Summary
Alternative polyadenylation (APA) is emerging as a widespread mechanism used to control gene expression. Like alternative splicing, usage of alternative poly(A) sites allows a single gene to encode multiple mRNA transcripts. In some cases, this changes the mRNA coding potential; in other cases, the code remains unchanged but the 3’UTR length is altered, influencing the fate of mRNAs in several ways, for example, by altering the availability of RNA binding protein sites and microRNA binding sites. The mechansims governing both global and gene-specific APA are only starting to be deciphered. Here we review what is known about these mechanisms and the functional consequences of alternative polyadenlyation.
Introduction
Regulation of mRNA processing is well known to play a fundamental role in determining the outcome of gene expression, but alternative polyadenylation has only recently gained attention as a major player influencing the dynamics of gene regulation. The maturation of 3’ ends of mRNA precursors (pre-mRNAs), although a relatively simple process, has been known for some time to require a complex set of protein factors. One explanation for this has been that the complexity reflects the importance of regulating 3’ end formation. It is well established that polyadenylation can contribute in several ways to gene control (Colgan and Manley, 1997; Barabino and Keller, 1999); however, in the past few years it has become clear that regulation of APA is considerably more widespread than previously thought, and can affect gene expression in multiple ways. In this review, we discuss both the mechanisms and the consequences of APA, and how regulated mRNA 3’ processing contributes to cell growth control and disease. We begin by providing some background and a brief overview of 3’ processing and its regulation.
The mature 3’ ends of nearly all eukaryotic mRNAs, with the exception of replication-dependent histone transcripts, are created by a two-step reaction that involves an endonucleolytic cleavage of the pre-mRNA, followed by synthesis of a polyadenylate tail onto the upstream cleavage product. This relatively simple reaction requires numerous protein factors that are directed to the correct cleavage site by sequence elements within the pre-mRNA (reviewed in Colgan and Manley, 1997; Mandel et al., 2008; Millevoi and Vagner, 2010; Zhao et al., 1999). The core molecular machinery responsible for 3’ end formation in mammals includes four multi-subunit protein complexes, CPSF (Cleavage and Polyadenylation Specificity Factor), CstF (Cleavage stimulation Factor), CFI and CFII (Cleavage Factor I and II), as well as additional accessory factors and the single subunit poly(A) polymerase (PAP). RNA polymerase II (RNAP II), and specifically the C-terminal domain of its largest subunit, also plays an important role in processing. The assembly of the 3’ end processing complex on the pre-mRNA begins with the cooperative interaction of CPSF and CstF with specific sequences, the canonical poly(A) signal AAUAAA located upstream of the cleavage site, recognized by CPSF (specifically by the CPSF160 subunit), and a less defined downstream U/GU-rich region that constitutes the binding site for CstF (through the CstF64 subunit). Usage of one poly(A) site over another is often attributed to the relative “strength” of these core elements, but in fact auxiliary sequences and protein factors play a role in influencing poly(A) site choice in different contexts. Indeed, today we know that many more proteins than previously thought are involved in the fine-tuning of 3’ end formation (Shi et al., 2009), and a good number of these likely mediate crosstalk between pre-mRNA maturation and other nuclear events.
Polyadenylation influences many aspects of mRNA metabolism. Transcription termination by RNAP II, mRNA stability, mRNA export to the cytoplasm and the efficiency of translation are all dependent on 3’ processing. These topics have all been reviewed recently, and won’t be discussed here (Ji et al., 2011; Richard and Manley, 2009; Vinciguerra and Stutz, 2004; Zhang et al., 2010).
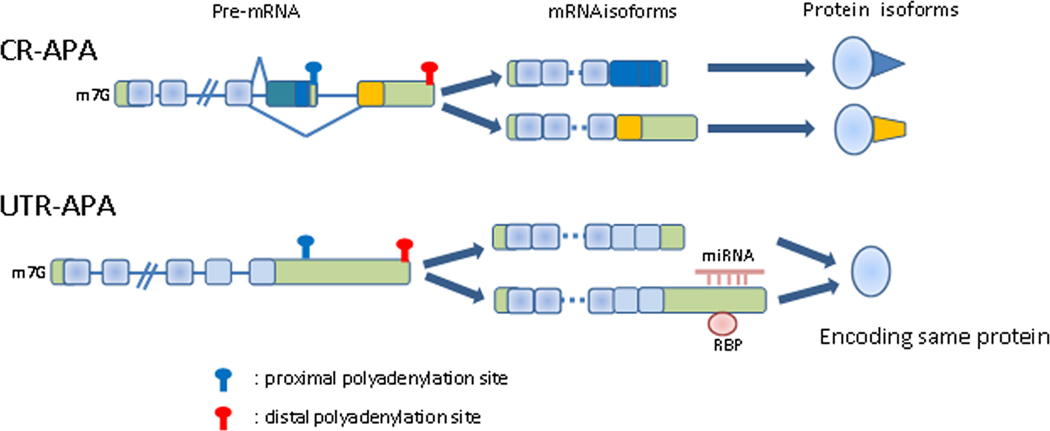
In recent years it has become increasingly evident that APA is extensively used to regulate gene expression. For example, 50% or more of human genes encode multiple transcripts derived from APA (Tian et al., 2005). We will consider here two general classes of APA. In some cases the alternative poly(A) sites are located in internal introns/exons and therefore APA events will produce different protein isoforms, we will refer to this type as CR-APA (Coding Region-APA). In other cases, APA sites are all located in the 3’ untranslated region (3’UTR), resulting in transcripts with 3’UTRs of different length but encoding the same protein; we refer to this type of APA as UTR-APA (Figure 1).
Figure 1. Schematic representation of CR-APA and UTR-APA.
CR-APA produces mRNA isoforms with distinct C-terminal coding regions, resulting in distinct protein isoforms. UTR-APA produces distinct mRNA isoforms with different length 3’UTRs, but encode the same protein. Longer 3’UTRs usually contain cis-regulatory elements, such as miRNA and/or protein binding sites, which often bring about mRNA instability or translational repression. CR-APA, coding region-alternative polyadenylation; UTR-APA, 3’UTR-alternative polyadenylation. Light green boxes, untranslated regions; blue light blue boxes, shared coding regions; dark blue and yellow boxes, unshared coding regions; lines, introns.
While CR-APA can affect gene expression qualitatively by producing distinct protein isoforms, UTR-APA has the potential to affect expression quantitatively. 3’UTRs often harbor microRNA (miRNA) binding sites and/or other regulatory sequences, such as AU-rich elements (AREs) (Barreau et al., 2005; Fabian et al., 2010). Longer 3’UTRs will more likely possess such signals, or more of them, and the mRNA will therefore likely be more prone to negative regulation. Indeed, the amount of protein generated by an mRNA has been showed to depend on its 3’UTR length, such that transcripts with shorter 3’UTRs produce higher levels of protein (Mayr and Bartel, 2009; Sandberg et al., 2008). Furthermore, as discussed below, the length of the 3’UTR can affect not only the stability but also the localization, transport and translational properties of the mRNA.
Differential processing at multiple poly(A) sites can be influenced by physiological conditions such as cell growth, differentiation and development, or by pathological events such as cancer. The mechanisms that regulate such global events are mostly unknown, and intense research is currently being carried out in order to better understand this phenomenon at the molecular level. In this review, we will provide an overview of current studies on APA, both on genome-wide analyses and specific examples, focusing on the possible mechanisms of regulation and the functional consequences of differential poly(A) site usage.
Genome-wide analyses of APA
Analyses of APA at the global level have been largely responsible for the appreciation that APA constitutes a significant contributor to gene regulation across species. Genome-wide studies carried out in humans, mice, worms, yeast, plants and algae revealed that the number of genes encoding transcripts with multiple poly(A) sites ranges from 10–15% in S. cervisiae (Nagalakshmi et al., 2008) to ~54% in humans (Tian et al., 2005). Significantly, orthologous human and mouse genes were found to have a high similarity in the numbers of 3’ ends mapped for each gene (Ara et al., 2006; Tian et al., 2005), indicating that APA sites have been actively selected during evolution. Interestingly however, as shown by a genome-wide bioinformatic analysis, the majorities of tissue-specific and non-canonical poly(A) sites seem to be species-specific and are not themselves conserved (Ara et al., 2006). This suggests that gain or loss of APA sites is a frequent event in mammalian genomes, implying that very often novel sites would be quickly lost if their presence is either neutral or deleterious.
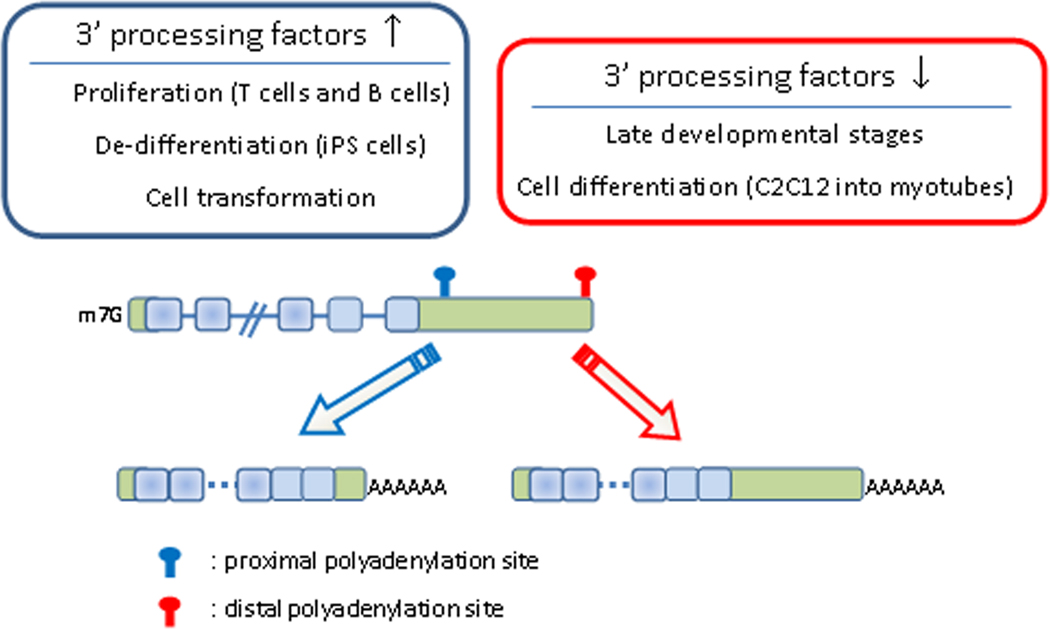
Through genome-wide analysis of APA it has been possible to define a pattern that relates the proliferation and differentiation status of cells with the length of 3’UTRs (Figure 2). Specifically, states of increased proliferation, de-differentiation and disease (i.e., cancer) are associated with a general shortening in 3’UTR length, while 3’UTRs tend to be longer during late developmental stages and cell differentiation (Ji et al., 2009; Ji and Tian, 2009; Mayr and Bartel, 2009; Sandberg et al., 2008; Shepard et al., 2011; Wang et al., 2008a). In C. elegans, the length of the 3’UTRs correlates inversely with animal age (Mangone et al., 2010). Interestingly, a considerable number of miRNAs diminish in expression over adult life in C. elegans (Ibanez-Ventoso and Driscoll, 2009), suggesting that a relaxation in miRNA-3’UTR control of mRNA stability/translation might be a general feature of advancing adult life.
Figure 2. Connecting APA to cellular proliferative and developmental states.
Enhanced proliferation such as during de-differentiation (e.g., in the generation of iPS cells), T cell activation, or cellular transformation are associated with upregulation in expression of certain 3’ processing factors and with increased usage of proximal poly(A) sites. Late developmental stages and cellular differentiation (e.g. differentiation of C2C12 into myotubes) are associated with downregulation of expression of 3’ processing factors and increased usage of distal poly(A) sites.
Given the fact that different types of APA exist, it is interesting to note that CR-APA and UTR-APA can be differently regulated. During T cell activation for example, CR-APA events occur at both early and late stages of activation (Sandberg et al., 2008). Moreover, proximal-to-distal and distal-to-proximal shifts in APA were similarly represented, whereas changes in UTR-APA were mostly evident during late stages of activation with a clear pattern of increased usage of the proximal site. This suggests that the regulation of different types of APA (CR-APA versus UTR-APA) may rely at least in part on different mechanisms. For example, CR-APA often occurs in conjunction with splicing of an overlapping intron, and it is thus possible that splicing regulation may also affect APA. Also, the observation that changes in UTR-APA is not an early event during T cell activation suggests that it is perhaps necessary to enhance the expression or activity of basal or auxiliary 3’ processing factors, which then function at later times. A consequence of the differential temporal behavior of CR-APA versus UTR-APA in T cell activation is that during early activation there will be more APA events affecting the protein isoform produced while during later stages, APA will lead to transcripts that differ in the length of their 3’UTR and therefore the main effect will be changes in the abundance of the proteins produced.
To understand how 3’UTR lengthening is related to regulation of biological processes, the association of selected genes with gene ontology (GO) terms was examined during different developmental/differentiation states. 3’UTR lengthening during mouse embryonic development coincides with upregulation of genes involved in morphogenesis and differentiation, such as cell morphogenesis and extracellular structure organization, and with downregulation of genes involved in proliferation, such as DNA replication and cell cycle phase (Ji et al., 2009). The same pattern is detected during differentiation of proliferative C2C12 muscle cells into myotubes. In contrast, during generation of human and mouse induced pluripotent stem (iPS) cells, most of the same GO terms displayed regulation in the opposite direction. It is of particular interest that 3’ processing factors, such as CPSF and CstF components, were found to be strongly upregulated during generation of iPS cells (Ji and Tian, 2009). This may hint at a regulatory mechanism where the abundance of 3’ processing factors in undifferentiated cells (such as iPS) facilitates the usage of the proximal poly(A) site, which usually has a “weaker” consensus than the distal site (see below), thereby generating transcripts with shorter 3’UTRs. Since both early embryonic and iPS cells are rapidly proliferating, a significant question is whether differentiation per se affects APA in a system where proliferation and differentiation could be uncoupled. For example, the leukemic cell line HL60 is capable of differentiating into neutrophils or monocytes (in response to different stimuli) even when the cell cycle is blocked in early G1 or S phase, indicating that differentiation and proliferation can be regulated independently (Brown et al., 2002). It would be of interest to compare changes in usage of APA sites before and after differentiation, independently from alterations in the proliferation rate.
Cancer cells provide an important subset of proliferating cells. In this regard, it is remarkable that in primary tumor samples from a mouse leukemia/lymphoma model (Singh et al., 2009), APA seems to define molecular signatures that can distinguish similar tumor subtypes with high accuracy. Mice lacking p53 and the core NHEJ factor DNA Ligase IV develop pro-B-cell lymphomas with frequent genomic amplification of c-Myc (designated LPC), while mice lacking p53 and the accessory NHEJ factor Artemis develop lymphomas with either c-Myc or N-Myc amplification (APC or APN, respectively). While LPC, APC and APN lymphomas are histologically and immunophenotypically indistinguishable, using microarray analysis, specific sets of transcripts with differential 3’UTR processing were identified between these lymphoma subtypes. The diagnostic capacity of these assignments was confirmed by analysis of unknown samples, which were correctly assigned at rates of 100% for LPC, 92% for APC and 74% for APN. These results anticipate the possibility of future usage of APA as a molecular biomarker with prognostic potential. In accordance with previously findings (Mayr and Bartel, 2009), shortening of 3’UTRs in the cancer cells compared to normal (pro-B) cells was the most common pattern observed, although some transcripts with elongated 3’UTRs were also detected. In addition, levels of a number of mRNAs encoding 3’ processing factors, notably those encoding CstF subunits, were upregulated in the lymphomas. This data supports a model in which changes in expression and/or stoichiometry of 3’ processing factors leads to changes in poly(A) site selection, for instance enhanced expression of these factors might help increase utilization of suboptimal proximal poly(A) signals and thereby contribute to the tumor-specific shortening of 3’UTRs (see below for further discussion of this hypothesis).
Bioinformatic approaches and genomic studies have also been used to shed light on the link between differential usage of APA sites in relation to tissue specificity (Wang et al., 2008a; Zhang et al., 2005). Using expressed sequence tag data, 42 distinct human tissue types were analyzed, revealing considerable tissue-specific APA. For example, retina, placenta, blood and ovary were more likely to use proximal poly(A) sites, while tissues from bone marrow, uterus, brain and nervous system showed increased usage of the distal poly(A) sites. These tissue-specific preferences are observed on a global rather than gene-specific level, indicating that the mechanism may lie in tissue-specific regulation or expression of polyadenylation factors. It will be of interest to compare the proliferation potential of these two groups of tissues and access whether it correlates with the usage of the proximal/distal poly(A) sites. For example brain tissues are known to have low mitotic activity, suggesting that decreased proliferation is associated with tissues harboring transcripts with longer 3’UTRs
Another important role of large-scale studies is the contribution they have made to our understanding of the role that cis-sequences play in APA. Computational analyses have indicated that variations of the canonical AAUAAA sequence are relatively frequent, occurring in more than 30% of 3’ends (Tian et al., 2005). Interestingly, while the canonical sequence predominates in genes with a unique poly(A) site, the less-conserved, variant poly(A) sites occur frequently in genes with multiple poly(A) sites. In these cases, the variant sites are usually located promoter proximal, whereas canonical poly(A) signal often appears downstream of variant sites (Beaudoing et al., 2000). This suggests that the efficient utilization of the proximal alternative poly(A) signals is likely dependent on additional auxiliary factors, on different abundance of core 3’ processing factors, and/or auxiliary surrounding RNA sequences. Indeed, genome-wide analyses recently identified conserved motifs, mostly around alternative poly(A) sites, that might help explain, at least in part, how the choice between the usage of the different sites is made (Nunes et al., 2010; Ozsolak et al., 2010). For example, a genome-wide analysis of over 10,000 human poly(A) sites shows that about one-third of non-canonical, proximal, poly(A) signals tend to have higher frequency of U and GU nucleotides downstream of the poly(A) site compared with canonical poly(A) signals, implying that a strong CstF binding site might compensate for the absence of a consensus hexanucleotide (Nunes et al., 2010). How these variations in core sequences, as well as other signals, contribute to APA is discussed below.
Specific examples of APA
There are now a growing number of examples of specific APA events for which the function and/or mechanism is at least reasonably well understood. In this section we discuss several of these, highlighting those that play important roles in cell growth and differentiation or disease. In the following sections, we discuss in more detail the known mechanisms and functions of APA.
Immunoglobulin (Ig) M heavy chain
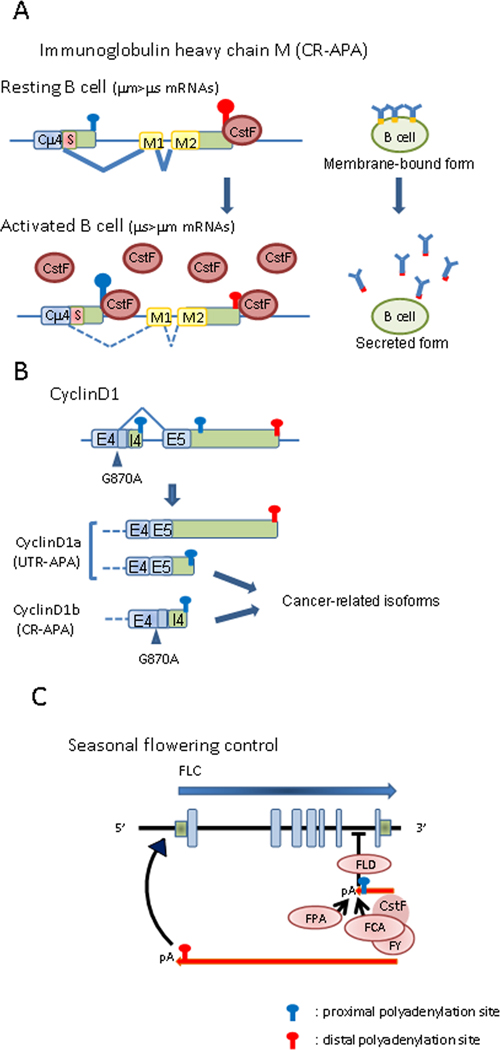
The immunoglobulin (Ig) M heavy chain gene provided perhaps the first example of APA, specifically of CR-APA, as a regulatory mechanism (Alt et al., 1980; Early et al., 1980; Rogers et al., 1980; reviewed in Peterson, 2007). During the transition of a B cell to a plasma cell, the IgM protein switches from a membrane-bound form to a secreted form. This switch is caused in large part by the selection of one of two poly(A) sites. The secreted form is produced by using a proximal poly(A) site, while the membrane-bound form is produced from the spliced Cu4-M1 mRNA by using distal poly(A) site (Figure 3A). The switch from membrane-bound to secreted form IgM in LPS-induced mouse primary B cells was shown to be accompanied with a specific increase of CstF64 protein levels (Takagaki et al., 1996). Moreover, overexpression of CstF64 in a B cell line was enough to induce the switch from membrane-bound to secreted form by preferentially using the proximal poly(A) site. In the same context, conditional knockdown of CstF64 also showed a relative enhancement of distal poly(A) site usage (Takagaki and Manley, 1998).
Figure 3. Examples of gene regulation by APA.
(A) The immunoglobulin heavy chain M gene is partly shown; a constant regions (Cμ4) is shared by both µm and µs mRNAs, while exons M1 and 2 (yellow boxes) and S (red boxes) are specific to µm and µs mRNAs, respectively. In resting B cells, the amount of CstF is limiting, and the distal poly(A) site, which binds CstF more avidly, is preferentially used, resulting in production of the membrane-bound form of IgM (µm). In activated B cells, the concentration of CstF is elevated and no longer limiting, so the proximal, first transcribed poly(A) site is preferentially selected, leading to production of secreted-form IgM (µs). Additional factors, such as the transcription factor Ell2 (see text), may also contribute to the switch. (B) CyclinD1 is subject to both UTR-APA and CR-APA. Two major isoforms are created by CR-APA: cyclin D1a (full-length isoform) and cyclin D1b (truncated isoform). The truncated isoform is associated with a polymorphism at the end of exon 4 (E4) (G870A, arrowhead), resulting in increased usage of the poly(A) site located in intron 4 (I4). This isoform is retained in the nucleus and is associated with increasing transforming capability. UTR-APA of CyclinD1 leads to increased usage of the weak proximal poly(A) site in cancer cells generally, or in the usage of a newly mutational-derived proximal poly(A) site in Mantle Cell Lymphoma. In both, mRNAs with shorter 3’UTRs are generated. Light green boxes, untranslated regions; blue light blue boxes, shared coding regions; lines, introns. (C) Seasonal flowering control by antisense-RNA transcript. FPA and FCA promote selection of the proximal poly(A) site of an antisense transcript that initiates downstream of the FLC gene (red arrows) by stimulating 3’ end formation at that site. 3’ end processing at the proximal poly(A) site recruits the histone demethylase, FLD, which induces histone modifications on internal nucleosomes that result in silencing the sense FLC transcript (blue arrow). In the absence of FPA and FCA, the distal poly(A) site of the antisense transcripts is selected. This may facilitate the recruitment of positive transcription factors to the FLC promoter, resulting in activation of FLC transcription.
Subsequent investigations of IgM switching mechanisms revealed that ELL2, a protein related to the transcription elongation factor ELL, may also contribute to selection of the proximal poly(A) site. Martincic et al. (2009) provided evidence that ELL2 and CstF64 track together with RNAP II across the IgM gene. Like CstF64, ELL2 levels were induced in LPS activated B cells. This may provide an additional mechanism to enhance CstF levels at the proximal poly(A) site, increasing the efficiency with which it is utilized.
Germ cell-specific APA
Mammalian testis have unique APA processing characteristics. The canonical AAUAAA sequence is infrequent in testis-specific mRNAs, which often use proximal poly(A) sites that are not efficiently polyadenylated in somatic cells. (Liu et al., 2007a; MacDonald and Redondo, 2002; McMahon et al., 2006; Zhang et al., 2005). A CstF64 variant (τCstF64) is highly expressed in male germ cells compared to other tissues (Monarez et al., 2007) and it may contribute to the different cleavage specificity observed in germ cells. In agreement with this hypothesis, knockout of Cstf2t, the gene encoding τCstF64, in mice resulted in a spermatogenetic defect, but no significant influence on somatic cells(Dass et al., 2007; Hockert et al., 2011). In addition, microarray experiments showed that transcripts encoding a number of core polyadenylation factors were significantly more abundant in germ cells than somatic cells (Liu et al., 2007a). Furthermore, during spermatogenesis, τCstF64 levels were found to increase while those of CstF64 decreased (Liu et al., 2007a). Germ cell-specific and stage-specific APA events may thus be induced by altered expression levels of 3’ processing factors, including τCstF64.
One interesting example of germ cell-specific APA is provided by transcripts encoding a transcription factor, BZW1, which exist as three mRNA isoforms created by UTR-APA. The two longer isoforms are expressed ubiquitously at low levels, while the shortest is expressed at high levels only in testis, especially spermatogonia. Expression of EGFP-BZW1 fusion genes with distinct BZW1 3’UTRs showed that the shortest transcript had the lowest translation efficiency, suggesting that BZW1 expression is fine-tuned through 3’UTR length in a cell type-specific manner (Yu et al., 2006). This result is contrary to the expectation that shorter 3’UTRs produce more protein than those with longer 3’UTRs. In this case, low expression of the short isoform of BZW1 may be due to its unusually short 3’UTR, which is ~25 times shorter than the average (500nt) 3’UTR in testis germ cells (Sood et al., 2006) and may negatively affect translational efficiency (Tanguay and Gallie, 1996).
Disease related APA
Only a few studies have focused on the pathophysiology of diseases related to APA (Chen et al., 2006). However, it is well established that 3’UTRs play an important role in various diseases and their progression (Conne et al., 2000). We describe two disease-related examples reflecting changes in APA caused by mutated poly(A) signals; one is the equivalent of a loss-of-function mutation, and the other to a gain-of-function mutation.
IPEX (Immune dysfunction, Polyendocrinopathy, Enteropathy, X-linked), a disease characterized by dysfunction of regulatory T-cells and subsequent autoimmunity, is caused by mutations in the FOXP3 gene, which encodes a transcription factor containing a forkhead DNA binding domain. Most of the reported mutations affect the forkhead domain, resulting in disruption of DNA binding. However, a rare mutation lies within the poly(A) signal (AAUAAA→AAUGAA). This mutation leads to skipping of the first poly(A) signal and usage of the next signal, located 5.1 kb downstream. This appears to result in an unstable mRNA, leading to a decrease of FOXP3 protein and in this way leading to IPEX (Bennett et al., 2001).
The loss of controlled cell-cycle progression is a critical event in tumorigenesis. Cyclin D1 regulates progression through G1-S phase by its association with cyclin-dependent kinase 4 or 6 (Knudsen et al., 2006). Two major isoforms, cyclin D1a and b, are created by alternative splicing/polyadenylation (CR-APA) (Figure3B). CyclinD1a mRNA is full-length, whereas cyclin D1b mRNA is cleaved at an APA site within an intron. Cyclin D1b protein is constitutively nuclear, resulting in increased transforming capability (Lu et al., 2003; Solomon et al., 2003). High expression of cyclin D1b is observed in several human cancers, including breast and prostate cancer (Burd et al., 2006; Wang et al., 2008b). A G870A polymorphism at the end of exon 4 has been associated with production of the cyclin D1b isoform (Comstock et al., 2009; Knudsen et al., 2006). This polymorphism may cause impaired recognition by the splicing machinery, resulting in APA using the intron 4 poly(A) signal (Betticher et al., 1995) .
Cyclin D1 levels can also be elevated by UTR-APA. Wiestner et al. (2007) investigated cyclin D1 expression in positive Mantle Cell Lymphoma (MCL) patients. They found that patients who have isoforms of cyclin D1a mRNA with short 3’UTRs had a median survival shorter than patients not expressing this isoform. Sequencing revealed that these short 3’UTR-containing isoforms all contained mutated polyadenylation signals that would be predicted to strengthen the poly(A) signal (e.g., AAUAAUCAA→AAUAAA, 3 base pair deletion; AAUAAU→AAUAAAU, an A insertion). Since full-length cyclin D1a mRNA contains mRNA destabilizing elements, the truncated mRNAs will be more stable. In the same context, Mayr and Bartel (2009) showed that cancer cell lines preferentially expressed shorter 3’UTR mRNAs of some oncogenes, including cyclin D1, whose shorter 3’UTR mRNA isoform was produced by usage of a proximal poly(A) signal (AAGAAA), and not by mutations that creates a new proximal poly(A) signal, as in the case of MCL. Although the mechanism of generating cyclin D1 isoforms with shorter 3’UTRs are different in the two cases described here, the final outcome, increased expression of cyclin D1, is the same..
Mechanisms regulating APA
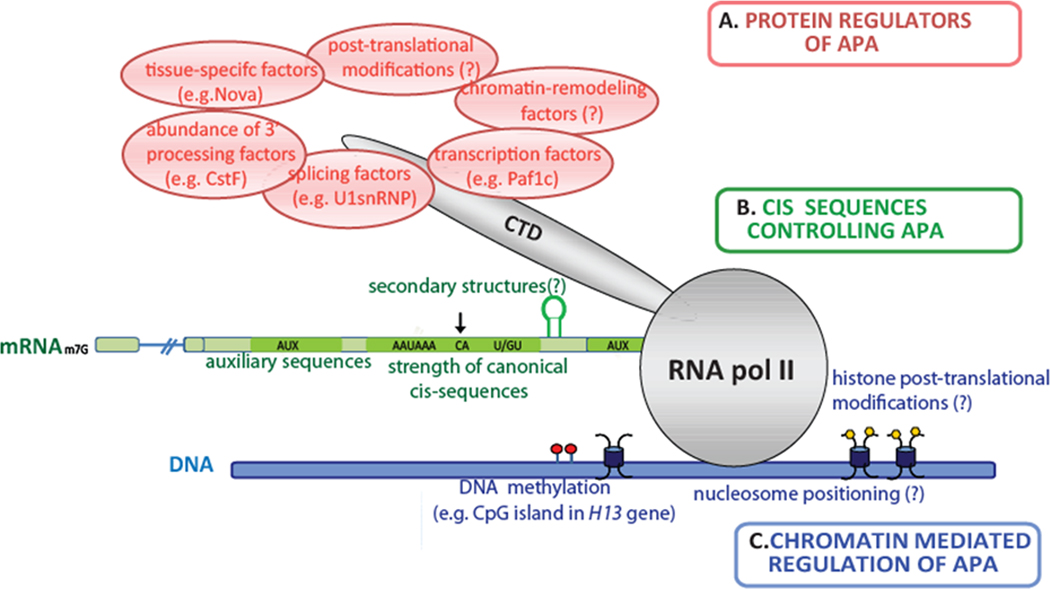
As discussed above, variations in the levels or activity of core polyadenylation factors can determine APA patterns. Another mechanism for regulating APA involves gene/tissue-specific RNA binding proteins. Interestingly, this is in many ways analogous to the control of alternative splicing (Chen and Manley, 2009) and, as with splicing, it is probably the combined effect of multiple trans-acting factors that determines the probability of using each poly(A) site (Figure 4A). Additionally, and again analogous to splicing, regulation likely involves cis-acting elements not only on the nascent mRNA but also at the DNA/chromatin level (Figure 4B and 4C). Here we discuss what is known about the regulation of APA and speculate about additional possible mechanisms.
Figure 4. Mechanisms regulating APA.
The choice of using one poly(A) site over another is dictated by a combination of several features. (A) Variations in the abundance or activity of trans-acting factors such as core 3’ processing proteins and tissue-specific RNA-binding proteins, as well as through interaction with splicing and transcription factors. (B) Combinations of cis-acting RNA elements, such as the strength of binding sites for core 3’ processing factors, auxiliary sequences and/or new motifs directing the interaction of protein components with the mRNA, and perhaps RNA secondary structures. (C) APA is likely also influenced by chromatin, including nucleosome positioning around the poly(A) site, DNA methylation and histone post-translational modifications.
Regulation of APA by trans-acting factors
One way to regulate the choice of alternative poly(A) sites is by differential expression of general polyadenylation factors. This mechanism could for example promote the usage of an APA site that inefficiently recruits the 3’ processing machinery due to the presence of sub-optimal cis-acting elements by increasing the concentration of one or more limiting processing factor. A well-known example of this model of action occurs during B-cell differentiation. As discussed above, upregulation of CstF64, and indeed the CstF complex, results in a switch from distal to proximal poly(A) site selection, resulting in conversion of IgM heavy chain from membrane-bound to secreted form (Takagaki and Manley, 1998; Takagaki et al., 1996). This was shown to reflect a greater affinity of the purified CstF complex for the distal GU-rich downstream element relative to the corresponding promoter-proximal site, leading to a model in which the stronger, high-affinity site is utilized under conditions of limiting CstF, while at high concentrations of CstF, the first site encountered during transcription, i.e, the proximal site, is preferentially used. This model not only explains the switch in IgM pre-mRNA APA during B cell activation, but also provides a mechanistic explanation for the more recent global observations, also discussed above, that promoter proximal APA sites are frequently “weaker” than downstream sites. Thus the switch to proximal sites that occurs generally in proliferating cells could be brought about by increased levels of CstF or other processing factors, which as we discussed is indeed frequently observed. It is also noteworthy that the global studies revealed that variations in the AAUAAA sequence frequently characterized the proximal sites, but the studies with IgM indicate that the nature of the GU-rich sequence can also influence APA.
Another example of regulation of APA by CstF is provided by control of the mRNA encoding the transcription factor NF-ATc during T cell activation. The transcription factor NF-ATc can be synthesized in three prominent isoforms, two long forms that are expressed in naïve T cells and a shorter form, arising from usage of a proximal poly(A) site during differentiation to effector T cells. Analogous to the situation in B cells, CstF64 levels are low in naïve T cells when the distal poly(A) site is used but increase during T cell activation when APA switches to the proximal site. Again, this switch appears to exploit the relatively low affinity of the proximal site for CstF64 (Chuvpilo et al., 1999).
The 3’ processing factor CFI has also been shown capable of influencing APA site choice, at least in human tissue culture cells. As opposed to the examples of CstF-mediated regulation of APA where low protein levels promote the usage of the distal poly(A) site, reduced levels of CFI-25, achieved by siRNA knock down, resulted in an upstream shift in poly(A) site selection in transcripts of several genes tested (Kubo et al., 2006). These results suggest that CFI may be selectively recruited to the distal poly(A) site, perhaps by sequence-specific RNA binding. Indeed, previous studies have shown that CFI preferentially binds to RNA sequences containing UGUAN (Brown and Gilmartin, 2003; Venkataraman et al., 2005). Additional work is required to determine if alterations in CFI levels is a physiological mechanism of APA control.
An important question in considering the role of general poly(A) factors in APA regulation is whether the levels of any of these factors change in a systematic way in response to changes in proliferation and/or during differentiation. Indeed, genome-wide studies have found that expression of most 3’ processing factors does change in ways that correlate with APA changes. For example, expression of most of the core polyadenylation factors, including CstF and CPSF subunits, RBBP6 (Shi et al. 2009) and symplekin, was found to be upregulated during generation of iPS cells derived from different cell types, correlating with a general trend of 3’UTR shortening, while the same factors were downregulated in differentiated embryonic tissues where longer 3’UTRs are observed (Ji and Tian, 2009). In agreement with this, many of these same genes are downregulated during differentiation of C2C12 myoblasts into myotubes, when 3’ UTRs are lengthened (Ji et al., 2009). Moreover, Mayr and Bartel (2009) found that genes encoding several 3’ processing factors were upregulated in cancer cells, correlating with shorter 3’UTRs (Figure 2). The most striking difference was in levels of CstF64 and CPSF160, which as mentioned directly recognize the GU-rich region and AAUAAA hexanucleotide, respectively. These findings together support the view that changes in concentrations of core poly(A) factors in conjunction with relatively weak proximal poly(A) sites indeed plays an important, general role in controlling APA. However, as we discussed above with respect to the IgM gene, it is likely that other factors can contribute to APA control, perhaps providing redundancy, functioning together with the core factors, and/or allowing more gene-specific regulation. Consistent with this, proximal APA sites displaying higher variation of usage in different human tissues tend to be flanked by sequences with higher conservation rate (Wang et al., 2008a).
A number of RNA binding proteins have been implicated in APA control. An example of a tissue-specific factor, initially characterized as a splicing factor but which also controls APA, is Nova2 (Licatalosi et al., 2008). RNAs extracted from brains of wt versus Nova2 knockout mice were hybridized to exon arrays and the pattern of APA was found to be altered in ~300 transcripts. A Nova2 binding site, YCAY, was identified flanking the Nova2-regulated alternative poly(A) sites; moreover, the position of Nova2 binding was found to determine whether the protein acts to promote or inhibit poly(A) site use. In transcripts where Nova2 enhances poly(A) site use, it binds to more distal elements, where it possibly antagonizes the action of (unknown) auxiliary factors. In cases where Nova2 has an inhibitory effect, binding sites are located within 30 nt of the poly(A) signal sequences and therefore binding likely interferes with the formation of the 3’ processing complex. Therefore the position of Nova2 binding may determine the outcome of poly(A) site selection in a manner analogous to its action on splicing regulation (Ule et al., 2006). Another example of a “splicing factor” that can regulate polyadenylation is the polypyrimidine tract binding protein, PTB. PTB can compete with CstF binding to the downstream sequence element (Castelo-Branco et al., 2004) or can stimulate 3’ processing (Moreira et al., 1998) by increasing the binding of heterogeneous nuclear ribonucleoprotein H (hnRNP H, like Nova2 better known as a splicing factor) to the G-rich auxiliary element, which in turn stimulates cleavage by recruiting CstF and PAP (Danckwardt et al., 2007; Millevoi et al., 2009).
Additional genome-wide analysis also implicates hnRNP H in APA regulation. Specifically, Katz et al. (2010) used a statistical model to infer isoform regulation from RNA-seq data. The results showed that upon hnRNP H knockdown, preferential use of distal poly(A) sites was observed. This effect could be due to either hnRNP H-mediated inhibition of distal poly(A) sites or by direct activation of proximal sites. The authors found that genes with higher expression of shorter 3’ UTRs in the presence of hnRNP H displayed higher binding of hnRNP H near the proximal poly(A) site, implying that the second mechanism is the one used. Since this would imply a role of hnRNP H in recruitment of 3’ processing factors, this finding is in agreement with the fact that hnRNP H has been previously shown to exert a stimulatory role by interacting with PAP (Millevoi et al., 2009). High levels of hnRNP H have been observed in certain cancers (Honore et al., 2004), suggesting that this protein contributes to the shortening of 3’UTRs observed in cancer cells.
Several bone fide splicing factors are also known to influence 3’ processing (reviewed in Millevoi and Vagner, 2010). For example, the splicing factor U2AF65 binds to the polypyrimidine tract at the last intron 3’ splice site, stimulating both cleavage and polyadenylation by recruiting the CFI complex to the polyA site (Millevoi et al., 2006). Likewise, the SF3B component of U2 snRNP and the SR-related protein SRm160 have both been reported to influence 3’ processing by interacting with the CPSF complex (Kyburz et al., 2006; McCracken et al., 2002). It is an intriguing possibility that the interplay between factors involved in splicing of the 3’-terminal exon and polyadenylation factors in the 3’ UTR, as well as the physical distance between these two protein complexes, contributes to APA. U1 snRNP has also been shown to affect poly(A) site utilization, but independent of its role in splicing (Kaida et al., 2010). When binding of U1 snRNP to 5’splice sites was blocked using an antisense morpholino oligonucleotide (AMO), premature polyadenylation in many pre-mRNAs at cryptic poly(A) sites, frequently in introns near the start of the transcript, was detected. This effect was proved to be specific to U1 snRNP and not dependent on splicing, since splicing inhibition by using an AMO to U2 snRNP did not have the same effect. Binding of U1 snRNP in the proximity of cryptic poly(A) sites likely blocks their use by inhibiting recruitment of core 3’ processing factors to these sites. Whether this provides a mechanism to regulate APA remains to be determined.
As with other gene regulatory mechanisms, APA is likely to be modulated by cell-signaling pathways. Although little is so far known about this, a potentially interesting example is provided by the mechanism that upregulates the levels of the protease thrombin under conditions of stress, which is achieved through 3’ end processing regulation (Danckwardt et al., 2011). Stress conditions, such as inflammation, activate the kinase p38 MAPK, which on the one hand, phosphorylates the RNA-binding proteins FBP2 and FBP3. Once phosphorylated, FBP2/3 no longer bind to a highly conserved upstream sequence element (USE) in the thrombin mRNA. On the other hand, activation of p38 MAPK signaling also upregulates the levels of 3’ processing factors, as well as of proteins involved in splicing regulation. Interestingly, USE-RNP complexes where shown to include CPSF/CstF components and splicing regulators. The data suggest that p38 MAPK activation during stress leads to dissociation of FBP2/3 from the USE so that the USE is now able to counterbalance the relatively inefficient 3’ cleavage site by recruiting 3’ processing factors, leading to polyadenylation of the thrombin pre-mRNA. This finding has important implications since deregulated thrombin expression, leading to the pathogenesis of thrombophilia, can result from point mutations in the 3’UTR that improve the strength of the cleavage site (Gehring et al., 2001). Although not a direct example of APA, this emphasizes the existence of potential mechanisms by which 3’ processing of a weak poly(A) site, such as typical proximal poly(A) sites, can be selectively enhanced under specific physiological conditions.
Core 3’ processing factors are also regulated by post-translational modification (reviewed in Ryan and Bauer, 2008). The best studied example of this to date is provided by PAP. For example, during mitosis PAP is hyperphosphorylated by Cdc2/Cyclin B, which reduces its activity and contributes to a general repression of mRNA and protein production during mitosis (Colgan et al., 1996). PAP was also shown to be sumoylated, a modification that is important both for its nuclear localization and its stability (Vethantham et al., 2008). Examples of post-translational modifications of core processing factors that influence APA have not yet been reported but are likely to exist.
The process of transcription, and transcription-related proteins, appear capable of affecting the choice of APA site. The coupling between transcription and 3’ processing is well-established (reviewed in Hirose and Manley, 2000; Perales and Bentley, 2009; Proudfoot et al., 2002). The CTD of RNAP II is necessary for efficient 3’ processing in vivo and in vitro (Hirose and Manley, 1998; McCracken et al., 1997); the CTD interacts with 3’ processing factors such as CPSF and CstF (Glover-Cutter et al., 2008; Licatalosi et al., 2002), CPSF interacts with the transcription factor TFIID, (Dantonel et al., 1997); 3’ processing factors have been detected by ChIP assays at both ends of genes (Glover-Cutter et al., 2008; Rozenblatt-Rosen et al., 2009; Venkataraman et al., 2005); and the 3’ processing factor symplekin binds to and stimulates the CTD phosphatase Ssu72, which is necessary for efficient transcription-coupled polyadenylation in vitro (Xiang et al., 2010). It has also recently been shown that a transcriptional activator can enhance the efficiency of transcription-coupled 3’ processing, in a manner that requires the transcription elongation complex PAF1c (Nagaike et al., 2011). PAF1c is a multifunctional complex implicated in various aspects of transcription (Rosonina and Manley, 2005), and known to associate with 3’ processing factors (Rozenblatt-Rosen et al., 2009).
The above findings indicate multiple mechanisms by which 3’ end formation can be coupled to transcription. An explanation for this extensive coupling is that it serves to increase the efficiency by which nascent transcripts are cleaved, by facilitating recruitment of processing factors to the site of processing. But how might this influence APA? As discussed by Naigaike et al. (2011), an attractive model is that increasing the efficiency of 3’ processing along transcribed genes will tend to favor use of proximal poly(A) sites. Given that transcriptional activators can enhance processing efficiency, use of proximal poly(A) sites has the potential to further enhance expression of activated genes, by removing repressive elements from the 3’UTR. In support of this mechanism, knockdown of a PAF1c subunit led to increased accumulation of 3’ extended transcripts of a PAF1c target gene (Rozenblatt-Rosen et al., 2009). It will be of interest to determine whether this provides a general mechanism of APA control.
An additional recent study emphasizes the potential connection between transcription elongation rate and APA. Pinto et al. (2011) found that a mutant Drosophila strain with a reduced elongation rate (because of a mutation in the RNAP II largest subunit) displays increased usage of proximal poly(A) sites in a number of alternatively polyadenylated transcripts, suggesting that RNAP II elongation may have an important role in poly(A) site selection. A mechanistic explanation for these findings is simply that a slower RNAP II would enable the proximal poly(A) signal to be exposed to the 3’ processing complex for a longer time before the second poly(A) site is transcribed, increasing the efficiency with which it is used. This scenario is analogous to the effect that lower transcriptional rate has on alternative splicing: a human RNAP II carrying the equivalent of the above-mentioned Drosophila mutation, when introduced into human cells, was shown to lead to the inclusion of otherwise skipped alternative exons in several transcripts (de la Mata et al., 2003)
RNA signals that modulate APA
As mentioned in the Introduction, specific RNA sequences in the pre-mRNA define the binding sites for different components of the 3’ processing complex, dictating the precise site where cleavage will occur. These are usually termed the “core” polyadenylation elements, while there are also less-defined auxiliary downstream and upstream elements. As discussed below the “strength” of the core elements in combination with auxiliary elements is likely to play a critical role in selection of APA sites.
Large-scale computational analyses of 3’UTRs have revealed interesting features of cis-acting elements in regulating usage of alternative poly(A) sites. As expected, poly(A) sites containing the consensus sequence AAUAAA are used more frequently than other variants. Nonetheless, usage of variant hexamers is not uncommon (Hu et al., 2005; Jan et al., 2011; Tian et al., 2005). Importantly, these variant sequences are usually found in a promoter-proximal position within the 3’UTR, and the ones used more often are characterized by increased sequence conservation around the poly(A) site. This suggests that appropriate context can compensate for lack of a strong poly(A) site, probably by enhanced recruitment of 3’ processing factors, such as CstF, to these sites.
Analysis of APA in 15 human tissues using deep-sequencing found a set of heptanucleotides showing high conservation located in the region between APA sites (Wang et al. 2008). These include seed matches to a number of miRNAs, as expected, but also a consensus binding motif for FOX1/FOX2 (or other proteins with the same RNA-binding specificity). FOX1/FOX2 are well-characterized tissue-specific splicing factors (reviewed in Kuroyanagi, 2009), but such a strong conservation of their binding sequence in 3’UTR regions suggests that they have additional roles. It will be interesting to determine if such roles are connected to regulation of APA and/or to determining mRNA localization and stability.
To examine the sequence patterns governing APA, Ozsolak et al. (2010) used Direct RNA Sequencing to analyze RNA samples extracted from human liver, human brain, and yeast. Three new motifs were identified near human poly(A) sites: a TTTTTTTTT motif positioned ~21 nt upstream of the poly(A) site, a AAWAAA motif (where W represents either A or T) positioned upstream of the poly(A) site, and a palindromic sequence CCAGSCTGG (S=C/G) found downstream of the poly(A) site. The palindromic sequence strongly co-occurs with the TTTTTTTTT motif and with another sequence that was later found using a less-stringent scan (RGYRYRGTGG, where R=A/G and Y=C/T). These sequences are present in intragenic and newly found intergenic poly(A) sites (likely to represent novel mRNAs), whereas they do not co-occur, and actually anticorrelate, with the canonical AATAAA signal localization. The anticorrelation hints at a possible role for these sequences in coordinating APA events. An interesting analogy is with TATA-less promoters, which use the same set of core transcription factors but involving different interactions with promoter sequences (Juven-Gershon et al., 2008; Sikorski and Buratowski, 2009). Another possibility is that these new motifs function by directing the binding of yet unknown proteins which in turn affect the recruitment and formation of the 3’processing factors. A third possibility is that, under certain conditions, the affinity of CPSF and CstF complexes to RNA sequences might be modulated by mechanisms such that post-translational modifications or association with other factors shift their binding from the canonical sequences to these new motifs, thereby affecting APA.
Finally, although not yet documented, it is possible that secondary structures and stem-loop motifs in 3’UTRs may affect APA. Such structures in 3’UTRs have been shown to regulate stability and other aspects of mRNA metabolism (Erlitzki et al., 2002; Liu et al., 2010b), and it is possible that they could also enhance or inhibit the binding of protein factors involved in 3’ processing, and, as a result modulate APA.
Chromatin and epigenetic-mediated regulation of APA
An important recent discovery is that chromatin structure and epigenetic marks can act as regulators of alternative splicing (Fox-Walsh and Fu, 2010; Luco et al., 2011; Luco et al., 2010). Recent data suggest that 3’ end processing might be similarly modulated by chromatin and histone modifications. While much attention has focused on nucleosome organization around the promoter regions, little is known about their organization at the end of genes. The first evidence for a connection between polyadenylation and histone positioning was reported in S. cervisae, where antibodies against tagged histones H3 and H4 where used to perform a ChIP-seq analysis. Significantly, the 3’ region near the poly(A) site was shown to be depleted of nucleosomes (Mavrich et al., 2008; Shivaswamy et al., 2008; Spies et al., 2009). This effect could reflect the nucleotide sequence itself, which might have lower intrinsic affinity for nucleosomes (as shown for poly(dA:dT) streches), or by the presence of a nucleosome-excluding DNA-binding protein that binds near poly(A) site.
Sites of mRNA polyadenylation and transcription termination by RNAP II are closely spaced in yeast genes (reviewed in Richard and Manley, 2010), so it could be that the nucleosome-free regions are related to transcription and not 3’ processing. More recently, however, a confirmation of strong nucleosome depletion around human poly(A) sites was obtained, suggesting that these regions are indeed connected to 3’processing. Spies et al. (2009) analyzed two previously published ChIP-Seq data sets from human T cells (Barski et al., 2007; Schones et al., 2008) and found that the dip in nucleosome density observed at the AATAAA sequence (and variants) was even more pronounced around actively used poly(A) sites (in genes with multiple poly(A) sites), suggesting that either additional sequences around the poly(A) signal, such as T-rich stretches, may play a role in nucleosome positioning or that a yet unknown nucleosome-excluding DNA binding protein maybe be commonly bound near the poly(A) sequence. Moreover, higher downstream nucleosome density, from approximately +75 to +375 downstream of the poly(A) signal, was observed to be associated with higher poly(A) site usage. Whether nucleosome positioning affects APA, for example by influencing the rate of polymerase elongation, or if the opposite is true, via a 3’complex-dependent recruitment of a chromatin remodeling factor, remains to be clarified.
Genomic imprinting has also been implicated in APA regulation. Alternative poly(A) sites on transcripts of the mouse imprinted gene H13 (encoding for a signal peptide peptidase) have been found to be utilized in an allele-specific manner, such that two proximal poly(A) sites are used in the maternal allele while, a distal poly(A) site (one of three distal sites) is preferentially used in the paternal derived allele (Wood et al., 2008). The two clusters of poly(A) signals are separated by a CpG island, which is located 0.5 to 3 Kb downstream of the first cluster and ~20 Kb upstream of the second cluster of poly(A) sites. This CpG island has been shown to be specifically methylated only on the maternal allele. Since the maternal and paternal alleles are exposed to the same array of trans-regulatory factors, allelic differences in APA of this imprinted locus must be the result of epigenetic regulation. A possible explanation for this is that methylation of the CpG island on maternally derived alleles recruits an inhibitory factor that prevents binding of polyadenylation factors to the upstream poly(A) sites and therefore, the distal poly(A) site is used. CpG binding proteins have been showed to be able to indirectly change chromatin structure. For example the protein CFP1 binds specifically to non-methylated CpGs and changes chromatin by recruiting a methyltrasferase, which leads to increased H3K4me3 (Thomson et al., 2010). Similarly, changes in chromatin induced by the specific state of CpG methylation, if it occurs proximal to poly(A) sites, could affect their utilization.
While additional work is required, it seems that nucleosome positioning and epigenetic marks can affect the outcome of gene expression through regulation of APA. The precise mechanisms involved are not yet known but in theory could influence APA either indirectly, for example by influencing the transcription rate and therefore allowing more time for the assembly of the 3’ complex, or directly, by facilitating recruitment of components, or modulators, of the 3’ processing machinery. The latter case would be analogous to the mechanism by which recognition of H3K4me3 by CHD1 functions, at the 5’ ends of actively transcribed genes, to recruit core spliceosomal components, therefore facilitating the efficiency of pre-mRNA splicing (Sims et al., 2007). However, at this point it is difficult to establish a cause or effect relationship between APA and epigenetic marks (as well as nucleosome postioning). It is possible that poly(A) site selection may induce specific chromatin marks, perhaps through 3’ processing complex-dependent recruitment of chromatin modifiers, rather than chromatin marks acting to promote particular APA patterns.
Biological functions of APA
UTR-APA produces mRNA isoforms that either contain or lack a full complement of cisregulatory elements (e.g., AREs or miRNA binding sites), depending on the choice of proximal versus distal poly(A) sites. Thus the landscape of such sequences throughout 3’ UTRs can determine the robustness of APA as a regulatory mechanism. In this regard, Legendre et al.(2006) carried out a systematic examination of 3’UTRs produced by APA and found that 52% of miRNA target sites are located downstream of the first poly(A) site. Sandberg et al. (2008) also found that in T cells mRNAs with longer 3’UTRs have a 2.1-fold higher number of miRNA target sites than those with shorter 3’UTRs. AREs have been estimated to be present in ~10–15% of all transcripts (Halees et al., 2008), and were shown to interact with several proteins, some of which contribute to mRNA stability (reviewed in Barreau et al., 2005) and others control translation (reviewed in Espel, 2005). In addition, cooperation between miRNAs and ARE binding proteins has been documented in ARE-mediated mRNA degradation (Jing et al., 2005).
As previously discussed, states of increased cell proliferation are associated with generation of transcripts having shorter 3’UTRs. This results in increased gene expression, consistent with the need of faster proliferating cells to produce more proteins. Indeed, Sandberg et al. (2008) showed that luciferase reporters with short 3’UTRs from several genes produced about twice as much luciferase than those with longer 3’UTRs. For example, one of the tested genes, Hip2, contains conserved binding sites for miR-21 and miR-155. Expression of the longer Hip2 3’UTR isoform is decreased during T cell activation while protein levels of Hip2 are increased. Mutation of these sites resulted in the same luciferase levels as the reporter with the shorter Hip2 3’UTR produced. Likewise, Mayr and Bartel (2009) also showed that the longer 3’UTRs of IMP-1, Cyclin D2 or DICER1 genes negatively affected expression of similar luciferase constructs, and that this could be partially reversed by specific deletions of miRNA sites (let-7 in IMP-1, miR103/107 and/or let7 in DICER1, and miR15/16 in Cyclin D2). Significantly, some miRNAs, including let-7 and miR15/16, have been reported to act as tumor suppressors (Calin et al., 2002; Yu et al., 2007). Extending this notion, escape from miRNA-mediated regulation can induce increased oncogene protein synthesis, suggesting that loss of 3’UTR regulatory elements by APA contributes to oncogenic transformation. Notably, deletions of miRNA binding sites within the full-length 3’UTRs caused only a quarter to two-thirds increase in protein levels compared to levels observed with the shortened 3’ UTRs produced by APA (Mayr and Bartel, 2009). Thus, other regulatory factors, such as RNA binding proteins, likely influence this process.
Another mechanism by which UTR-APA can influence protein expression is via regulating mRNA localization. Since localization is mainly dictated by cis-elements found within the 3’UTR (Kislauskis and Singer, 1992; Andreassi and Riccio, 2009), this process can be modulated by APA. Examples include ASH1 mRNA in budding yeast (Takizawa et al., 1997), bicoid mRNA in Drosophila embryos (Johnstone and Lasko, 2001), VegT1 mRNA in Xenopus oocytes (King et al., 2005), β-actin mRNA in human fibroblasts (Condeelis and Singer, 2005), and MBP mRNA in oligodendrocytes (Smith, 2004). Strikingly, high-resolution in situ hybridization techniques revealed that more than 70% of transcripts in Drosophila embryos are expressed in spatially distinct patterns (Lecuyer et al., 2007). Thus, mRNA localization is a global phenomenon, conserved from yeast to mammals. Asymmetric localization is observed in highly polarized cells like differentiated neurons where APA events are often observed and where mRNA localization is used to promote rapid local protein synthesis.
Several examples illustrate the role of APA in mRNA localization. One is the brain-derived neurotrophic factor BDNF. The brain produces two BDNF transcripts encoding the same protein, with either a short or long 3’ UTR (Timmusk et al., 1993). The long BDNF mRNA was found to be preferentially targeted to dendrites in cultured rat neurons. In addition, a significant reduction of dendritic BDNF mRNA was observed in hippocampal and cortical neurons of mutant mice that lack the long 3’UTR mRNA isoform due to the insertion of three strong poly(A) sites after the first BDNF poly(A) site (An et al., 2008). Furthermore, the long and the short 3’UTRs are differently regulated in translation: while the short 3’UTR BDNF mRNA is predominantly associated with polyribosomes, the long 3’UTR BDNF mRNA is largely sequestered into translationally dormant ribonucleoprotein particles. After neuronal stimulation, polyribosome association with the long 3’UTR mRNA was increased accompanied with increased BDNF protein, although levels of BDNF mRNAs were not changed. These observations show that the long 3’UTR mRNA specifically undergoes robust translational activation in the hippocampus before transcriptional up-regulation of BDNF, while the short 3’UTR mRNA mediates active translation to maintain basal levels (Lau et al., 2010). Another example of 3’UTR-mediated localization is provided by the calmodulin-dependent protein kinase II, CaMKIIα. CaMKIIα mRNAs also have different length 3’UTRs (Bulleit et al., 1988) and again, the longer isoform specifically localizes in dendrites (Blichenberg et al., 2001), suggesting that a similar mechanism might exist as with BDNF mRNA. Indeed, a number of mRNAs localized in dendrites possess 3’UTR sequences required for localization (Andreassi and Riccio, 2009), although in most of these the role of APA has not been investigated. It is possible that 3’UTR signals regulated by APA may provide a general mechanism for localizing mRNAs to soma and dendrites, as well as to other subcellular destinations.
APA also plays a role in control of gene expression in plants. For example, the control of seasonal flowering has a complex but unique gene-regulation mechanism that involves APA (Figure 3C) (Hornyik et al., 2010; Liu et al., 2010a). Flowering time is negatively regulated by expression of the FLC gene. Two RNA binding proteins, FPA and FCA, act independently to repress FLC expression and thereby allow flowering. Both FPA and FCA have been shown to repress FLC expression by mediating APA of a non-coding antisense transcript. A promoter situated downstream of the poly(A) site of FLC and on the opposite strand generates anti-sense transcripts that have alternative poly(A) sites: one cluster of poly(A) sites (proximal) is located opposite the terminal intron of FLC and another cluster (distal) is located opposite the FLC promoter. Both FPA and FCA promote usage of the proximal poly(A) sites. Interestingly, mutants of CstF components (CstF64 and CstF77) showed elevation of sense FLC transcripts and reduction of antisense FLC transcripts, suggesting FLC antisense transcripts are sensitive to CstF activity (Liu et al., 2010a). However, FLD, a histone H3 Lys 4 (H3K4me2) demethylase, is also required for effective FLC silencing (Baurle and Dean, 2008; Liu et al., 2010a; Liu et al., 2007b). In addition, another layer of complexity is added by the fact that FCA interacts with FY (the homolog of the 3’ processing factor WDR33; Shi et al., 2009) to promote proximal poly(A) site selection in its own pre-mRNA, resulting in production of a non-functional, truncated FCA-mRNA (Simpson et al., 2003). How does selection of the proximal poly(A) site in the antisense RNA transcript promote silencing of FLC? Perhaps, as suggested by Rosonina and Manley (2010), when the proximal poly(A) sites in the anti-sense transcript is used, the recruited FLD demethylase catalyzes removal of the transcriptionally active chromatin mark H3K4me2 in the body of the FLC gene, leading to FLC silencing, while utilization of the distal poly(A) site of the anti-sense transcript facilitates recruitment of positive factors to the FLC promoter, resulting in enhanced FLC mRNA expression.
Regulation by antisense APA has the potential to extend beyond plants. A genome-wide analysis of polyadenylated RNAs from both yeast and human liver cells (Ozsolak et al., 2010) demonstrated that antisense transcription is very common in eukaryotes, being present in at least ~60% of yeast annotated open reading frames and as much as 30% in human liver. It is therefore possible that gene regulation through APA of anti-sense transcripts may occur also in mammalian genes, in a way analogous to the FLC gene in yeast.
Concluding remarks
While the existence of APA has been known for some time, whether or not it constitutes a general mechanism of gene control has until recently been unclear. In the last several years however, genome-wide analyses have shown that APA is in fact widespread in mammalian cells, is regulated during development and differentiation, and can become deregulated in disease. One of the most interesting questions is how mechanistically alternative poly(A) sites are selected. As we have discussed, this will likely involve core polyadenylation factors, such as CstF-64 in B cell activation (Takagaki et al., 1996). Indeed, since CPSF-160 and CstF-64 are upregulated in cancer cells (Mayr and Bartel, 2009), and several 3’ processing factors are downregulated during myoblast differentiation (Ji et al., 2009), regulation of APA by varying the levels of core factors may be a general mechanism. However, given how widespread we now know APA to be, it is likely that other factors are involved. Similar to control of alternative splicing (Chen and Manley, 2009), this is likely to reflect a combination of core processing factors and gene-specific RNA binding proteins. Indeed, APA is coupled with splicing events as well as with transcription, suggesting that numerous auxiliary factors involved in these processes may affect APA. And as we have seen, chromatin modifications also play a role in APA. In keeping with this, analysis of the poly(A) “proteome” revealed over 80 proteins (Shi et al., 2009), potentially linking polyadenylation efficiency, and hence APA regulation, with multiple cellular processes.
So far, only a few examples linking aberrant APA directly with known diseases have been documented. However, mutations in 3’UTRs, including poly(A) signal sequences, have been associated with a number of medically relevant issues (reviewed in Danckwardt et al., 2008), including the early examples of alpha- and beta-thalassemias (Higgs et al., 1983; Orkin et al., 1985) as well as the above-mentioned examples of thrombophilia (Gehring et al., 2001), IPEX (Bennett et al., 2001) and cyclinD1-related cancers (Burd et al., 2006; Wang et al., 2008b; Wiestner et al., 2007). It is therefore likely that, similar to diseases reflecting aberrant splicing (Cooper et al., 2009), more examples of diseases caused by changes in APA will emerge.
Acknowledgments
We would like to thank Dr. Emanuel Rosonina and Dr Tristan Coady for critical reading of the manuscript. Work from the authors’ lab was supported by NIH grant R01 GM28983.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980;20:293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Ara T, Lopez F, Ritchie W, Benech P, Gautheret D. Conservation of alternative polyadenylation patterns in mammalian genes. BMC Genomics. 2006;7:189. doi: 10.1186/1471-2164-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Baurle I, Dean C. Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS One. 2008;3:e2733. doi: 10.1371/journal.pone.0002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Brunkow ME, Ramsdell F, O'Briant KC, Zhu Q, Fuleihan RL, Shigeoka AO, Ochs HD, Chance PF. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA-->AAUGAA) leads to the IPEX syndrome. Immunogenetics. 2001;53:435–439. doi: 10.1007/s002510100358. [DOI] [PubMed] [Google Scholar]
- Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–1011. [PubMed] [Google Scholar]
- Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur J Neurosci. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- Brown G, Drayson MT, Durham J, Toellner KM, Hughes PJ, Choudhry MA, Taylor DR, Bird R, Michell RH. HL60 cells halted in G1 or S phase differentiate normally. Exp Cell Res. 2002;281:28–38. doi: 10.1006/excr.2002.5654. [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3' processing by human cleavage factor Im. Mol Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- Bulleit RF, Bennett MK, Molloy SS, Hurley JB, Kennedy MB. Conserved and variable regions in the subunits of brain type II Ca2+/calmodulin-dependent protein kinase. Neuron. 1988;1:63–72. doi: 10.1016/0896-6273(88)90210-3. [DOI] [PubMed] [Google Scholar]
- Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA, Lu S, Fenoglio-Preiser CM, Li J, Knudsen ES, et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A. 2006;103:2190–2195. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN. A systematic analysis of disease-associated variants in the 3' regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3' UTR variants. Hum Genet. 2006;120:301–333. doi: 10.1007/s00439-006-0218-x. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, et al. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity. 1999;10:261–269. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Comstock CE, Augello MA, Benito RP, Karch J, Tran TH, Utama FE, Tindall EA, Wang Y, Burd CJ, Groh EM, et al. Cyclin D1 splice variants: polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin Cancer Res. 2009;15:5338–5349. doi: 10.1158/1078-0432.CCR-08-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- Conne B, Stutz A, Vassalli JD. The 3' untranslated region of messenger RNA: A molecular 'hotspot' for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Gantzert AS, Macher-Goeppinger S, Probst HC, Gentzel M, Wilm M, Grone HJ, Schirmacher P, Hentze MW, Kulozik AE. p38 MAPK controls prothrombin expression by regulated RNA 3' end processing. Mol Cell. 2011;41:298–310. doi: 10.1016/j.molcel.2010.12.032. [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 3' end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3' end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3' end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, Macdonald CC. Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci U S A. 2007;104:20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980;20:313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Erlitzki R, Long JC, Theil EC. Multiple, conserved iron-responsive elements in the 3'-untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J Biol Chem. 2002;277:42579–42587. doi: 10.1074/jbc.M207918200. [DOI] [PubMed] [Google Scholar]
- Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh K, Fu XD. Chromatin: the final frontier in splicing regulation? Dev Cell. 2010;18:336–338. doi: 10.1016/j.devcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE. Increased efficiency of mRNA 3' end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halees AS, El-Badrawi R, Khabar KS. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36:D137–D140. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hockert KJ, Martincic K, Mendis-Handagama SM, Borghesi LA, Milcarek C, Dass B, MacDonald CC. Spermatogenetic but not immunological defects in mice lacking the tauCstF-64 polyadenylation protein. J Reprod Immunol. 2011;89:26–37. doi: 10.1016/j.jri.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honore B, Baandrup U, Vorum H. Heterogeneous nuclear ribonucleoproteins F and H/H' show differential expression in normal and selected cancer tissues. Exp Cell Res. 2004;294:199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell. 2010;18:203–213. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Driscoll M. MicroRNAs in C. elegans Aging: Molecular Insurance for Robustness? Curr Genomics. 2009;10:144–153. doi: 10.2174/138920209788185243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3'UTRs. Nature. 469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3'UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Kong J, Liebhaber SA. An RNA-protein complex links enhanced nuclear 3' processing with cytoplasmic mRNA stabilization. EMBO J. 2011 doi: 10.1038/emboj.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. Reprogramming of 3' untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Singer RH. Determinants of mRNA localization. Curr Opin Cell Biol. 1992;4:975–978. doi: 10.1016/0955-0674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3'-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3' end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Lau AG, Irier HA, Gu J, Tian D, Ku L, Liu G, Xia M, Fritsch B, Zheng JQ, Dingledine R, et al. Distinct 3'UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF) Proc Natl Acad Sci U S A. 2010;107:15945–15950. doi: 10.1073/pnas.1002929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Legendre M, Ritchie W, Lopez F, Gautheret D. Differential repression of alternative transcripts: a screen for miRNA targets. PLoS Comput Biol. 2006;2:e43. doi: 10.1371/journal.pcbi.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3' end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3'-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007a;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010a;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillen P, Baurle I, Swiezewski S, Dean C. The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell. 2007b;28:398–407. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang Y, Russell JE. A potential regulatory role for mRNA secondary structures within the prothrombin 3'UTR. Thromb Res. 2010b;126:130–136. doi: 10.1016/j.thromres.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63:7056–7061. [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CC, Redondo JL. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol Cell Endocrinol. 2002;190:1–8. doi: 10.1016/s0303-7207(02)00044-8. [DOI] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3'-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of C. elegans 3'UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3'-end cleavage. Mol Cell Biol. 2002;22:148–160. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KW, Hirsch BA, MacDonald CC. Differences in polyadenylation site choice between somatic and male germ cells. BMC Mol Biol. 2006;7:35. doi: 10.1186/1471-2199-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Decorsiere A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, Vagner S. A physical and functional link between splicing factors promotes pre-mRNA 3' end processing. Nucleic Acids Res. 2009;37:4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3' end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3' end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monarez RR, MacDonald CC, Dass B. Polyadenylation proteins CstF-64 and tauCstF-64 exhibit differential binding affinities for RNA polymers. Biochem J. 2007;401:651–658. doi: 10.1042/BJ20061097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3' end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH., Jr Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985;4:453–456. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. "Cotranscriptionality": the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ML. Mechanisms controlling production of membrane and secreted immunoglobulin during B cell development. Immunol Res. 2007;37:33–46. doi: 10.1007/BF02686094. [DOI] [PubMed] [Google Scholar]
- Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ, et al. RNA polymerase II kinetics in polo polyadenylation signal selections. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Early P, Carter C, Calame K, Bond M, Hood L, Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980;20:303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Manley JL. From transcription to mRNA: PAF provides a new path. Mol Cell. 2005;20:167–168. doi: 10.1016/j.molcel.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3' mRNA processing factors. Proc Natl Acad Sci U S A. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Bauer DL. Finishing touches: post-translational modification of protein factors involved in mammalian pre-mRNA 3' end formation. Int J Biochem Cell Biol. 2008;40:2384–2396. doi: 10.1016/j.biocel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3' processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3' untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. Moving molecules: mRNA trafficking in Mammalian oligodendrocytes and neurons. Neuroscientist. 2004;10:495–500. doi: 10.1177/1073858404266759. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, Senderowicz AM, Conti CJ, Knudsen ES. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Tanguay RL, Gallie DR. Translational efficiency is regulated by the length of the 3' untranslated region. Mol Cell Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 2008;22:499–511. doi: 10.1101/gad.1628208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P, Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr Opin Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008a;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dean JL, Millar EK, Tran TH, McNeil CM, Burd CJ, Henshall SM, Utama FE, Witkiewicz A, Rui H, et al. Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res. 2008b;68:5628–5638. doi: 10.1158/0008-5472.CAN-07-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, Liu H, Rosenwald A, Muller-Hermelink HK, Ott G, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu M, Sha H, Gao Y, Zeng H, Zhu M, Gao X. Alternative 3' UTR polyadenylation of Bzw1 transcripts display differential translation efficiency and tissue-specific expression. Biochem Biophys Res Commun. 2006;345:479–485. doi: 10.1016/j.bbrc.2006.04.113. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Virtanen A, Kleiman FE. To polyadenylate or to deadenylate: that is the question. Cell Cycle. 2010;9:4437–4449. doi: 10.4161/cc.9.22.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]