Abstract
Natural Killer (NK) cells play an important role in the control of viral infections, recognizing virally infected cells through a variety of activating and inhibitory receptors1–3. Epidemiological and functional studies have recently suggested that NK cells can also contribute to the control of HIV-1 infection through recognition of virally infected cells by both activating and inhibitory Killer Immunoglobulin-like receptors (KIRs)4–7. However, it remains unknown whether NK cells can directly mediate antiviral immune pressure in vivo in humans. Here we describe KIR-associated amino acid polymorphisms in the HIV-1 sequence of chronically infected individuals on a population level. We show that these KIR-associated HIV-1 sequence polymorphisms can enhance the binding of inhibitory KIRs to HIV-1-infected CD4+ T cells, leading to reduced antiviral activity of KIR+ NK cells. These data demonstrate that KIR+ NK cells can place immunological pressure on HIV-1, and that the virus can evade such NK cell mediated immune pressure by selecting for sequence polymorphisms, as previously described for virus-specific T cells and neutralizing antibodies8. NK cells might therefore play a previously underappreciated role in contributing to viral evolution.
We hypothesized that HIV-1 can evade NK cell recognition through the selection of sequence polymorphisms within regions targeted by KIRs, and that KIR-associated polymorphisms within the HIV-1 sequence can be identified on a population level. To test this hypothesis, we examined the relationship between KIR genotypes and HIV-1 polymorphisms in a cohort of 91 untreated chronically HIV-1-infected individuals (Supplemental Table 1) in whom full-length HIV-1 sequences were determined, and HLA class I associated polymorphisms were previously described9. We used a decision-tree approach which corrects for phylogenetic structure among the sequences and allows for a multivariate analysis to identify KIR-associated sequence polymorphisms10. This analysis led to the identification of 22 positions within the HIV-1 genome in which amino acid polymorphisms were significantly associated with the presence of a specific KIR gene (Table 1 and supplemental figure 1). Taken together, these data show that HIV-1 can adapt to host KIR genotypes on a population level.
Table 1.
KIR footprints in HIV-1 sequence
| Protein | aa position | KIR-association | Consensus aa* | q-value | |
|---|---|---|---|---|---|
| 1 | Gag | 93 | KIR2DS3 | E | 0.1389 |
| 2 | Gag | 138 | KIR2DL2 | L | 0.1852 |
| 3 | Gag | 138 | KIR2DS2 | L | 0.1852 |
| 4 | Gag | 371 | KIR2DS5 | T | 0 |
| 5 | Gag | 389 | KIR3DS1 | T | 0 |
| 6 | Gag | 479 | KIR2DS1 | I | 0 |
| 7 | Vpr | 37 | KIR2DS3 | I | 0.0909 |
| 8 | Tat | 3 | KIR2DL2 | S | 0.0246 |
| 9 | Tat | 3 | KIR2DS2 | S | 0 |
| 10 | Tat | 3 | KIR3DS1 | S | 0.1311 |
| 11 | Tat | 9 | KIR2DS3 | P | 0.1311 |
| 12 | Tat | 28 | KIR2DS1 | V | 0.1311 |
| 13 | Tat | 28 | KIR2DS5 | V | 0.1339 |
| 14 | Vpu | 3 | KIR2DL3 | S | 0.0833 |
| 15 | Vpu | 71 | KIR2DL2 | M | 0.125 |
| 16 | Vpu | 74 | KIR2DL2 | H | 0.1354 |
| 17 | Env | 17 | KIR2DL2 | W | 0 |
| 18 | Env | 20 | KIR2DL2 | M | 0.1667 |
| 19 | Env | 46 | KIR3DS1 | K | 0.1667 |
| 20 | Env | 347 | KIR2DS1 | L | 0.2 |
| 21 | Env | 595 | KIR2DS1 | I | 0.2 |
| 22 | Nef | 9 | KIR2DL2 | S | 0.0833 |
HIV-1 consensus sequence in 91 study subjects
To assess the consequences of these KIR-associated amino acid sequence polymorphisms within HIV-1 on NK cell mediated recognition of HIV-1 infected cells, we initially evaluated observed polymorphisms that occurred within a region of HIV-1 encoding for an overlapping segment that spans the C-terminal end of Vpu and the N-terminal end of Env (Polymorphisms #15–18 in Table 1 and Supplemental Figure 1). We selected this region as these polymorphisms were present within both reading frames at significantly higher frequencies in individuals that possessed at least one copy of the KIR2DL2 gene compared to individuals that did not (Table 2, and Supplemental Tables 2–3). In amino acid positions 71 and 74 of Vpu, HIV-1 sequences derived from KIR2DL2+ individuals encoded for a methionine (M) in position Vpu71 and a histidine (H) in position Vpu74 in over 70% of cases. Because of the overlapping Vpu/Env coding region, these positions of Vpu corresponded to a tryptophan (W) in position Env17 and a methionine (M) in position Env20, respectively. The Vpu71M/74H (Env17W/20M) sequence was significantly less frequent in HIV-1-infected individuals that did not encode for KIR2DL2 (p < 0.0001). Furthermore, the presence of the Vpu71M polymorphism was in strong linkage disequilibrium with the Vpu74H polymorphism (p = 3.17−12). Taken together, these data demonstrate a significant enrichment for HIV-1 viruses containing the Vpu71M/74H (Env17W/20M) polymorphism in individuals encoding for KIR2DL2.
Table 2.
Frequency of amino acid polymorphisms among KIR2DL2+ and KIR2DL2- subjects
|
VPU aa position |
E 70 |
M | G | H | H | A | P | W | D |
V 79 |
Percent of indivdiuals |
Number of Individuals |
| KIR2DL2+ | . | . | . | . | . | . | . | . | . | . | 72 | (34/47) |
| . | L/Q/- | . | . | D | . | . | . | . | . | 24 | (11/47) | |
| . | R | . | . | L | . | . | . | . | . | 4 | (2/47) | |
| KIR2DL2- | ./D | . | ./E | ./Q | . | ./D | . | . | . | . | 32 | (14/44) |
| ./H/M | N/V/Q/G | ./A | . | D/R | . | ./L | ./G | ./V | ./I | 34 | (15/44) | |
| ./H | R | . | ./R | L | . | ./L | ./G/R/L | ./V | ./I | 34 | (15/44) | |
|
ENV aa position |
R 16 |
W | G | T | M | L | L | G | M |
L 25 |
Percent of indivdiuals |
Number of Individuals |
| KIR2DL2+ | . | . | . | . | . | . | . | . | . | . | 85 | (40/47) |
| ./K | R/. | . | I | T/. | . | . | . | . | . | 11 | (5/47) | |
| ./K | G | . | I/A | L | ./F | . | . | ./L/I | . | 4 | (2/47) | |
| KIR2DL2- | . | . | . | ./I | . | . | . | . | . | . | 43 | (19/44) |
| . | R/M | . | ./I | ./V | . | . | . | . | . | 23 | (10/44) | |
| ./K | G | . | ./I | L | . | . | . | . | . | 34 | (15/44) | |
|
GAG aa position |
I 134 |
V | Q | N | L | Q | G | Q | M |
V 143 |
Percent of indivdiuals |
Number of Individuals |
| KIR2DL2+ | . | . | . | . | . | . | . | . | . | ./I | 68 | (32/47) |
| ./V | . | . | . | M/V/A | . | . | . | . | ./I | 32 | (15/47) | |
| . | . | . | . | I | . | . | . | . | . | 0 | (0/47) | |
| KIR2DL2- | . | . | ./R | . | . | . | . | . | . | . | 57 | (25/44) |
| . | . | ./R | . | ./M/V/A | . | . | . | . | . | 25 | (11/44) | |
| . | . | . | . | I | . | . | . | . | . | 18 | (8/44) | |
|
NEF aa position |
W 5 |
S | K | R | S | V | V | G | W |
P 14 |
Percent of indivdiuals |
Number of Individuals |
| KIR2DL2+ | ./X | . | . | ./X | . | ./X | ./X | ./X | . | ./X | 75 | (35/47) |
| ./X | . | . | ./X | E/R/N/C/L/- | ./X | ./X | ./E | . | ./X | 23 | (11/47) | |
| . | . | . | . | K | E | N | . | . | S | 2 | (1/47) | |
| KIR2DL2- | ./C | . | ./R | ./X | . | ./X | ./X | ./X | . | ./X | 52 | (23/44) |
| ./C | . | ./R | ./X | ./P/R/I/G/W/ | ./X | ./X | ./X | . | ./X | 27 | (12/44) | |
| . | . | . | ./X | K | ./X | ./X | ./D | . | ./S | 21 | (9/44) | |
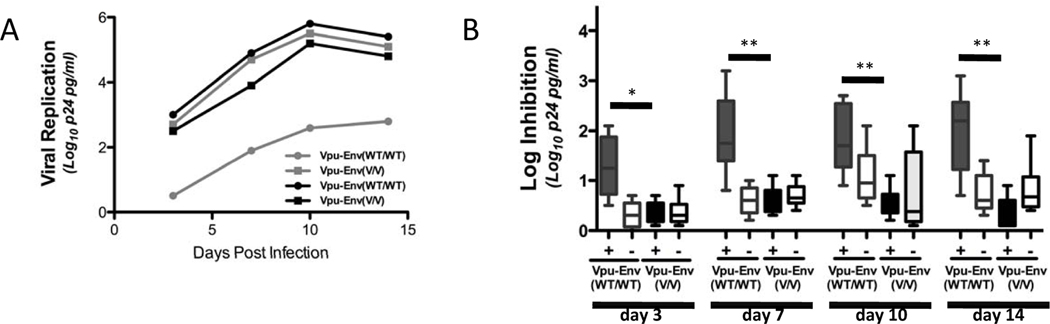
To determine the functional consequences of these KIR2DL2-associated polymorphisms within HIV-1 Vpu/Env, we constructed HIV-1 viral variants, using an NL4-3 backbone11–12, that either encoded the Vpu71M/74H (Env17W/20M) sequence observed in KIR2DL2+ individuals (referred to as Vpu-EnvV/V, with “V” standing for variant), or the Vpu71R/74L (Env17G/20L) sequence that was most commonly observed in KIR2DL2neg subjects (referred to as Vpu-EnvWT/WT). No significant differences were observed in the ability of the viral variants to replicate in primary CD4+ T cells in vitro (Supplemental Figure 2A). We subsequently assessed whether the different viral variants had any impact on NK cell recognition and/or antiviral activity. Primary CD4+ T cells were infected with viruses either containing the Vpu-EnvV/V or the Vpu-EnvWT/WT sequence, and then placed in coculture with autologous NK cells derived from KIR2DL2+ or KIR2DL2neg HIV-1 negative individuals (Figure 1). Both viruses replicated well in the presence of KIR2DL2neg NK cells, with less than half a log inhibition of viral replication in the presence of NK cells compared to replication in CD4+ T cells alone (Figure 1A and 1 B). In contrast, the Vpu-EnvWT/WT virus was significantly inhibited by NK cells derived from KIR2DL2+, but not KIR2DL2neg subjects (Figure 1B). Taken together, the Vpu-EnvV/V virus containing polymorphisms that were strongly associated with the presence of KIR2DL2 on the population level was not inhibited by KIR2DL2+ NK cells in vitro, while the “wild-type” variant (Vpu-EnvWT/WT) rarely observed in KIR2DL2+ individuals was strongly inhibited by KIR2DL2+ NK cells, consistent with the selection of Vpu-EnvV/V viruses in KIR2DL2+ individuals.
Figure 1. KIR2DL2 associated sequence polymorphisms result in a loss of NK cell inhibition of HIV replication in vitro.
(A) The Vpu-EnvWT/WT virus was inhibited more robustly by NK cells derived from a KIR2DL2+ individual (gray lines) compared to the Vpu-EnvV/V virus. NK cells derived from a KIR2DL2neg (black lines) individual did not inhibit either virus. (B) Similarly, the Vpu-EnvWT/WT virus was inhibited significantly more strongly by NK cells derived from individuals that expressed KIR2DL2 (dark grey bars, n=6) than the Vpu-EnvV/V virus (black bars). NK cells derived from individuals that did not express KIR2DL2 (white bars, n=6) did not significantly inhibit either virus. * p < 0.05; ** p < 0.005; + KIR2DL2pos; - KIR2DL2neg.
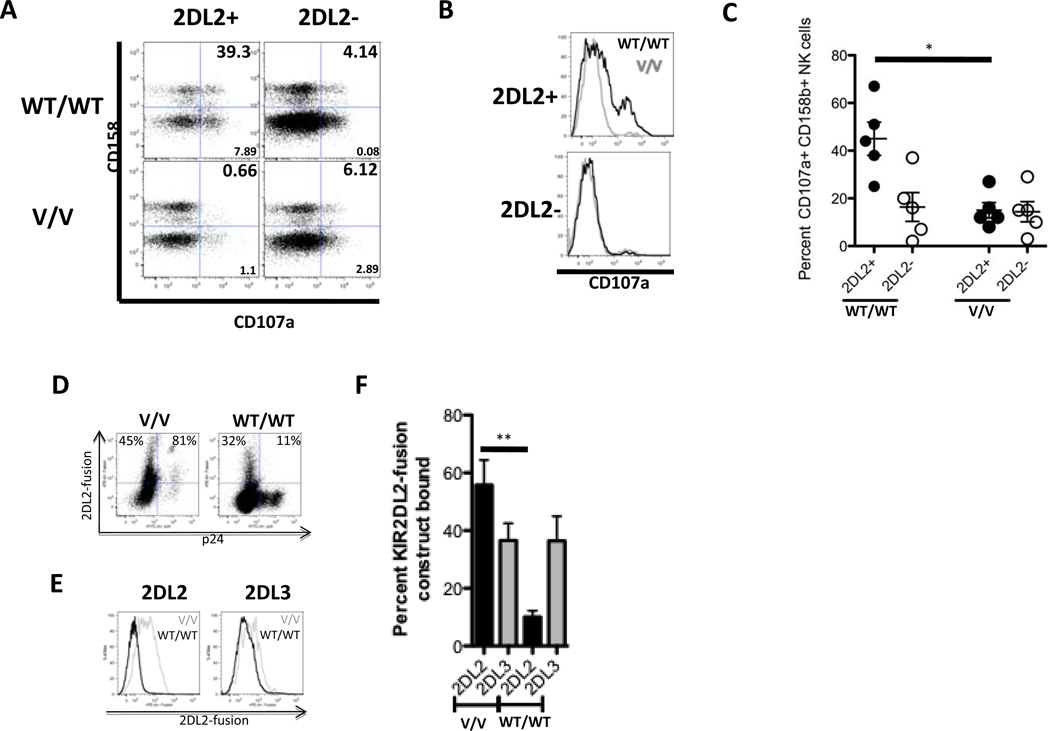
To further elucidate the mechanism of KIR2DL2+ NK cell-mediated inhibition of replication of Vpu-EnvWT/WT viruses, we monitored the induction of CD107a expression on NK cells in vitro following stimulation with autologous CD4+ T cells infected with either the Vpu-EnvWT/WT or the Vpu-EnvV/V virus (Figure 2A–C). In line with the viral inhibition data, CD158b+ NK cells from KIR2DL2+ individuals were strongly activated by CD4+ T cells infected with the Vpu-EnvWT/WT virus, but not by CD4+ T cells infected with the Vpu-EnvV/V virus (Figure 2A–C). Taken together, these data are consistent with a model in which the inhibitory NK cell receptor KIR2DL2 does not bind to cells infected with HIV-1 strains containing the Vpu-EnvWT/WT sequence, but can bind to cells infected with HIV-1 Vpu-EnvV/V, providing a strong inhibitory signal to KIR2DL2+ NK cells thereby protecting cells infected with Vpu-EnvV/V viruses from NK cell lysis.
Figure 2. Amino acid polymorphisms at positions 71/74 in VPU inhibit KIR2DL2, but not KIR2DL3, recognition and binding.
The flow cytometric plots depict the frequency of total CD158b (KIR2DL2/2DL3/2DS2)+ NK cells that degranulated following coculture with autologous CD4+ T cells infected with the Vpu-EnvWT/WT virus or the Vpu-EnvV/V virus for 2 representative subjects (left 2DL2+ subjects; right 2DL2- subject; dot plots (A)). Furthermore, the frequency of degranulating CD158+ NK cells of the total CD158+ NK cell population is also represented for both the KIR2DL2+ (top) and KIR2DL2- (bottom) donor in histograms for both the WT/WT (black line) and the V/V virus (grey line) (B). Figure 2 C shows the combined data for NK cell degranulation in 2DL2+ (n=5) and 2DL2- (n=5) individuals. *p < 0.05. The dot plots in figure 2D depict the staining pattern of a KIR2DL2-fusion construct on HIV-infected CD4+ T cells from an HLA-C1/C2 heterozygous donor, and the histogram in figure 2E demonstrates the staining of KIR2DL2- and KIR2DL3-IgG fusion constructs on the same donors for the 2 viral variants. In figure 2F KIR2DL2-IgG- (black) and KIR2DL3-IgG-fusion construct (grey) binding data are summarized for a total of 5 different HLA-C1 heterozygous CD4+ T cell donors for the 2 viral variants in the bar graph. ** p < 0.005.
KIR2DL2 segregates as an allele of the same locus with KIR2DL313, and is in strong linkage disequilibrium with KIR2DS2 (Wn = 0.976, p < 0.001). All but three individuals in our cohort that expressed KIR2DL2 also expressed KIR2DS2; however, these three individuals also encoded for the Vpu-EnvV/V polymorphism, suggesting that KIR2DL2, and not KIR2DS2 is responsible for the association with this polymorphism. To test whether KIR2DL2 was directly involved in the recognition of the viral variants, a KIR2DL2-Ig fusion construct was used to assess whether the Vpu-EnvV/V polymorphism modulated the interaction of KIR2DL2 with HIV-1-infected CD4+ T cells. The KIR2DL2 fusion construct bound robustly to all HIV-uninfected CD4+ T cells and also stained CD4+ T cells infected with the HIV-1 Vpu-EnvV/V variant significantly better than CD4+ T cells from the same donor infected with the Vpu-EnvWT/WT variant (Figure 2D–F). In contrast, the binding of a KIR2DL3 fusion construct to HIV-1-infected CD4+ T cells was not affected by the KIR2DL2-associated polymorphism (Figures 2E–F). HLA-C group 1 and group 2 molecules serve as the ligands for the inhibitory receptor KIR2DL2, which has been shown to bind with greater affinity for HLA-C group 113. Staining with the KIR2DL2 fusion construct was consistent with these results, and highest using cells from individuals homozygous for HLA-C1 group (Supplemental Figure 3). In line with these binding data, the Vpu-EnvV/V polymorphisms were significantly enriched in KIR2DL2+ individuals homozygous for HLA-C group 1 (p = 0.008 for Vpu71M and p = 0.01 for Vpu74H). Taken together, these data suggest that the Vpu-EnvV/V polymorphism enhances the ability of the inhibitory receptor KIR2DL2 to bind to HIV-1-infected cells, in particular those expressing the higher affinity ligands for KIR2DL2.
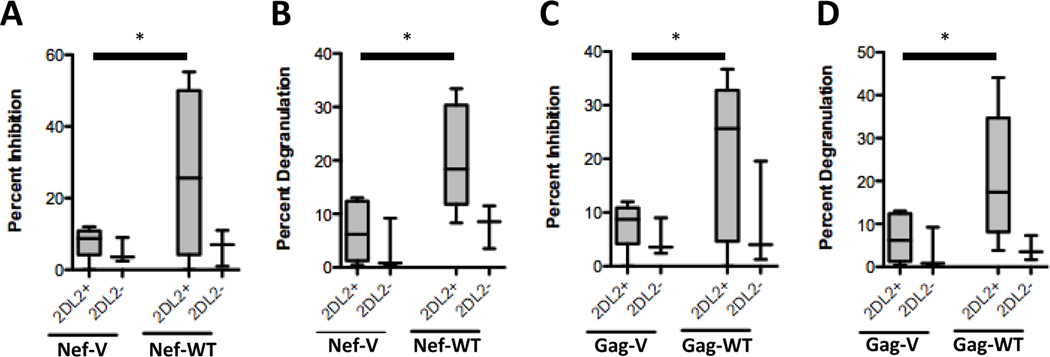
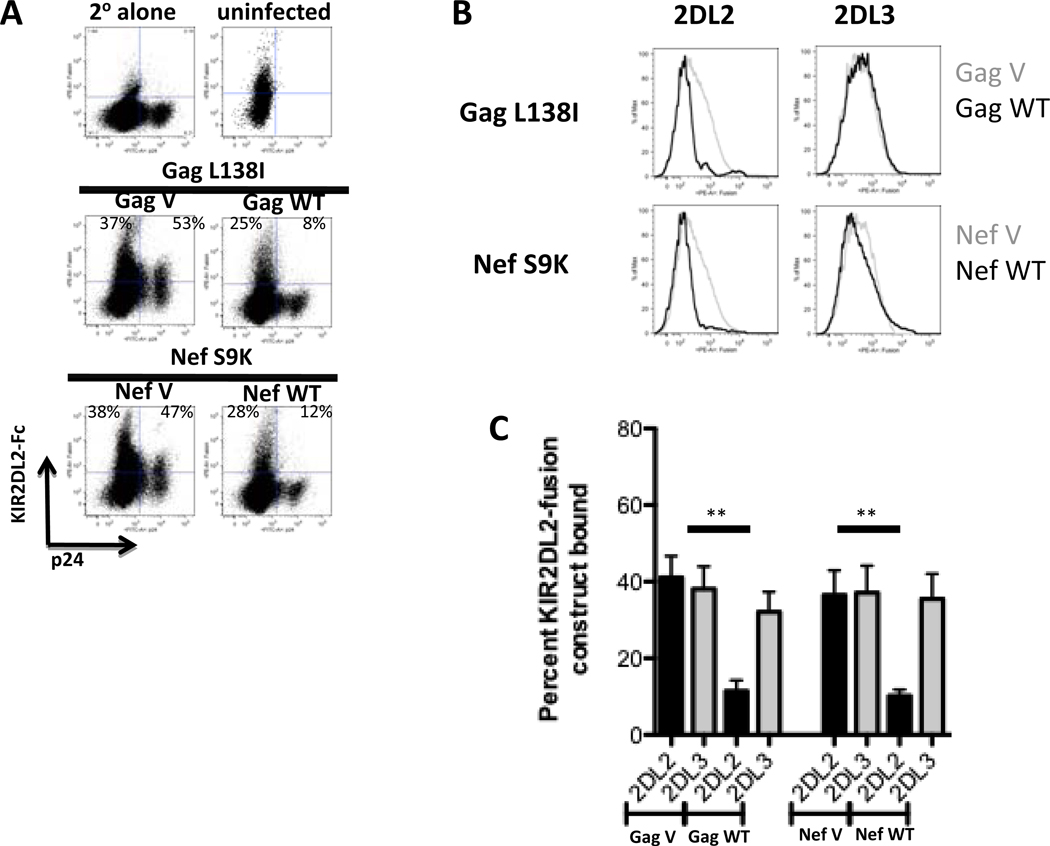
In addition to the Vpu71M/74H polymorphism, two additional amino acid polymorphisms (Gag138L/I and Nef9S/K) were associated with the presence of the KIR2DL2 gene in the study population (Table 1, polymorphisms # 2 and #22). We next determined whether these KIR2DL2-associated polymorphisms were also associated with differential recognition of HIV-1-infected cells by KIR2DL2+ NK cells. Viruses containing the respective polymorphisms replicated similarly in CD4+ T cells (Supplemental Figure 2B). As observed for the Vpu/Env variants, viruses containing the amino acids that were rarely observed in KIR2DL2+ individuals (GagWT and NefWT, Table 2 and Supplemental Tables 4 and 5) were inhibited significantly stronger by NK cells derived from KIR2DL2+ individuals than were viruses containing the variants selected in KIR2DL2+ subjects (Figure 3A and C). KIR2DL2+ NK cells furthermore degranulated more robustly in response to CD4+ T cells infected with the GagWT and NefWT viruses (Figure 3B and 3D), and KIR2DL2-IgG fusion constructs bound significantly less to CD4+ T cells infected with the GagWT and NefWT viruses (Figure 4A–C). Taken together, these data demonstrate that HIV-1 may evolve in KIR2DL2+ individuals to exclude particular amino acid polymorphisms in an effort to escape recognition by KIR2DL2+ NK cells.
Figure 3. Two additional KIR2DL2-associated amino acid polymorphisms reduce KIR2DL2-mediated NK cell recognition of virally infected cells.
NK cells from KIR2DL2+ individuals (gray bars, n=5) inhibited the replication of the Gag-WT virus and the Nef-WT virus significantly better than the replication of the Gag-V and Nef-V virus (A and C). Similarly, NK cells derived from KIR2DL2+ individuals (gray bars, n=5) were activated significantly more by cells infected with the Gag-WT However, NK cells derived from KIR2DL2- individuals did not inhibit viral replication or degranulate in the response to cells infected with either of the WT or V viruses. * p < 0.05.
Figure 4. KIR2DL2-associated amino acid polymorphisms affect KIR2DL2-, but not KIR2DL3-binding to infected CD4+ T cells.
CD4+ T cells infected with the respective variant viruses were stained with KIR2DL2-IgG and KIR2DL3-IgG fusion constructs. The dot plots (A) and histograms (B) depict the staining pattern of KIR2DL2-IgG and KIR2DL3-IgG fusion constructs of HIV-1-infected CD4+ T cells from a HLA-C1/C2 heterozygous donor. The bar graph summarizes KIR2DL2- (black) or KIR2DL3- (grey) IgG fusion construct binding data for 5 different HLA-C1/C2 heterozygous CD4+ T cell donors for the viral variants, as indicated (C). ** p < 0.005
Increasing evidence suggests that NK cells play an important role in the control of HIV-1 infection4–6. Here we report a number of amino acid polymorphisms within the HIV-1 clade B sequence that are significantly associated with the expression of specific KIR genes on the population level, and demonstrate in functional studies that these “KIR footprints” can modulate the interaction of KIR+ NK cells with HIV-1-infected CD4+ T cells. The selection of particular amino acid residues that result in enhanced binding of inhibitory KIRs to infected cells represents a novel approach by which HIV-1 can evade NK cell-mediated immunity. The molecular mechanisms and precise receptor/ligand interactions involved in this evasion from NK cell recognition require further investigation. Previous in vitro studies have demonstrated that sequence variations in HLA class I presented epitopes14–23 and small changes in the peptide repertoire presented on HLA class I molecules24 can both modulate the binding of KIR, providing a potential mechanism for virus sequence-dependent recognition of infected cells by NK cells. The KIR2DL2-associated sequence polymorphisms studied here had no impact on KIR2DL2-binding to HLA-Cw*0102-expressing TAP-deficient T2 cells (Supplemental Figure 4), despite some degree of HLA-Cw*0102 stabilization. However, several different HLA-C group 1 molecules might present epitopes within these regions of HIV-1, resulting in differential recognition by KIR2DL2. Additionally other mechanisms than the modulation of KIR-binding to HLA class I by viral sequence polymorphisms might account for the observed reduction in recognition of variant virus infected cells by KIR2DL2+ NK cells. KIR-associated sequence polymorphisms within HIV-1 proteins might directly modulate the ability of these proteins to be processed and presented25, subtly alter hydrostatic interactions with KIR, or change the profile of NK cell receptor ligands expressed on infected cells, as described for HIV-1 Nef, Vpu and Vpr 26–27.
Protective effects of specific KIRs or KIR/HLA class I combinations have been described for many infectious disease, including HIV, HCV, and HPV. While KIR2DL2 has not been identified as a protective KIR gene in any of these viral infections, homozygosity of its allotypic counterpart, KIR2DL3, has been associated with the resolution of HCV infection when co-expressed with its ligand, HLA-C group 128. This protective KIR2DL3/HLA-C group 1 combination provides a weaker inhibitory signal, resulting in weaker inhibition of NK cells28. Here we describe a mechanism by which HIV-1 selects for sequence polymorphisms in KIR2DL2+ individuals that lead to an enhanced binding of this inhibitory KIR to infected cells, resulting in the inhibition of NK cell function, and thereby escaping the potential protective role of this KIR. Overall, the data from different viral infections are consistent with a model in which enhanced NK cell activity can contribute to the control of viral replication, and suggest that viruses can evade this NK cell-mediated immune pressure by selecting for variants that modulate the recognition of infected cells by KIR.
Methods summary
Viral sequencing: Genomic DNA was extracted from peripheral blood mononuclear cell samples and nested PCR protocols were used to amplify HIV-1 genomes9. Phylogenetic analysis of KIR-associated sequence polymorphisms: A decision-tree approach, followed by adjustment for multiple comparisons10, was used to identify KIR-associated sequence polymorphisms. Construction of viruses containing sequence polymorphisms: Mutations of interest were inserted into the HIV-1 NL4-3 backbone by GeneTailor site-directed mutagenesis system11–12. Viral Inhibition Assay: Viral inhibition assays were performed following infecting CD4+ T cells with the viral constructs, as indicated. The level of viral inhibition was then calculated as the difference in viral production (p24 Gag) in wells containing autologous NK cells compared to those containing CD4+ T cells alone6. NK cell degranulation assay: KIR2DL2+ NK cell degranulation was examined by flow-cytometry following NK cell co-culture with autologous CD4+ T cells infected the viral construct as indicated in the presence of Golgi-stop and anti-CD107a-PECy5 for 6 h29. NK cells were stained with anti-CD3, -CD56, -CD16, and -CD158b (KIR2DL2/2DL3/2DS2) antibodies, and the level of degranulation was assessed as the proportion of CD107a+ NK cells belonging to the CD158b+ NK cells.
Methods
Study subjects
Ninety-one chronic untreated HIV-1 subtype B-infected subjects for which HLA class I and KIR genotypes were available (Supplemental Table 1) were included in this study9. In addition, 100 HIV-1 negative controls were genotyped for KIR/HLA genotypes. Of this large cohort of uninfected controls, a group 15 subjects that were KIR2DL2+ (KIR2DL2+/KIR2DL3+) and 15 subjects that were KIR2DL2- (KIR2DL3+/KIR2DL3+) were enrolled to provide samples for the generation of NK cells and autologous CD4+ target cells (Supplemental Table 6). For these in vitro studies, only individuals that did not encode for KIR3DS1 and HLA-B Bw4-80I were selected, as we had previously observed very strong inhibition of HIV-1 replication in vitro in individuals encoding this combined KIR/HLA genotype6. All study subjects were enrolled in Boston through the Massachusetts General Hospital, the Lemuel-Shattuck Hospital, and the Fenway Community Health Center. The study was approved by the Massachusetts General Hospital Review Board, and all subjects gave written informed consent.
Viral sequencing
Genomic DNA was extracted from peripheral blood mononuclear cell samples and nested PCR protocols were used to amplify HIV-1 genomes as described previously9. Five independent PCR products of each sample were pooled and directly population sequenced at the Massachusetts General Hospital DNA Sequencing Core facility using clade B consensus sequencing primers as previously described9.
HLA class I and KIR typing
High resolution HLA class I typing and KIR genotyping were performed as described previously6.
Phylogenetic analysis of KIR-associated sequence polymorphisms
We used the decision-tree approach, which corrects for phylogenetic structure among the sequences and allows for a multivariate analysis10, to identify KIR-associated sequence polymorphisms. All results were adjusted for multiple comparisons (both p- and q-values are assigned to each result). For each protein analyzed, a maximum likelihood phylogenetic tree was constructed from the corresponding sequences. For every KIR/HLA gene, amino-acid position, and amino acid at that position, two generative, or directed graphical, models of the observed presence or absence of the amino acid in each sequence were created - one representing the null hypothesis that the observations are generated by the phylogenetic tree alone, and the other representing the alternative hypothesis that additional escape or reversion takes place due to KIR/HLA pressure in the subjects for which the sequences are observed. The likelihood of the observations was then maximized over the parameters of both models using an Expectation-Maximization algorithm, and a p value was computed using a likelihood ratio test based on those likelihoods10. To increase power, the tests were binarized, such the presence or absence of a given KIR/HLA gene was correlated with the presence or absence of a given amino acid. In addition, KIR-polymorphism pairs were analyzed only when the actual or expected count in every cell of the corresponding two-by-two contingency table was greater than or equal to three. For every amino-acid at each position, the KIR/HLA gene with the strongest association (and its corresponding p-value) was added to the list of identified associations. The analysis was then repeated after removing individuals having or possibly having this KIR/HLA gene. This procedure was iterated until no KIR/HLA gene yielded an association with p-value less than 0.05. A q-value statistic, estimating the proportion of false positives among the associations identified, was computed for each association by repeating this analysis on null data (generated by permuting the KIR/HLA data). Correction for multiple comparisons was undertaken both using q<0.05 (estimating 5% false positives) and using q<0.2 (estimating 20% false positives).
Construction of viruses containing sequence polymorphisms
HIV-1 strain NL4-3 was modified to express one or two mutations in vpu/env, nef or gag using the GeneTailor site-directed mutagenesis system (Invitrogen) or the QuikChange Lightning Site-Directed Mutagenesis system (Stratagene)11–12. In brief, mutagenesis was performed using 5’ oligonucleotide primers Vpu_M71R-f (CTTGTGGAGATGGGGGTGGAAAGGGGGCACCAT [nt] 6279), Vpu_M71R-r (TTTCCACCCCCATCTCCACAAGTGCTGATACTTCT [nt] 6234), Vpu_H74L-f (ATGGGGGTGGAAATGGGGCACCTTGCTCCTTGG [nt] 6288), Vpu_H74L-r (GGTGCCCCATTTCCACCCCCATCTCCACAAG [nt] 6247), Vpu_ M71R/H74L-f (ATGGGGGTGGAAAGGGGGCACCTTGCTCCTTGG [nt] 6288), and Vpu_ M71R/H74L-r (GGTGCCCCCTTTCCACCCCCATCTCCACAAG [nt] 6247); Nef/S9K-f (GTGGTCAAAAAGTAAAGTGATTGGATGGCC), Nef/S9K-r (GGCCATCCAATCACTTTACTTTTTGACCAC); Gag/L138I-f (CCTATAGTGCAGAACATCCAGGGGCAAATGG), Gag/L138I-r (CCATTTGCCCCTGGATGTTCTGCACTATAGG). Mutated nucleotides are underlined, and primer positions are numbered according to those of NL-4-3 (GenBank accession number AF324493). The complete HIV-1 coding region of the variant proviruses was sequenced on an ABI3730 XL DNA analyzer. Propagation of provirus, and generation of viral stocks was performed as previously described11–12. While the full length sequence of the NL4-3 viruses differed from the autologous sequence of the respective study subjects, the areas flanking the Vpu, Gag and Nef sequences studied were identical between the NL4-3 virus and the respective areas of interest, and only differed in the respective amino acids indicated in the respective Tables and Supplemental Table.
Viral Inhibition Assay
Viral inhibition assays using NK cells were performed as previously described6. CD4+ T cells that were generated after 4 days in culture with a bispecific antibody to CD3/CD8 were infected with lab strains containing the amino acid polymorphisms as indicated at a multiplicity of infection of 0.01 for 4 h at 37°C. Cells were washed two times. Equal numbers of CD4+ T cells were plated at NK cell/CD4+ T cell ratios of 10:1, or alone for 14 days in the presence of 50 U/ml IL-2. Supernatant was collected every 3–4 days for quantification of p24 Gag production by ELISA (p24 ELISA; Perkin Elmer).
NK cell degranulation assay
To examine whether KIR2DL2+ NK cells specifically degranulated in response to autologous CD4+ T cells infected with either of the viral variants, we monitored the level of CD107a up-regulation on KIR2DL2+ or KIR2DL2- NK cells29. HIV-1 negative donors were selected that expressed KIR2DL2 in the absence of KIR2DS2 (referred to as KIR2DL2+), or that expressed KIR2DL3 in the absence of KIR2DL2 and KIR2DS2 (referred to as KIR2DL2neg). An anti-CD158b antibody was used to detect KIR2DL2-, KIR2DL3-, and KIR2DS2-expressing NK cells. The level of degranulation was assessed as the proportion of CD107a+ NK cells belonging to the CD158b+ NK cells. Thus purified NK cells were co-cultured in the presence of autologous CD4+ T cells infected in vitro with the respective HIV-1 strains for 7 days. Monensin was added to co-cultures on day 7 at 0.3 µg/ml in the presence of 20 µl CD107a–PE-Cy5 for 6 h. Cells were washed and stained with CD3-Pacific Blue, CD56-PECy7, CD16-APCCy7 (BD Biosciences), and CD158b-PE (Beckman Coulter) for 30 min, washed, and fixed in 1% paraformaldehyde until flow cytometric analysis was performed (FACSCalibur; BD Biosciences).
KIR-IgG-fusion construct binding assay
Differences in the ability of KIR2DL2 or KIR2DL3 to interact with CD4+ T cells infected with either of the viral variants were ascertained using a KIR2DL2- and KIR2DL3-fusion construct (kindly provided by Drs. Eric Long, Salim Khakoo and Ofer Mandelboim). Thus PBMCs were obtained from HLA-C1/C1 homozygotes, -C1/C2 heterozygotes, or -C2/C2 homozygotes and were treated with 0.3ug of a bispecific CD3/8-antibody in the presence of 50 units of IL-2/ml of complete medium. After 3 days, the cells were infected with one of the 3 KIR2DL2-variants (Vpu 71/74, Gag 138, or Nef 9) or wildtype for 2 days. The cells were then collected and stained with 2ul of the KIR2DL2- or KIR2DL3-Fc for 1 hour on ice. The cells were then washed and stained with a secondary APC-goat- anti-human antibody for an additional 20 minutes on ice. In parallel, of infected cells were stained with the secondary anti-human antibody alone to define the background level of staining. All tubes were then fixed with 100ul of Fix A (Invitrogen) solution for 10 minutes, washed and permeabilized using 100ul of Perm B (Invitrogen). The tubes were then stained for intracellular p24 using the KC-57-RD1 antibody for 20 minutes on ice, and then washed. The tubes were then fixed in 1% paraformaldehyde until flow cytometric analysis was performed (FACSCalibur; BD Biosciences).
HLA class I stabilization assays
HLA-C stabilization was assessed in 2×10’5 T2 cells that were incubated with 0.04mg/ml peptide, as indicated, overnight at 26°C. The following day peptide-pulsed T2 cells were stained with DT9 antibody (HLA-C/E) for 30mins at 4°C. Cells were then washed in PBS before staining with anti-mouse IgG-PE (Sigma) for 30mins at 4°C. Cells were washed twice with PBS and fixed in Perm A solution (BD). KIR2DL2-Fc binding to peptide-pulsed T2 cells was assessed as previously described (24).
Supplementary Material
Acknowledgements
These studies were supported by National Institute of Health / National Institutes of Allergy and Infectious Diseases grants R01 AI067031 (M. Altfeld) and PO1 AI074415 (M. Altfeld and T. Allen) and the Doris Duke Charitable Foundation (M. Altfeld). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. SIK is a recipient of a Wellcome Trust Senior Clinical Fellowship and M. Altfeld is a Distinguished Clinical Scientist of the Doris Duke Charitable Foundation. We thank Microsoft Research, the Bill & Melinda Gates Foundation, the Mark and Lisa Schwartz Foundation, and the Phillip T and Susan M Ragon Foundation for their support.
Footnotes
Author Contribution:
G.A. conducted the immunology experiments, and L. F. performed the KIR staining experiments on T2 cell lines. A. S. and C. O-N. constructed the viral variants, and D.H., C.K. and J.C. performed the data analysis identifying KIR associated polymorphisms. GA, DH, AS and LF contributed equally. B.L. and T. A. performed the viral sequencing, M.C. and M.M. performed the HLA and KIR typing, and L.F. and S.K. provided the KIR fusion construct. G.A. and M.A. planned the studies, prepared the manuscript and supervised the project.
Author Information:
Reprints and permissions information is available at www.nature.com/reprints. The authors do not have any competing financial interests.
References
- 1.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama WM. Specific and non-specific natural killer cell responses to viral infection. Adv Exp Med Biol. 2005;560:57–61. doi: 10.1007/0-387-24180-9_8. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. NK Cell Recognition. Annu Rev Immunol. 2004 doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 4.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 5.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alter G, et al. Differential natural killer cell mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007 doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altfeld M, Goulder P. 'Unleashed' natural killers hinder HIV. Nat Genet. 2007;39:708–710. doi: 10.1038/ng0607-708. [DOI] [PubMed] [Google Scholar]
- 8.Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 9.Wang YE, et al. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83:1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson JM, et al. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol. 2008;4:e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneidewind A, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moesta AK, et al. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 14.Stewart CA, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart-Jones GB, et al. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur J Immunol. 2005;35:341–351. doi: 10.1002/eji.200425724. [DOI] [PubMed] [Google Scholar]
- 16.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 17.Thananchai H, et al. Cutting Edge: Allele-Specific and Peptide-Dependent Interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malnati MS, et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 20.Peruzzi M, Parker KC, Long EO, Malnati MS. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J Immunol. 1996;157:3350–3356. [PubMed] [Google Scholar]
- 21.Mandelboim O, Wilson SB, Vales-Gomez M, Reyburn HT, Strominger JL. Self and viral peptides can initiate lysis by autologous natural killer cells. Proc Natl Acad Sci U S A. 1997;94:4604–4609. doi: 10.1073/pnas.94.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandelboim O, et al. The binding site of NK receptors on HLA-C molecules. Immunity. 1997;6:341–350. doi: 10.1016/s1074-7613(00)80336-2. [DOI] [PubMed] [Google Scholar]
- 23.Fadda L, et al. Common HIV-1 peptide variants mediate differential binding of KIR3DL1 to HLA-Bw4 molecules. J Virol. 2011 doi: 10.1128/JVI.00412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fadda L, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward J, et al. HIV-1 Vpr triggers natural killer cell-mediated lysis of infected cells through activation of the ATR-mediated DNA damage response. PLoS Pathog. 2009;5:e1000613. doi: 10.1371/journal.ppat.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward J, et al. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood. 2007;110:1207–1214. doi: 10.1182/blood-2006-06-028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 29.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.