Abstract
The P450cam monooxygenase system consists of three separate proteins: the FAD-containing, NADH-dependent oxidoreductase (putidredoxin reductase or Pdr), cytochrome P450cam and the 2Fe2S ferredoxin (putidaredoxin or Pdx), which transfers electrons from Pdr to P450cam. Over the past few years our lab has focused on the interaction between these redox components. It has been known for some time that Pdx can serve as an effector in addition to its electron shuttle role. The binding of Pdx to P450cam is thought to induce structural changes in the P450cam active site that couple electron transfer to substrate hydroxylation. The nature of these structural changes has remained unclear until a particular mutant of P450cam (Leu358Pro) was found to exhibit spectral perturbations similar to those observed in wild type P450cam bound to Pdx. The crystal structure of the L358P variant has provided some important insights on what might be happening when Pdx docks. In addition to these studies, many Pdx mutants have been analyzed to identify regions important for electron transfer. Somewhat surprisingly, we found that Pdx residues predicted to be at the P450cam-Pdx interface play different roles in the reduction of ferric P450cam and the ferrous P450-O2 complex. More recently we have succeeded in obtaining the structure of a chemically crosslinked Pdr-Pdx complex. This fusion protein represents a valid model for the noncovalent Pdr-Pdx complex as it retains the redox activities of native Pdr and Pdx and supports monooxygenase reactions catalyzed by P450cam. The insights gained from these studies will be summarized in this review.
Keywords: P450cam, putidaredoxin, putidaredoxin reductase, protein-protein interaction, electron transfer, crystallography, mutagenesis
Introduction
While P450 monooxygenase systems include a rich variety of redox partners, a majority of research has centered on the P450 protein. This is understandable considering that the complex redox chemistry and substrate specificity reside in the P450 active site. The interest in P450 catalysis is also fueled by the growing number of available genome sequences that has led to the discovery and characterization of many more P450s with novel properties (http://drnelson.uthsc.edu/cytochromeP450.html). The P450 redox partners and inter-protein electron transfer reactions, however, have remained somewhat in the background. Part of the problem is the difficulty in obtaining detailed structural information on electron transfer complexes, as they tend to be weak and possibly dynamic. For rapid turnover, the redox partner must bind, deliver an electron, and then dissociate. Such functional demands preclude formation of stable long-lived complexes, which makes protein co-crystallization challenging. This is why to date there is only one known crystal structure of a P450-redox partner complex [1]. Even so, our understanding of redox partner recognition, dynamics, and electron transfer is reasonably well advanced with P450cam. The direct electron donor to P450cam is the 2Fe2S ferredoxin, putidaredoxin (Pdx). Pdx receives electrons from the FAD-containing putidaredoxin reductase (Pdr) via NADH. Here we summarize our recent work in understanding structural changes that accompany the interaction between P450cam and Pdx, as well as crystallographic and biochemical studies on the Pdr-Pdx pair.
1. P450cam-Pdx Interaction: Comparison with the Homologous Redox Systems
While it is generally true that P450cam has been a paradigm for nearly all aspects of P450 function, the interaction between P450cam and Pdx has been particularly interesting since this interaction is highly specific and Pdx cannot be substituted by other redox partners [2-4]. Such specificity has long been thought to be coupled to an effector role of Pdx wherein the P450cam-Pdx interaction results in some structural changes that enable coupling of electron transfer to O2 activation and product formation. While any reductant with a suitable redox potential can convert the P450cam heme iron from Fe3+ to Fe2+, only Pdx can deliver the second electron and promote camphor hydroxylation.
Owing to some recent advances with P450s closely related to P450cam, we now have a better sense on the unusual selectivity exhibited by the P450cam-Pdx system. In the soil bacterium Citrobacter braakii, a close cousin to P450cam, P450cin, regio- and stereo-selectively hydroxylates the monoterpenoid cineole as the first step in the utilization of cineole as a carbon source [5]. Cineole is very similar to camphor and has the same number of carbon and oxygen atoms, although cineole is an ether while camphor is a ketone. As expected, the P450cin structure [6] closely resembles that of P450cam except near the substrate binding pocket. However, unlike P450cam, P450cin receives electrons from the FMN-containing flavodoxin called cindoxin (Cdx) rather than a 2Fe2S ferredoxin [5]. Furthermore, recombinant P450cin hydroxylates cineole during expression in E. coli and thus must be able to use E. coli redox proteins as partners [5]. This finding enabled the development of a P450cin activity assay utilizing Cdx and the E. coli reductase. In this reconstituted system, P450cin exhibits a NADPH oxidation rate of 15 s−1 with ~80% of NADPH coupled to product formation [5]. The electron transfer reaction between Cdx and ferric P450cin (conversion of Fe3+ to Fe2+) has been examined in detail using stopped flow methods [7] and gives a rate of 15 s−1. For comparison, the rates of electron transfer between other P450s and their respective redox partners are presented in Table 1. Pdx and adrenodoxin (Adx) also can reduce P450cin but at much lower rates, 0.6 s−1 and 0.03 s−1, respectively, while the FMN domain of human P450 reductase reduces P450cin at a rate of 0.7 s−1. Overall, P450cin appears to favor its natural redox partner, Cdx, which can be substituted by other electron carriers for delivering the first electron to the heme iron but with lesser success. Since P450cin exhibits excellent coupled activity with non-native partners, the P450cin system cannot be as selective as P450cam. Unfortunately, the only structural insights we have for the P450cin-Cdx complex are based on homology modeling of Cdx since the Cdx crystal structure is not known [7]. Although it is presently not possible to gain any significant knowledge on how the P450cam-Pdx complex differs from the P450cin-Cdx complex, we can compare the P450cam and P450cin structures. As noted in our earlier work [7], the overall topology of the surface to which redox partners bind are very similar in P450cin and P450cam. This similarity extends to the location of key basic side chains such as Arg112 in P450cam (Arg102 in P450cin), known to be important for interaction with Pdx [8, 9]. However, most of our new insights on the P450cam-Pdx complex and the effector role of Pdx are derived from mutagenesis work on Pdx [10]. Given that the Cdx structure is still unknown and the mutagenesis data on Cdx are scarce (this work is just beginning), it is not possible at the moment to extend the comparison between the P450cin and P450cam redox complexes and explain why P450cam is so selective but P450cin is not. Another recently characterized P450 system closely related to P450cam corroborates the notion on high redox partner selectivity of bacterial P450s. The photosynthetic bacteria Rhodopseudomonas palustris has seven P450 enzymes, four of which have been cloned and characterized [11]. The monooxygenase activity of one P450, CYP199A2, was reconstituted with cognate palustriredoxin reductase (PuR) and 2Fe2S palustriredoxin (Pux), homologous to Pdr and Pdx [12]. Despite low sequence identity, Pdr and PuR efficiently substitute each other in reactions catalyzed by P450cam and CYP199A2 [11, 12]. In contrast, Pdx and Pux (42% identity) cannot effectively deliver electrons to an alternative P450 (>100-fold decrease in the turnover) [11]. While the Pux [13] and Pdx structures are very similar, a comparison between the two illustrates some clear difference and similarities at the redox partner interface. Critical for Pdx activity Asp38 [10] corresponds to Glu in Pux. Despite this important similarity, there is a noticeable difference in the distribution of charged groups at the proposed docking surfaces, although the role of these residues has not been probed by mutagenesis as yet. The most important residue in Pdx is the C-terminal Trp106, critical for electron transfer to P450cam [14] and corresponding residue in Pux is Arg. This alone would eliminate the ability of Pux to serve as a redox partner for P450cam. Additionally, Trp106 may be a key player in the effector role of Pdx, a topic we consider further on.
Table 1.
Comparison of electron transfer rates in various P450s and various electron donors. PDX - 2Fe2S-containing putidaredoxin, CPR - cytochrome P450 reductase, CDX - FMN-containing cindoxin, ET - electron transfer, CO - carbon monoxide complex. Redox potentials for P450s are for the substrate complexes. For CPR and CDX the redox potentials are for the FMN semiquinone/hydroquinone couple.
| Organism | Electron donor | P450 | k (s−1) | Temp (°C) | References |
|---|---|---|---|---|---|
| Pseudomonas putida | PDX (−240mV) |
cam (−173mV) |
30 - 41 CO |
25 | [10][59, 60] |
| Citrobacter braakii | CDX (−226mV) |
cin (−202mV) |
15 CO |
21 | [7] |
| Cow | ADX (−252mV) |
scc | 0.7 - 1 CO |
15 | [61, 62] |
| Human | CPR (−269mV) |
2E1 | 23-32 CO |
37 | [63, 64] |
| Human | CPR | 1A2 | 13.3 CO |
37 | [63] |
| Human | CPR | 2C9 | 2.7 - 4.3 CO |
37 | [63] |
| Human | CPR | 2C19 | 16 CO |
37 | [63] |
| Human | CPR | 3A4 | 0.2-38 CO |
37 | [63] |
Somewhat lesser ferredoxin/P450 specificity is observed in Novosphingobium aromaticivorans. This oligotrophic bacterium encodes 15 P450s, most of which have been recombinantly expressed and purified [15], as well as several FAD-dependent reductases and 2Fe2S ferredoxins. One redox couple homologous to Pdr and Pdx, ArR and Arx, supports activity of at least five N. aromaticivorans P450s, with the product formation rate of 1,000-2,000 min−1 and 63-99% coupling [16]. Two of these P450 enzymes, CYP101D1 and CYP101D2, share high sequence identity with P450cam and hydroxylate camphor to the same 5-exo-hydroxycamphor product [17]. Again, Pdr can substitute for ArR and mediate NADH-dependent reduction of Arx. The Pdr/Arx hybrid pair can support the monooxygenase activity of several N. aromaticivorans P450s albeit less efficiently than ArR/Arx (product formation rate of 90-170 versus 140-750 min−1, respectively) [15]. Unfortunately, it has not been reported whether Arx and Pdx can substitute each other as well. Nonetheless, the ability of Arx to serve as an electron donor and support catalytic turnover of multiple P450s suggests that the N. aromaticivorans redox partners are less selective that their P. putida and R. palustris counterparts. Whether the ferredoxin components act as effectors in the aforementioned P450-dependent systems remains to be determined.
2. P450cam-Pdx Interaction: Computer Modeling
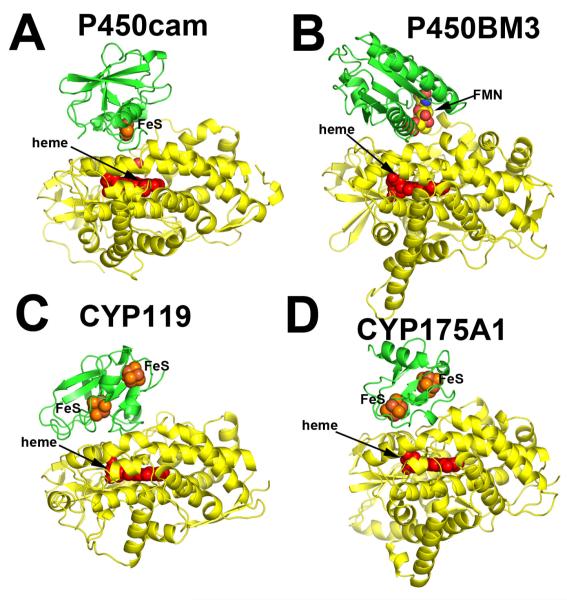
Significant advances in the NMR spectroscopy [18, 19] and the development of biophysical markers has helped unravel structural changes in P450cam caused by Pdx binding and further elucidate the effector role of Pdx. However, to understand the structural underpinnings we first need a structural model of the P450cam-Pdx complex. The currently available data indicate that the docking site in all P450s is to the proximal surface near the thiolate ligand side of the heme. This is the closest approach to the heme that affords the shortest electron transfer path to the heme iron. As already mentioned, there is only one known crystal structure of a P450-redox partner complex and that is formed between the heme- and FMN-domains of P450BM3 [1]. In this structure, the FMN module indeed docks on the proximal side of the heme (Fig. 1B). Experimental testing of this model strongly suggests that the crystal structure is a valid representation of the complex that forms in solution [20].
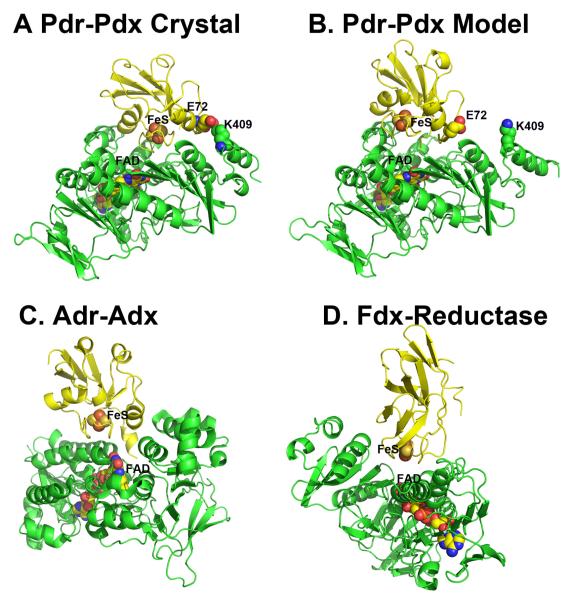
Figure 1.
Various P450 (yellow)-redox partner (green) complexes. Panels A, C, and D are based on computer docking and modeling while panel B represents the crystal structure [1].
Given that co-crystallization of redox proteins is challenging and has a very low success rate, computer modeling can be used as an alternative approach to obtain information on the docking orientations and interacting surfaces of the redox partners. We have routinely employed various programs to dock P450s with their respective electron donors. Some recent examples include two thermophilic P450s, CYP119 and CYP175A1. The redox partner of Thermus thermophilus CYP175A1 has been identified as a ferredoxin that contains one 3Fe4S and one 4Fe4S cluster [21]. Since the structure of this ferredoxin is known [22], we were able to use the docking program GRAMM [23, 24] to identify potential binding sites. GRAMM performs a computational search of all possible configurations to find protein complexes with the highest surface complementarity. The program output contains a list of docking models ranked in order from most to least favorable. The most favorable CYP175A1-ferredoxin structure is shown in Figure 1D. We went through a similar exercise with Sulfolobus acidocaldarius CYP119. Despite uncertainty about the correct redox partner, there is evidence indicating that CYP119 may use a 3Fe4S/4Fe4S ferredoxin as well [25]. Since no structural information on the S. acidocaldarius ferredoxin is available, a homology model was generated based on the structure of its close homologue [26] and then docked to CYP119. As shown in Figure 1C, the top ranking solution is remarkably similar to the CYP175A1-ferredoxin complex. Furthermore, the P450 docking surface in both hypothetical complexes is the same as we found for P450BM3. Finally, Figure 1A shows the hypothetical P450cam-Pdx complex. This model is based on the Pdx crystal structure [27], molecular modeling [39], and NMR data [28] with some additional refinement. There is substantial experimental data indicating that the computer-generated P450cam-Pdx structure closely resembles the actual complex [10, 19, 29-31] which will be summarized in section 4. This and high similarity of docking orientations in the hypothetical complexes give a certain level of confidence that all P450s will use the same site/molecular surface for interacting with the redox partners. Table 2 provides a list of ionic interactions in the P450cam-Pdx and other electron transfer complexes based on known crystal structures. Note that ionic interactions are not critical in all these complexes. For example, in the P450BM3 redox complex there is only one potential ion pair between His100 in P450 and Glu494 in the FMN domain. P450BM3 His100 is the homologue of Arg112 in P450cam, and a basic side chain at this position is highly conserved in P450s. Arg112 and His100 also pair with a heme propionate, so a basic side chain at this position may play a dual function in both heme and redox partner binding.
Table 2.
A list of ion pair interactions in various redox complexes. The P450cam-Pdx complex is based on computer docking models while the others are based on known crystal structures of the complexes.
| P450cam-Pdx complex | P450BM3-FMN domain complex | ||
|---|---|---|---|
| P450cam | Pdx | P450BM3 | FMN domain |
| R112 | Pdx D38 | H100 | E494 |
| K344 | Pdx E65 | ||
| R79 | Pdx C-term carboxyl | ||
| Pdr-Pdx complex | Adr-Adx complex | ||
|---|---|---|---|
| Pdr | Pdx | Adr | Adx |
| R310 | D38 | K27 | D39 |
| K409 | E72 | H28 | D41 |
| R240 | D79 | ||
| R244 | D76 | ||
| R211 | D76 | ||
3. P450cam-Pdx Interaction: Effector Role of Pdx
As noted, there are distinct biophysical signatures associated with Pdx binding to P450cam. The first to be observed was the IR stretching frequency of the P450cam Fe2+-CO band, shifting from 1940 cm−1 to 1932 cm−1 when Pdx binds [32]. Since CO coordinates to the iron on the distal side of the heme while Pdx binds on the proximal/thiolate side, the redox partner docking must induce some structural perturbation that is transmitted to the opposite side of the heme. A more structurally rich NMR study [19] shows that resonances of the distal side groups in P450cam, especially those of camphor, the key distal side residue Thr252, and the Cys ligand, are perturbed in the presence of Pdx. Once again this indicates that binding of Pdx changes the environment in the substrate pocket of P450cam. Resonance Raman studies also demonstrate that Pdx causes perturbations in stretching modes associated with CO and NO ligands, as well as in the heme electronic structure [33].
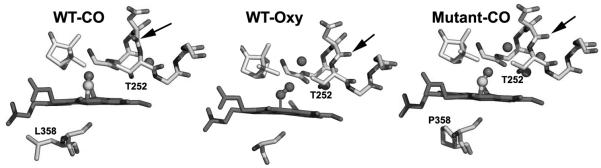
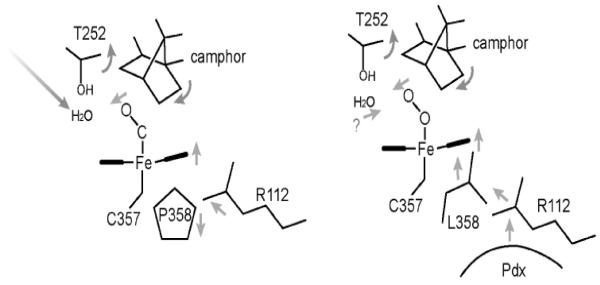
Obviously, the P450cam-Pdx crystal structure is needed to relate specific structural changes and the spectral results. While this has not yet been achieved, a serendipitous discovery provides some clues on what happens to P450cam when Pdx binds. The NMR properties of the Fe2+-CO complex of the L358P P450cam mutant are very similar to those of P450cam complexed with Pdx [18]. In addition, the L358P mutant binds Pdx more tightly than wild type (WT) and can catalyze the camphor hydroxylation reaction using artificial reductants as electron donors [18]. These results strongly suggest that mutating Leu358 to Pro results in structural changes that mimic what happens when Pdx binds. The big advantage here is that it has been possible to solve the L358P crystal structure [34]. In Figure 2, the CO complex of the L358P mutant is compared with the wild type CO and oxy complexes. In the wild type-O2 structure, there are substantial changes associated with O2 binding. The key change involves the I helix near Thr252 in the immediate vicinity of the O2 binding pocket. In the ferric state, the peptide group of Asp251 (indicated by an arrow in Fig. 2) is oriented perpendicular to the helical axis and, therefore, cannot form a normal helical H-bond. In the ferrous-O2 state, however, this peptide rotates to form a normal helical H-bond. This change widens the I helix near the bound O2 which enables key water molecules to be positioned into the active site [35, 36]. These highly ordered active site waters together with Thr252 are thought to play a critical role in O2 activation by providing protons to the distal O atom required for heterolytic O-O bond cleavage. None of this happens in the ferrous-CO complex. The only significant change is movement of the camphor substrate to make room for the CO molecule. However, in the L358P mutant CO binding causes the same changes as observed in the wild type-O2 complex: the Asp251 peptide rotates to form a normal helical H-bond thereby allowing the catalytic waters to be positioned into the active site. These changes take place because the rigid Pro358 side chain pushes on the bottom of the heme and repositions the camphor and the CO ligand similarly to the wild type-O2 complex (Figs. 2, 3). Since the properties of L358P-CO so closely resemble the P450cam-CO-Pdx complex, we have postulated that Pdx binding lowers the energy barrier to structural rearrangements required for O2 activation. The effector role of Pdx, therefore, is to arm the catalytic O2 activation machinery by “pushing” on the proximal face of the heme, which can be mimicked by the L358P mutation. Important to this scenario is that electron transfer and O2 activation are coupled to proton transfer. Without proper protonation of the iron-linked O2, electron transfer and O-O bond cleavage cannot proceed. Only when the proton transfer machinery is properly in place as a result of Pdx binding will electron transfer occur. Although P450cam is the only P450 system known to operate by such a complex regulatory mechanism, it would be quite surprising if other P450s do not employ their redox partners to control turnover. In mammalian systems, for instance, cytochrome b5 can form a complex with P450 and alter activity possibly as an effector redox partner [37].
Figure 2.
Crystal structures of the O2 and CO complexes of wild type and the L358P P450cam mutant. The arrow indicates the carbonyl oxygen atom of Asp251 that flips upon formation of the WT-O2 but not WT-CO complex. The peptide bond flip does occur in the L358P mutant-CO complex [36].
Figure 3.
Schematic diagram showing the chain of events taking place in the L358P P450cam mutant, which we hypothesize also occurs in wild type P450cam when Pdx binds. The binding of Pdx “pushes” on the proximal surface of the heme which transmits structural changes to the distal pocket. These changes are required to enable catalytic water molecules to be correctly positioned for O2 activation.
4. P450cam-Pdx Interaction: Experimental Test of a Hypothetical Model
The P450cam-Pdx interaction has been extensively studied using a variety of techniques and these results have been interpreted in light of the favored hypothetical model shown in Figure 4. In one of the earliest, perhaps first, protein engineering studies of the P450cam system (pre-cloning days), Sligar et al. found that removal of Trp106 in Pdx by carboxypeptidase results in a major loss in the P450cam monooxygenase activity [14]. This initial study was followed up by more detailed investigations where Trp106 was substituted with a variety of residues [38]. It was found that Trp106 is critical for binding but not kET, the electron transfer rate, for the reduction of Fe3+ P450cam. Our more recent studies are qualitatively similar as we demonstrated that the W106A mutant lowers kET for the first electron transfer step to Fe3+-P450cam by 7-fold but increases Kd by 15-fold [10]. In contrast, the W196F mutant has no effect on kET and increases Kd by only 4-fold. This implies that the first reductive step requires an aromatic or possibly nonpolar residue (i.e. Leu, Ile) at position 106 for binding but not electron transfer. In sharp contrast, kET for the second electron transfer step, reduction of the Fe2+-oxy complex, specifically requires Trp106. The W106A mutant lowers kET for the second step by over 1000-fold while having little effect on Kd [10]. This could possibly mean that the structure of the P450cam-Pdx complex is dependent on the P450cam redox state. Indeed, computational docking methods indicate that subtle structural differences on the proximal side of Fe3+ and Fe2+-oxy P450cam may affect the manner of Pdx binding [10]. Ligation of oxygen to the heme and structural perturbations in Thr252 and I-helix induce subtle but noticeable changes on the proximal face of P450cam (Fig. 6 in [10]) and reshape the hydrophobic pocket where Pdx is expected to dock. Since Pdx exerts its effector role on the second and not the first electron transfer step and Trp106 is predicted to directly contact the proximal surface of P450cam (Fig. 4 and Fig. 7 in [10]), we proposed based on the mutational effects that Trp106 has a direct role in the Pdx-induced changes in the P450cam distal pocket required for O2 activation.
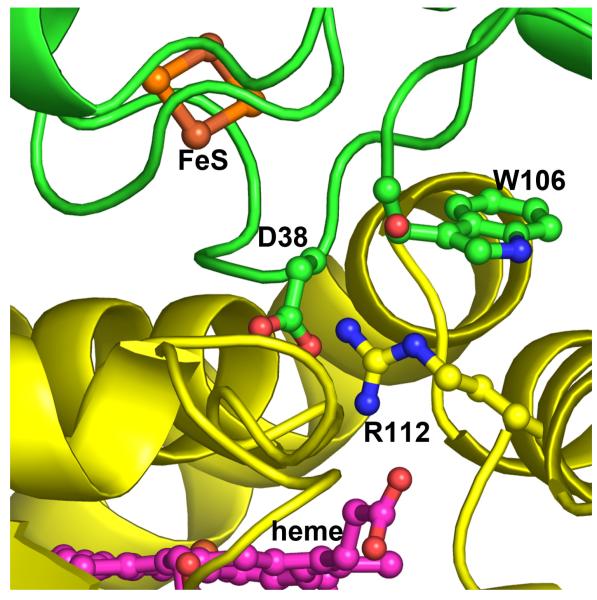
Figure 4.
Close up view at the P450cam (yellow)-Pdx (green) hypothetical complex. The key interaction involves Asp38 in Pdx and Arg112 in P450cam.
Another set of residues that has received considerable attention is the salt link predicted to form between Arg112 in P450cam and Asp38 in Pdx (Fig. 4). Both Arg112 [8, 9] and Asp38 [31] have been shown to be critical residues. One problem with the Arg112 mutants is that Arg112 also ion pairs with one P450cam heme propionate so it is difficult to separate the structural/redox potential effects of a residue at position 112 from its impact on electron transfer. Asp38 does not present such a problem because it is a surface residue that makes no intramolecular contacts. Thus, Asp38 replacements should have little effect on the Pdx structure. Our work on Asp38 is consistent with the earlier studies by other groups except, as with the Trp106 mutants, we studied the effects of Asp38 substitutions on both the first and second electron transfer steps, as well as the redox properties of the 2Fe2S cluster [10, 39]. Similar to W106A, the D38A mutation has no effect on the first kET but increases Kd, while kET for the second step decreases by ca. 360-fold with little change in Kd [10]. So like Trp106, Asp38 plays quite different roles in each electron transfer step. Although at first glance the Asp38-Arg112 ion pair appears to be critical, the D38N mutant suggests a more complex picture. The D38N replacement in Pdx lowers kET for the second step by only 6-fold, much less than the 360-fold decrease in kET observed for the D38A mutant. The Asn38 side chain is of similar length, can H-bond with Arg112 and, possibly, assist the effector role of Pdx in the second electron transfer step. Further, the Asp38-Arg112 pair is predicted by mixed quantum mechanics/molecular mechanics methods [40] to be part of the electron transfer pathway, although this study was restricted to Fe3+ P450cam. While the role of Asp38 in the first reductive step seems clear, its role in the second step is less so primarily because of the effector role of Pdx. Since Asp38 is so close to the P450cam-Pdx interface, it could directly participate in the Pdx-induced restructuring and/or play a more direct role as part of the electron transfer path. Finally, the complexity of the Asp38 mutational effects may arise from perturbations in the redox properties of the 2Fe2S cluster itself because, as we found, the D38N substitution affects the kinetic reversibility of redox transition in Pdx more profoundly than D38A or any other tested mutation [39].
5. Pdx-Pdr Redox Pair
Electron delivery to the P450 is, however, only half the story. The complete system involves another protein, Pdr. The electron flow is from NADH to the FAD of Pdr followed by two single electron transfers to the Pdx 2Fe2S center and finally from Pdx to the P450cam heme iron. Thus, Pdx must first pick up an electron from Pdr, dissociate, and then deliver an electron to P450cam or, in other words, operate as a shuttle. The Pdr-Pdx reaction has received much less attention than P450-redox partner interactions. For quite some time, studies have been limited to electron transfer kinetics [41], chemical modification to probe surface residues involved in complex formation [2, 29], and the energetics of binding using calorimetric methods [30], and could not be evaluated due to lack of structural information on Pdr and Pdx. Having solved both Pdr and Pdx x-ray structures [27, 42], we explored the Pdr-Pdx binding and electron transfer reaction in more detail. We first developed a hypothetical model of the Pdr-Pdx complex was generated and evaluated by mutating several interface residues of Pdx and analyzing the mutational effects on the Pdr to Pdx electron transfer [39]. Good agreement between the theoretical and experimental data allowed us to conclude that steric complementarity may be one of the factors assisting the Pdr-Pdx complex formation, and that bulky Tyr33, Arg66 and Trp106 in Pdx prevent its tight binding and facilitate dissociation upon reduction. Our results suggested also that the inter-protein electron transfer could proceed via different pathways that likely include Asp38 and Cys39 but do not involve Trp106. This initial effort has been followed by successful production of a functional covalent Pdr-Pdx complex [43], whose crystal structure was determined to 2.6 Å resolution [44] and experimentally tested [45].
In Figure 5, the Pdr-Pdx crystal structure is compared to our initial hypothetical model of the Pdr-Pdx electron transfer complex and two other known homologous crystal structures. The hypothetical model (Fig. 5B) does closely resemble the crystal structure (Fig. 5A) although the success of the docked model depends on one’s perspective. The computer-generated model has the correct docking orientation and inter-cofactor distance (12.3 vs. 12.0 Å in the x-ray structure) although the 2Fe2S cluster is translated about 5.0 Å. Perhaps part of the success in the computer docking approach is that Pdr and Pdx experience very little structural change upon complex formation. Figure 5 also shows the crystal structures of the covalently crosslinked Adr-Adx complex [46] and the complex formed between a ferredoxin and its reductase from Acidovorax sp. [47].
Figure 5.
Crystal structures of redox complexes analogous to the Pdr-Pdx complex. Pdr and its homologues are green while the redox partner is yellow. All complexes lead to very similar 2Fe2S-FAD distances but quite different protein-protein interfaces.
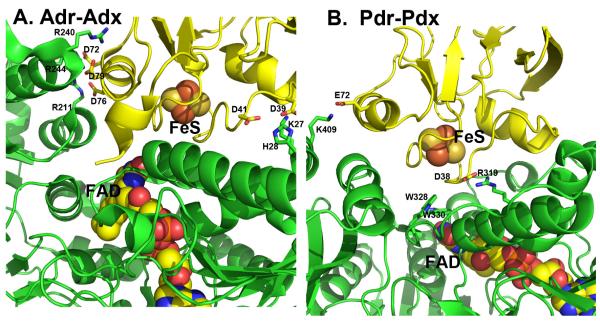
Since Adr-Adx and Pdr-Pdx are the closest homologues, it is instructive to compare their complexes in more detail to better understand the structural basis for some of the established biochemical differences. One of the most clearly defined differences in the two redox pairs is that the Adr-Adx interaction is strongly dependent on ionic strength [48] while the Pdr-Pdx interaction is not nearly as dependent [49]. A detailed thermodynamic analysis of the Pdr-Pdx complex formation led to the conclusion that the Pdr-Pdx interaction is dominated by non-bonded interactions and possibly H-bonds [30]. These results can be rationalized on the basis of the crystal structures. As shown in Figure 6A, Adx establishes 5 salt links with Adr, whereas the Pdr-Pdx pair has only two ionic interactions (Fig. 6B, Table 2). Also, the Pdr-Pdx interface is dominated by two Trp residues of Pdr, Trp328 and Trp330, which are about 3.7 Å from one of the 2Fe2S Cys ligands in Pdx and form a direct link between the flavin and iron-sulfur cofactors (Fig. 6B). The Adr-Adx complex has no aromatic groups at the interface and no similar direct connections between redox centers. Thus, one would predict from the structures that the Adr-Adx complex is dominated by ionic interactions while ionic interactions are not so important in the Pdr-Pdx system, precisely what the biochemical studies have shown.
Figure 6.
A comparison of the Adr-Adx and Pdr-Pdx complexes. Pdr and Adr are green while Pdx and Adx are yellow. Note that Adr-Adx has many more inter-protein ionic interactions while only Pdr-Pdx has aromatic groups bridging between 2Fe2S and FAD.
The Adr to Adx electron transfer rate is 12 s−1 [48] and from Pdr to Pdx is 235 s−1 [39]. Given the quite different experimental conditions for these estimates, a 20-fold variation in kET values is not large, especially considering that this translates into a modest 2 kcal/mol difference in activation energy. This indicates that the very distinct interface structures in these complexes do not contribute much to the difference in electron transfer rates. This perhaps is somewhat surprising since the Pdr-Pdx interface is rich in aromatic groups, often considered as excellent conduits for electron transfer. The more telling comparison is the theoretical electron transfer rate estimate for the Pdr-Pdx reaction, 2.7 × 105 s−1, which is three orders of magnitude larger than the experimental value [44]. This difference has been attributed to the large reorganizational energy and structural changes that accompany Pdx reduction [39, 44, 50] which are not taken into account in the theoretical kET estimates.
6. Experimental Evaluation of the Pdr-Pdx Crystal Structure
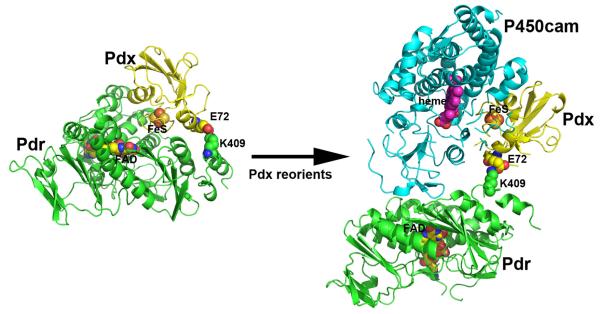
While the cross-linked Pdr-Pdx complex crystal structure is consistent with the mutagenesis and thermodynamic binding data, it also is important to test the various types of activities associated with the P450cam monooxygenase system. One of the most frequently used is the cytochrome c reductase activity assay, where the electron flow is NADH-to-Pdr-to-Pdx-to-cyt c. Functional comparison of the cross-linked complex with the free proteins is not straightforward since the cross-linked complex exhibits Michaelis-Menten type kinetics while a mixture of the proteins exhibits non-saturating second order kinetics. Therefore, it is necessary to compare the kcat/Km for the cross-linked complex with k2nd for the free proteins. Such comparison shows that the cross-liked complex reduces cyt c about 160-fold faster [43]. This increased rate is most likely due to the more efficient flow of electrons from NADH to Pdx in the cross-linked complex since binding and dissociation are not required. The cross-linked complex also supports the highly coupled hydroxylation of camphor catalyzed by P450cam but unlike the natural system, the rate of NADH oxidation and camphor consumption continues to increase with P450cam concentration and shows no signs of saturation. In the natural system, the camphor hydroxylation rate saturates at 15 μM min−1 at about 5 μM P450cam, while the rate for the cross-linked complex increases to 40 min−1 at the highest concentrations of P450cam tested, 15 μM. These results show that the cross-linked complex is functional but the electron transfer kinetics is quite different than in the natural system. This is expected because the establishment of only one bimolecular complex with P450cam is required for the cross-linked Pdr and Pdx, whereas in the natural system two different bimolecular complexes must be formed. Since there is strong evidence that the same surface of Pdx interacts with both Pdr and P450cam, the crosslinked Pdx must be able to reorient in order to bind P450cam. The schematic model shown in Figure 7 illustrates how this might work. The covalent bond formed between Glu and Lys is at the edge of the Pdr-Pdx complex interface so it is not difficult to envision how Pdx rotates in order to dock to P450cam or cyt c. Given the very high rate of cyt c reduction, the crosslink is not effecting the Pdx-cyt c interaction. In sharp contrast, the crosslinked complex does not support high rates of camphor hydroxylation unless the concentration of P450cam is high. This indicates that electron transfer to P450cam and the effector role of Pdx are not impaired by the covalent bond to Pdr but the Pdx-P450cam Kd is probably much higher.
Figure 7.
Possible model showing how Pdx might reorient in the covalent Pdr-Pdx complex in order to access the distal side of P450cam. Pdr is depicted in green, Pdx yellow, and P450cam cyan. The model was generated by rotation of Pdx around the crosslinked bond.
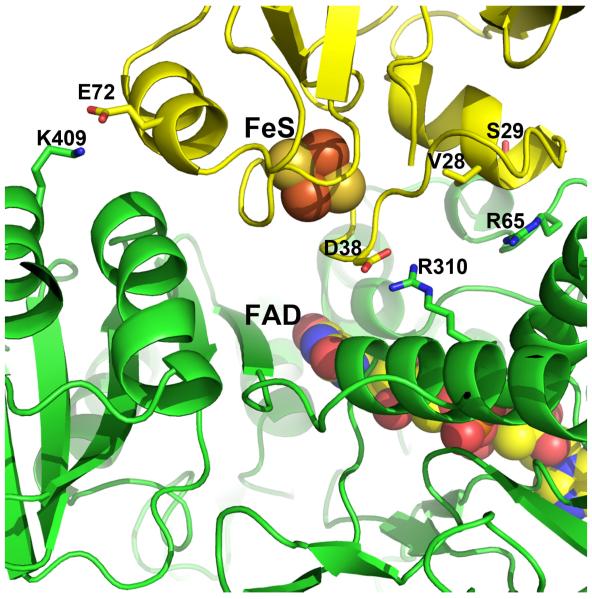
The specific interface interactions observed in the Pdr-Pdx crystal structure have been probed by mutagenesis [45]. The most interesting results were obtained with the Arg310 and Arg65 mutants of Pdr. As shown in Figure 8, both residues are predicted by the crystal structure to form direct ionic and/or H-bonding interactions with Pdx. Arg310 forms a bidentate salt bridge with Asp38 while the Arg65 guanidine group is H-bonded to the backbone atom of Val28 in Pdx. Changing Arg310 to either Ala or Glu lowers the rate of Pdr to Pdx electron transfer by 40- and 70-fold, respectively, but only the R310E mutation increases Kd, by a factor of 8, which is understandable since this substitution should cause charge repulsion with Asp38 in Pdx. The R65A mutation also lowers the rate of electron transfer (~20-fold) but does not effect Kd. These data indicate that the Arg310-Asp38 ionic interaction is not important in complex formation but is important in electron transfer. Since Arg310 is positioned too far from the flavin to be part of the electron transfer route, we suggested that this basic residue provides electrostatic steering during docking and optimally orients Pdx for electron transfer via charge-charge interactions with Asp38, whose side chain length and the negative charge are critical for establishing a productive Pdr-Pdx electron transfer complex [39]. Additionally, Asp38 is on the 2Fe2S-binding loop, which places the carboxyl group 6-7 Å from the metal center. This close proximity of a negative charge will very likely influence the redox properties of the 2Fe2S cluster. Simple electrostatic consideration suggests that Asp38 will stabilize the oxidized form of 2Fe2S. Ion pairing with Arg310 should significantly decrease such stabilization thereby lowering the activation free energy for 2Fe2S reduction. This might be one possible role of the Arg310-Asp38 pairing. That the D38N substitution in Pdx significantly affects the kinetic reversibility of the metal cluster oxidoreduction and drastically decreases the inter-protein electron transfer rate [39] suggests also that the carboxyl group may change the protonation state and couple electron and proton transfer. The effect of Arg65 mutants is a bit more difficult to understand since no charge-charge interactions are involved. However, positioned only 8.8 Å away from Asp38 in Pdx and buried at the interface free of bulk solvent, Arg65 could help attenuate the negative charge on Asp38 thereby promoting 2Fe2S reduction. In addition to the modest polar interactions with Asp38 of Pdx (Fig. 8), another suggested role of Arg65 is to help shape and buffer the Pdr-Pdx interface [45]. Arg65 effectively serves as a gate to prevent Pdx from penetrating too deeply at the interface, which both facilitates dissociation required for rapid turnover and helps to properly orient the redox partners for efficient electron transfer.
Figure 8.
Detailed view of the Pdr-Pdx interface. Key residues targeted for mutagenesis are highlighted.
7. Concluding Remarks
Although there is a paucity of crystal structures of P450 redox complexes, the available x-ray data together with extensive mutagenesis, kinetic, spectral, and computational approaches have provided some important advances and insights. We now are much closer to understanding the effector role of Pdx in the P450cam system. That the L358P substitution mimics the various spectral perturbations that occur when Pdx binds to P450cam suggests that the L358P mutant may be a good model for the P450cam-Pdx complex. The picture that emerges from this assumption is fairly simple. The binding of Pdx “pushes” on the proximal side of the heme resulting in structural changes in the distal O2 and substrate binding sites that allow the critical catalytic waters to be positioned for proper proton transfer to the Fe-linked O2. This effector role is only important in the second electron transfer step, reduction of the Fe2+ P450-O2 complex. It thus is noteworthy that certain key mutants in Pdx behave very differently in the first and second electron transfer steps [10]. In particular, the Asp38 to Ala conversion was found to have little effect on the first electron transfer event but nearly eliminates the second, whereas Pdx D38N retains about 20% of wild type rates for the Fe2+P450cam-O2 reduction. The W106A mutation also inactivates the second reductive step but the W106F variant retains about 60% of this activity. These results make sense in light of the proposed effector role of Pdx. If Pdx binding results in distal pocket restructuring via heme movement, then it is important to have close contacts between Pdx and P450cam at it enables Pdx to “push” on the proximal side of P450cam and, consequently, induce structural changes in the distal pocket. In this scenario, Asp38 and Trp106 effectively fill in the space required for direct contacts at the interface and thus are key players in the Pdx-induced reorganization.
The available Pdr-Pdx and Adr-Adx complex structures, on the other hand, provide important insights on inter-protein electron transfer. The structures correlate very well with the biochemical studies. The Adr-Adx complex is much richer in intermolecular ionic interactions and, as might be expected, is more dependent on ionic strength than the Pdr-Pdx complex. The dissimilarities in the protein-protein interface affect the FAD-to-2Fe2S electron transfer rates (20-fold difference). That the experimental electron transfer rates for both complexes are orders of magnitude lower that the computed values [39, 44, 48, 51] implies that some process other than electron transfer is limiting. One such process could be redox-linked conformational reorganization in Pdx [50]. The interesting possibility here is that the redox partner binding event itself might help to trigger the conformational changes required for electron transfer, and that the interface designed by Nature might promote the required rearrangements with the precise electron transfer pathway being of secondary importance, as long as the redox cofactors are close enough.
If subtle structural changes are indeed critical in redox complexes, then what we need are very accurate structures that carefully define the conformational differences between various redox states. Here we face a serious conundrum since it has become increasingly clear that x-rays generate hydrated electrons that rapidly reduce redox centers in the proteins even at cryogenic temperatures [52]. Nevertheless, using careful data collection protocols with single crystal spectroscopy it is possible to obtain structures of metalloproteins in a well defined redox state [50][53]. As these techniques and approaches become more wide spread, we are optimistic that it will be possible to probe in more detail the role of gating and redox dependent structural changes in inter-protein electron transfer reactions.
Another practical outcome of our work is demonstration that chemical cross-linking can be successfully used to trap unstable redox complexes in a catalytically active state. We should admit though that there is a certain amount of luck involved. In the Pdr-Pdx complex, a peripheral Glu and Lys pair just happened to be in an ideal position and close enough to form an EDC-mediated cross-link [43]. Suppose only Arg residues are involved in the inter-protein contacts or the interacting charged groups are in the vicinity of redox centers? In this case, simple cross-linking via carboxyl and primary amine groups will not be successful and when it is, the conjugates could be non-functional due to motional restrictions. A possible solution in this situation could be utilization of computationally docked models for designing specific cross-linking sites. Incorporation of Cys is an alternative choice since there are many Cys homo-bifunctional conjugating reagents of varying length. To avoid formation of non-productive conjugates, the hypothetical models should be experimentally tested prior cross-linking attempts, as we have done with the Pdr-Pdx complex [39][43][45].
Perhaps the most promising experimental approach to studying electron transfer complexes is NMR spectroscopy. Chemical shift mapping requires only HSQC data and not a full NMR structure to pinpoint which surface residues are most perturbed by complex formation. This approach has been successfully utilized to study several redox pairs [54-57]. It also is possible to use relaxation techniques to estimate the distance from surface residues to the heme iron. With enough such restraints it is possible to develop a reasonable model of the complex. Perhaps the major drawback to this approach is that regions near paramagnetic metal centers usually are not NMR visible so the most interesting part of the complex, the interface, may be NMR silent. One way around this problem is to label one of the partners with a paramagnetic lanthanide and map out distances using pseudocontact shift and paramagnetic relaxation enhancement methods. This strategy has recently been applied for the Adr-Adx noncovalent complex [58] and the structure derived from this study is quite close to the crystal structure of the crosslinked complex. This bodes well for the use of chemical crosslinking, NMR, and computational docking models for studying redox complexes.
P450cam monooxygenase system
Crystal structure of the putidaredoxin (Pdx)-putidaredoxin reductase (Pdr) complex
The structural basis for the effector role of Pdx on P450cam camphor hydroxylation
Mutagenesis and kinetics to test the validity of structural models
Acknowledgements
Work in our lab has been supported by NIH grants GM33688 (TLP) and GM067637 (IFS)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Proc Natl Acad Sci U S A. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Geren L, Tuls J, O’Brien P, Millett F, Peterson JA. J Biol Chem. 1986;261:15491–15495. [PubMed] [Google Scholar]
- [3].Tsai RL, Gunsalus IC, Dus K. Biochem Biophys Res Commun. 1971;45:1300–1306. doi: 10.1016/0006-291x(71)90160-4. [DOI] [PubMed] [Google Scholar]
- [4].Tyson CA, Lipscomb JD, Gunsalus IC. J Biol Chem. 1972;247:5777–5784. [PubMed] [Google Scholar]
- [5].Hawkes DB, Adams GW, Burlingame AL, de Montellano P.R. Ortiz, De Voss JJ. J Biol Chem. 2002;277:27725–27732. doi: 10.1074/jbc.M203382200. [DOI] [PubMed] [Google Scholar]
- [6].Meharenna YT, Li H, Hawkes DB, Pearson AG, De Voss J, Poulos TL. Biochemistry. 2004;43:9487–9494. doi: 10.1021/bi049293p. [DOI] [PubMed] [Google Scholar]
- [7].Kimmich N, Das A, Sevrioukova I, Meharenna Y, Sligar SG, Poulos TL. J Biol Chem. 2007;282:27006–27011. doi: 10.1074/jbc.M703790200. [DOI] [PubMed] [Google Scholar]
- [8].Unno M, Shimada H, Toba Y, Makino R, Ishimura Y. J Biol Chem. 1996;271:17869–17874. doi: 10.1074/jbc.271.30.17869. [DOI] [PubMed] [Google Scholar]
- [9].Koga H, Sagara Y, Yaoi T, Tsujimura M, Nakamura K, Sekimizu K, Makino R, Shimada H, Ishimura Y, Yura K, et al. FEBS Lett. 1993;331:109–113. doi: 10.1016/0014-5793(93)80307-g. [DOI] [PubMed] [Google Scholar]
- [10].Kuznetsov VY, Poulos TL, Sevrioukova IF. Biochemistry. 2006;45:11934–11944. doi: 10.1021/bi0611154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bell SG, Hoskins N, Xu F, Caprotti D, Rao Z, Wong LL. Biochem Biophys Res Commun. 2006;342:191–196. doi: 10.1016/j.bbrc.2006.01.133. [DOI] [PubMed] [Google Scholar]
- [12].Xu F, Bell SG, Peng Y, Johnson EO, Bartlam M, Rao Z, Wong LL. Proteins. 2009;77:867–880. doi: 10.1002/prot.22510. [DOI] [PubMed] [Google Scholar]
- [13].Bell SG, Xu F, Johnson EO, Forward IM, Bartlam M, Rao Z, Wong LL. J Biol Inorg Chem. 2010;15:315–328. doi: 10.1007/s00775-009-0604-7. [DOI] [PubMed] [Google Scholar]
- [14].Sligar SG, Debrunner PG, Lipscomb JD, Namtvedt MJ, Gunsalus IC. Proc Natl Acad Sci U S A. 1974;71:3906–3910. doi: 10.1073/pnas.71.10.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bell SG, Wong LL. Biochem Biophys Res Commun. 2007;360:666–672. doi: 10.1016/j.bbrc.2007.06.119. [DOI] [PubMed] [Google Scholar]
- [16].Yang W, Bell SG, Wang H, Zhou W, Hoskins N, Dale A, Bartlam M, Wong LL, Rao Z. J Biol Chem. 2010 doi: 10.1074/jbc.M110.118349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bell SG, Dale A, Rees NH, Wong LL. Appl Microbiol Biotechnol. 2010;86:163–175. doi: 10.1007/s00253-009-2234-y. [DOI] [PubMed] [Google Scholar]
- [18].Tosha T, Yoshioka S, Ishimori K, Morishima I. J Biol Chem. 2004;279:42836–42843. doi: 10.1074/jbc.M404216200. [DOI] [PubMed] [Google Scholar]
- [19].Tosha T, Yoshioka S, Takahashi S, Ishimori K, Shimada H, Morishima I. J Biol Chem. 2003;278:39809–39821. doi: 10.1074/jbc.M304265200. [DOI] [PubMed] [Google Scholar]
- [20].Sevrioukova IF, Hazzard JT, Tollin G, Poulos TL. J Biol Chem. 1999;274:36097–36106. doi: 10.1074/jbc.274.51.36097. [DOI] [PubMed] [Google Scholar]
- [21].Mandai T, Fujiwara S, Imaoka S. Febs J. 2009;276:2416–2429. doi: 10.1111/j.1742-4658.2009.06974.x. [DOI] [PubMed] [Google Scholar]
- [22].Macedo-Ribeiro S, Martins BM, Pereira PJ, Buse G, Huber R, Soulimane T. J Biol Inorg Chem. 2001;6:663–674. doi: 10.1007/s007750100243. [DOI] [PubMed] [Google Scholar]
- [23].Tovchigrechko A, Vakser IA. Nucleic Acids Res. 2006;34:W310–314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Proc Natl Acad Sci U S A. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Puchkaev AV, Wakagi T, de Montellano P.R. Ortiz. J Am Chem Soc. 2002;124:12682–12683. doi: 10.1021/ja0282036. [DOI] [PubMed] [Google Scholar]
- [26].Frazao C, Aragao D, Coelho R, Leal SS, Gomes CM, Teixeira M, Carrondo MA. FEBS Lett. 2008;582:763–767. doi: 10.1016/j.febslet.2008.01.041. [DOI] [PubMed] [Google Scholar]
- [27].Sevrioukova IF, Garcia C, Li H, Bhaskar B, Poulos TL. J Mol Biol. 2003;333:377–392. doi: 10.1016/j.jmb.2003.08.028. [DOI] [PubMed] [Google Scholar]
- [28].Pochapsky TC, Lyons TA, Kazanis S, Arakaki T, Ratnaswamy G. Biochimie. 1996;78:723–733. doi: 10.1016/s0300-9084(97)82530-8. [DOI] [PubMed] [Google Scholar]
- [29].Aoki M, Ishimori K, Morishima I. Biochim Biophys Acta. 1998;1386:157–167. doi: 10.1016/s0167-4838(98)00094-6. [DOI] [PubMed] [Google Scholar]
- [30].Aoki M, Ishimori K, Fukada H, Takahashi K, Morishima I. Biochim Biophys Acta. 1998;1384:180–188. doi: 10.1016/s0167-4838(98)00017-x. [DOI] [PubMed] [Google Scholar]
- [31].Holden M, Mayhew M, Bunk D, Roitberg A, Vilker V. J Biol Chem. 1997;272:21720–21725. doi: 10.1074/jbc.272.35.21720. [DOI] [PubMed] [Google Scholar]
- [32].Nagano S, Shimada H, Tarumi A, Hishiki T, Kimata-Ariga Y, Egawa T, Suematsu M, Park SY, Adachi S, Shiro Y, Ishimura Y. Biochemistry. 2003;42:14507–14514. doi: 10.1021/bi035410p. [DOI] [PubMed] [Google Scholar]
- [33].Unno M, Christian JF, Sjodin T, Benson DE, Macdonald ID, Sligar SG, Champion PM. J Biol Chem. 2002;277:2547–2553. doi: 10.1074/jbc.M108917200. [DOI] [PubMed] [Google Scholar]
- [34].Nagano S, Tosha T, Ishimori K, Morishima I, Poulos TL. J Biol Chem. 2004;279:42844–42849. doi: 10.1074/jbc.M404217200. [DOI] [PubMed] [Google Scholar]
- [35].Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- [36].Nagano S, Poulos TL. J Biol Chem. 2005;280:31659–31663. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- [37].Schenkman JB, Jansson I. Pharmacol Ther. 2003;97:139–152. doi: 10.1016/s0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]
- [38].Davies MD, Qin L, Beck JL, Suslick KS, Koga H, Horiuchi T, Sligar SG. J Amer Chem Soc. 1990;112:7396–7398. [Google Scholar]
- [39].Kuznetsov VY, Blair E, Farmer PJ, Poulos TL, Pifferitti A, Sevrioukova IF. J Biol Chem. 2005;280:16135–16142. doi: 10.1074/jbc.M500771200. [DOI] [PubMed] [Google Scholar]
- [40].Wallrapp F, Masone D, Guallar V. J Phys Chem A. 2008;112:12989–12994. doi: 10.1021/jp803538u. [DOI] [PubMed] [Google Scholar]
- [41].Roome PW, Peterson JA. Arch Biochem Biophys. 1988;266:41–50. doi: 10.1016/0003-9861(88)90234-2. [DOI] [PubMed] [Google Scholar]
- [42].Sevrioukova IF, Li H, Poulos TL. J Mol Biol. 2004;336:889–902. doi: 10.1016/j.jmb.2003.12.067. [DOI] [PubMed] [Google Scholar]
- [43].Churbanova IY, Poulos TL, Sevrioukova IF. Biochemistry. 2010;49:58–67. doi: 10.1021/bi901872s. [DOI] [PubMed] [Google Scholar]
- [44].Sevrioukova IF, Poulos TL, Churbanova IY. J Biol Chem. 2010;285:13616–13620. doi: 10.1074/jbc.M110.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sevrioukova IF, Poulos TL. Biochemistry. 49:5160–5166. doi: 10.1021/bi100626f. [DOI] [PubMed] [Google Scholar]
- [46].Muller JJ, Lapko A, Bourenkov G, Ruckpaul K, Heinemann U. J Biol Chem. 2001;276:2786–2789. doi: 10.1074/jbc.M008501200. [DOI] [PubMed] [Google Scholar]
- [47].Senda M, Kishigami S, Kimura S, Fukuda M, Ishida T, Senda T. J Mol Biol. 2007;373:382–400. doi: 10.1016/j.jmb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- [48].Lambeth JD, Seybert DW, Kamin H. J Biol Chem. 1979;254:7255–7264. [PubMed] [Google Scholar]
- [49].Roome PW, Jr., Philley JC, Peterson JA. J Biol Chem. 1983;258:2593–2598. [PubMed] [Google Scholar]
- [50].Sevrioukova IF. J Mol Biol. 2005;347:607–621. doi: 10.1016/j.jmb.2005.01.047. [DOI] [PubMed] [Google Scholar]
- [51].Muller EC, Lapko A, Otto A, Muller JJ, Ruckpaul K, Heinemann U. Eur J Biochem. 2001;268:1837–1843. [PubMed] [Google Scholar]
- [52].Hersleth HP, Hsiao YW, Ryde U, Gorbitz CH, Andersson KK. Chem Biodivers. 2008;5:2067–2089. doi: 10.1002/cbdv.200890189. [DOI] [PubMed] [Google Scholar]
- [53].Meharenna YT, Doukov T, Li H, Soltis SM, Poulos TL. Biochemistry. 2010;49:2984–2986. doi: 10.1021/bi100238r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Crowley PB, Otting G, Schlarb-Ridley BG, Canters GW, Ubbink M. J Am Chem Soc. 2001;123:10444–10453. doi: 10.1021/ja0112700. [DOI] [PubMed] [Google Scholar]
- [55].Guiles RD, Sarma S, DiGate RJ, Banville D, Basus VJ, Kuntz ID, Waskell L. Nat Struct Biol. 1996;3:333–339. doi: 10.1038/nsb0496-333. [DOI] [PubMed] [Google Scholar]
- [56].Morelli X, Dolla A, Czjzek M, Palma PN, Blasco F, Krippahl L, Moura JJG, Guerlesquin F. Biochemistry. 2000;39:2530–2537. doi: 10.1021/bi992306s. [DOI] [PubMed] [Google Scholar]
- [57].Ubbink M, Ejdeback M, Karlsson BG, Bendall DS. Structure. 1998;6:323–335. doi: 10.1016/s0969-2126(98)00035-5. [DOI] [PubMed] [Google Scholar]
- [58].Keizers PH, Mersinli B, Reinle W, Donauer J, Hiruma Y, Hannemann F, Overhand M, Bernhardt R, Ubbink M. Biochemistry. 2010;49:6846–6855. doi: 10.1021/bi100598f. [DOI] [PubMed] [Google Scholar]
- [59].Hintz MJ, Peterson JA. J Biol Chem. 1981;256:6721–6728. [PubMed] [Google Scholar]
- [60].Reipa V, Holden MJ, Mayhew MP, Vilker VL. Biochim Biophys Acta. 2000;1459:1–9. doi: 10.1016/s0005-2728(00)00108-0. [DOI] [PubMed] [Google Scholar]
- [61].Schiffler B, Zollner A, Bernhardt R. J Biol Chem. 2004;279:34269–34276. doi: 10.1074/jbc.M402798200. [DOI] [PubMed] [Google Scholar]
- [62].Huang YY, Hsu DK, Kimura T. Biochem Biophys Res Commun. 1983;115:116–122. doi: 10.1016/0006-291x(83)90977-4. [DOI] [PubMed] [Google Scholar]
- [63].Guengerich FP, Johnson WW. Biochemistry. 1997;36:14741–14750. doi: 10.1021/bi9719399. [DOI] [PubMed] [Google Scholar]
- [64].Munro AW, Noble MA, Robledo L, Daff SN, Chapman SK. Biochemistry. 2001;40:1956–1963. doi: 10.1021/bi001718u. [DOI] [PubMed] [Google Scholar]
- [65].Louerat-Oriou B, Perret A, Pompon D. Eur J Biochem. 1998;258:1040–1049. doi: 10.1046/j.1432-1327.1998.2581040.x. [DOI] [PubMed] [Google Scholar]