 15 μL of 2× Laemmli sample buffer containing 5% (vol/vol) β-mercaptoethanol. Final samples (20 μL) were analyzed without heating by 10% Tris-glycine SDS-PAGE. After electrophoresis, gels were soaked for 20 min in 2% (vol/vol) glycerol and dried for 1h at 80oC on a vacuum gel dryer (BioRad). Dried gels were exposed to a storage phosphor screen (GE Healthcare) and analyzed using a Typhoon Variable Mode Imager 9200 (Amersham) and ImageQuant 5.2 software (Molecular Dynamics).
Phosphoryltransfer of DegS-His6-P to DegU was carried out in combined reactions (Gutu et. al., 2010). DegS-His6 was autophosphorylated as described above for 10 min, after which DegU-His6 was added to reaction mixtures to a final concentration of 2.0 μM. Samples were taken at designated times between 30 sec and 20 min (Fig. 2S) and analyzed by SDS-PAGE as described above.
Electrophoretic mobility shift assay
Infrared-dye labeled probes (IDT) were amplified by PCR using B. subtilis 3610 genomic DNA using primers 346/1981 (Peps), 1782/1812 (Pfla/che), 1931/2009 (PflgM−393to−162), 1931/2010 (PflgM−393to−30) and 1931/2091 (PflgM−393to−80). Probes were purified by using a QIAquick PCR Purification Kit (Qiagen). Binding reactions were performed in 40 μl of binding buffer (50 mM Tris-HCl (pH 7.6), 0.1 mM EDTA, 10 mM MgCl2, 1 mM DTT, 50 mM KCl, 0.1 mg BSA/ml, 10% glycerol, 0.1 mM ATP, and 1μg poly(dI-dC)poly(dI-dC) (Sigma)). 0.2 pmole of labeled DNA probes were mixed with 0.08–5 μM DegU or DegU-P, which was phosphorylated by DegS as described above, in binding buffer and reaction mixtures were incubated at room temperature for 20 min. 15 μl of each reaction mixture was resolved in a 6% polyacrylamide gel containing 10% glycerol with 1 × Tris-Glycine-EDTA buffer (pH 8.0) for 1 h at 110 V at 4°C. Gels were scanned and bands were detected using an Odyssey Infrared Imaging System (Li-Cor Biosciences).
DNase footprinting
DNase footprinting of DegU binding to the PflgM site was performed as described previously (Ng et al., 2005) with the following modifications. Briefly, 62.5 pmole of primer 2221 was end-labeled in a 10 μl reaction with 5 pmole [γ-32P] ATP (Perkin-Elmer, BLU502Z) and T4 polynucleotide kinase (NEB, M0236) according to the manufacturer’s protocol. To produce end-labeled PflgM DNA fragment, 10 μl of 10× Pfu buffer (Stratagene), 0.8 μl of 100 mM of dNTPs, 2 μl of diluted 3610 genomic DNA, 6 μl of 10 μM primer 2221 (60 pmole final amount), 2 μl of Pfu Turbo (Stratagene, 600254), and 69.2 μl water were added to the labeled primer 2221. A standard PCR cycle was run (94° C for 30 s, 55° C for 30 s, 72° C for 30 s; 30 cycles), and amplicon purified with the QIAquick PCR purification kit (QIAGEN, 28106). An identical reaction was done in parallel without [γ-32P] ATP to check the yield and PCR specificity by agarose gel electrophoresis.
DegU was phosphorylated by DegS as described above. The binding condition of DegU or DegU~P to PflgM was the same as described for the electrophoretic mobility shift assay, except the total volume of the reaction was increased to 60 μl. 0.2–0.6 pmole of labeled PflgM DNA and 0–5 μM of DegU or DegU-P were added to each binding reaction. Reactions were incubated at 25o C for 20 min. 5 μl of diluted RQ1 DNase (Promega, M610A) were added, and the mixture was further incubated at 25o C for 5 min. 180 μl of STOP solution (0.4 M sodium acetate, 50 μg per ml sheered salmon sperm DNA (Ambion, AM9680), and 2.5 mM EDTA) was added to quench the digestion. Digested DNA was extracted with 240 μl phenol-chloroform-isoamyl alcohol (25:24:1, Fisher, BP1752I-100) at room temperature using a vortex mixer. The upper aqueous phase was transferred to a new tube, and DNA was precipitated by adding 3 volumes of ice cold ethanol and 1.0 μl of glycoblue (Ambion, 15 μg per μl). The pellet was collected by centrifugation at 16,100 g for 30 min at 4 oC, air dried for 10 min, and resuspended in 10 μl of 2× loading dye (Promega). A sequencing ladder was generated with 10 pmole of primer 2221 and 3610 genomic DNA, as described in the primer extension assay. The ladder and digested products were boiled for 5 min, and resolved by PAGE on an 8 % denaturing urea-gel followed by autoradiography.
Supplementary Material
Acknowledgments
We thank Kris Blair, Colleen Courtney, and Nicola Stanley-Wall for reagents. Y-HH was supported by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. This work was funded by an NIH Training Grant NIH T32 GM007757 to LMC, a Women in Science Fellowship to LMC, NIH grant AI060744 to MEW, and NIH grant GM093030 to DBK. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- Amati G, Bisicchia P, Galizzi A. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol. 2004;186:6003–6014. doi: 10.1128/JB.186.18.6003-6014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewski C, Savelli B, Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990;172:86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Chamberlin MJ. Secondary σ factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla D, Caramori T, Galizzi A. Coupling flagellin gene transcription to flagellar assembly in Bacillus subtilis. J Bacteriol. 1994;176:4558–4564. doi: 10.1128/jb.176.15.4558-4564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero MG, Gonzales B, Tarricone C, Ceciliani, Galizzi A. Overproduction and characterization of the Bacillus subtilis anti-sigma factor FlgM. J Biol Chem. 1999;274:12103–12107. doi: 10.1074/jbc.274.17.12103. [DOI] [PubMed] [Google Scholar]
- Calvio C, Celandroni F, Ghelardi E, Amati G, Salvetti S, Ceciliani F, Galizzi A, Senesi S. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J Bacteriol. 2005;187:5356–5366. doi: 10.1128/JB.187.15.5356-5366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramori T, Barilla D, Nessi C, Sacchi L, Galizzi A. Role of FlgM in σD-dependent gene expression in Bacillus subtilis. J Bacteriol. 1996;178:3113–3118. doi: 10.1128/jb.178.11.3113-3118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Guttenplan SB, Blair KM, Kearns DB. Role of the σD-dependent autolysins in Bacillus subtilis population heterogeneity. J Bacteriol. 2009;191:5775–5784. doi: 10.1128/JB.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Helmann JD. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis σ factor. Proc Natl Acad Sci USA. 1992;89:5123–5127. doi: 10.1073/pnas.89.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FFV, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa NE, Barker JR, Klose KE. The Vibrio cholerae FlgM homologue is an anti-σ28 factor that is secreted through the sheathed polar flagellum. J Bacteriol. 2004;186:4613–4619. doi: 10.1128/JB.186.14.4613-4619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozy LM, Kearns DB. Gene position in a long operon governs motility development in Bacillus subtilis. . Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07112.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst F, Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol. 1991;173:2539–2547. doi: 10.1128/jb.173.8.2539-2547.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartois V, Debarbouille M, Kunst F, Rapoport G. Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis. J Bacteriol. 1998;180:1855–1861. doi: 10.1128/jb.180.7.1855-1861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, σ28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Estacio W, Anna-Arriola S, Adedipe M, Márquez-Magaña LM. Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis. J Bacteriol. 1998;180:3548–3555. doi: 10.1128/jb.180.14.3548-3555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman MZ, Wiggs JL, Chamberlin MJ. Nucleotide sequences of two Bacillus subtilis promoters used by Bacillus subtilis sigma-28 RNA polymerase. Nuc Acids Res. 1981;9:5991–6000. doi: 10.1093/nar/9.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:75–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Gutu AD, Wayne KJ, Sham LT, Winkler ME. Kinetic Characterization of the WalRKSpn (VicRK) Two-Component System of Streptococcus pneumoniae: Dependence of WalKSpn (VicK) Phosphatase Activity on Its PAS Domain. J Bacteriol. 2010;192:2346–2358. doi: 10.1128/JB.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Van Werkhoven AF, Venema G, Dubnau D. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc Natl Acad Sci USA. 2000;97:9246–9251. doi: 10.1073/pnas.160010597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD, Márquez LM, Chamberlin MJ. Cloning, sequencing, and disruption of the Bacillus subtilis σ28 gene. J Bacteriol. 1988;170:1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Josenhans C, Niehus E, Amersbach S, Horster A, Betz C, Drescher B, Hughes KT, Suerbaum S. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol Microbiol. 2002;43:307–322. doi: 10.1046/j.1365-2958.2002.02765.x. [DOI] [PubMed] [Google Scholar]
- Karlinsey JE, Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007;66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- Le Breton Y, Mohapatra NP, Haldenwang WG. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1 a mariner-based transposon. Appl Environ Microbiol. 2006;72:327–333. doi: 10.1128/AEM.72.1.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levano-Garcia J, Verjovski-Almeida S, da Silva ACR. Mapping transposon insertion sties by touchdown PCR and hybrid degenerate primers. Biotechniques. 2005;38:225–229. doi: 10.2144/05382ST03. [DOI] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Mader U, Antelman H, Buder T, Dahl MK, Hecker M, Homuth G. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol Genet Genomics. 2002;268:455–467. doi: 10.1007/s00438-002-0774-2. [DOI] [PubMed] [Google Scholar]
- Margot P, Pagni M, Karamata D. Bacillus subtilis 168 gene lytF encodes a γ-D-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, σD. Microbiol. 1999;145:57–65. doi: 10.1099/13500872-145-1-57. [DOI] [PubMed] [Google Scholar]
- Márquez LM, Helmann JD, Ferrari E, Parker HM, Ordal GW, Chamberlin MJ. Studies of sigma D-dependent functions in Bacillus subtilis. J Bacteriol. 1990;172:3435–3443. doi: 10.1128/jb.172.6.3435-3443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Magaña LM, Chamberlin MJ. Characterization of the sigD transcriptional unit of Bacillus subtilis. J Bacteriol. 1994;176:2427–2434. doi: 10.1128/jb.176.8.2427-2434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel DB, Chamberlin MJ. The Bacillus subtilis flagellin gene (hag) is transcribed by the σ28 form of RNA polymerase. J Bacteriol. 1989;171:3095–3101. doi: 10.1128/jb.171.6.3095-3101.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel DB, Lustre VM, Chamberlin MJ. An operon of Bacillus subtilis motility genes transcribed by the σD form of RNA polymerase. J Bacteriol. 1992;174:4197–4204. doi: 10.1128/jb.174.13.4197-4204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel DB, Lauer P, Chamberlin MJ. Identification of flagellar synthesis regulatory and structural genes in a σD-dependent operon of Bacillus subtilis. J Bacteriol. 1994;176:4492–4500. doi: 10.1128/jb.176.15.4492-4500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msadek T, Kunst F, Henner D, Klier A, Rapoport G, Dedonder R. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J Bacteriol. 1990;172:824–834. doi: 10.1128/jb.172.2.824-834.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K, Kawata M, Tanaka T. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J Biol Chem. 1990;265:20000–20006. [PubMed] [Google Scholar]
- Ng WL, Tsui HC, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J Bacteriol. 2005;187:7444–7459. doi: 10.1128/JB.187.21.7444-7459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. DNA microarray analysis of the Bacillus subtilis DegU, ComA, and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucl Acids Res. 2001;29:3804–3813. doi: 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- Ohnishis K, Kutsukake K, Suzuki H, Iino I. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an anti-sigma factor inhibits the activity of the flagellum specific sigma factor, σF. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol. 2008;70:1166–1179. doi: 10.1111/j.1365-2958.2008.06469.x. [DOI] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J Bacteriol. 2009;191:7129–7133. doi: 10.1128/JB.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Montanez S, Tsui HC, Wayne KJ, Morris JL, Peters LE, Zhang F, Kazmierczak KM, Sham LT, Winkler ME. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol. 2008;67:729–746. doi: 10.1111/j.1365-2958.2007.06082.x. [DOI] [PubMed] [Google Scholar]
- Rust M, Borchert S, Niehus E, Kuehne SA, Gripp E, Bajceta A, McMurray JL, Suerbaum S, Hughes KT, Josenhans C. The Helicobacter pylori anti-sigma factor FlgM is predominantly cytoplasmic and cooperates with the flagellar basal body protein FlhA. J Bacteriol. 2009;191:4824–4834. doi: 10.1128/JB.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa M, Yamamoto H, Yamaguchi H, Fujita Y, Kobayashi K, Ogasawara N, Sekiguchi J. Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses. Gene. 2004;329:125–136. doi: 10.1016/j.gene.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Smits WK, Kuipers OP, Veening J-W. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol. 2008;4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- Tokunaga T, Rashid MH, Kuroda A, Sekiguchi J. Effect of degS-degU mutations on the expression of sigD, encoding an alternative sigma factor, and autolysin operon of Bacillus subtilis. J Bacteriol. 1994;176:5177–5180. doi: 10.1128/jb.176.16.5177-5180.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuhara K, Ogura M. Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol. 2008;8:8. doi: 10.1186/1471-2180-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behavior exhibited by Bacillus subtilis. Mol Microbiol. 2007;65:554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- Werhane H, Lopez P, Mendel M, Zimmer M, Ordal GW, Márquez-Magaña LM. The last gene of the fla/che operon in Bacillus subtilis, ylxL, is required for maximal σD function. J Bacteriol. 2004;186:4025–4029. doi: 10.1128/JB.186.12.4025-4029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JT, Estacio W, Márquez-Magaña L. Relative roles of the fla/che PA, PD-3, and PsigD promoters in regulating motility and sigD expression in Bacillus subtilis. J Bacteriol. 2000;182:4841–4848. doi: 10.1128/jb.182.17.4841-4848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasamura A, Abe S, Tanaka T. Involvement of nitrogen regulation in Bacillus subtilis degU expression. J Bacteriol. 2008;190:5162–5171. doi: 10.1128/JB.00368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.